Abstract
To investigate which domains of Nedd4 are responsible for the interaction with its substrate protein TMEPAI, the plasmids encoding GST-tagged WW domains of Nedd4 and His-HA-tagged TMEPAI were constructed, and the fusion proteins were expressed in E. coli BL21-CodonPlus (DE3)-RIL cells and purified. GST pull-down experiment indicated that Nedd4 directly interacts with His-HA-TMEPAI, and the WW2 and WW3 domains of Nedd4 are responsible for the interaction with TMEPAI, which represents a general mechanism for the interaction between Nedd4 and its substrate protein.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Ubiquitination is a common protein modification, which regulates protein degradation, as well as protein transport and a variety of metabolic pathways in the cells [1]. The process of ubiquitination involves the ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin-protein ligating enzymes E3. Nedd4 (neuronal precursor cell expressed developmentally down-regulated 4) belongs to the Nedd4 of HECT E3 ubiquitin ligase family including Nedd4, Nedd4-2, Smurf1, Smurf2, Itch, WWP1, WWP2, Nedl1, and Nedl2 [2, 3]. Nedd4 contains a C2 domain, four WW domains, and a HECT domain [4]. The C2 domain binds to phosphor-lipids. The WW domain is 35–40 amino acids long containing two conserved tryptophan (W) residues spaced 21 amino acids apart. The WW domains interact with a variety of proline-based motifs, and recognize proline-containing phosphor-serine/phosphor-threonine sequences on the substrate protein. The HECT domain is comprised of approximately 350 amino acids and is responsible for ubiquitin transfer from a conserved cysteine residue to a lysine residue in the substrate protein [5–7].
TMEPAI is a Nedd4-binding protein that is originally identified as a prostate abundant, highly androgen-induced protein, and the TMEPA1 gene is mapped to chromosome 20q13 [8]. TMEPAI protein contains two PY motifs (PPPY and PPTY), which are necessary for binding to the WW domains of Nedd4 ubiquitin ligase [8, 9].
It has been reported that TMEPAI is a substrate of Nedd4 through the interaction with the WW domains, however, which WW domains interact with TMEPAI is still unclear. In this study, we investigate the binding affinity of the four WW domains of Nedd4 with its substrate protein TMEPAI [10].
2 Materials and Methods
2.1 Construction of Plasmid
Four DNA fragments encoding the WW domains of Nedd4 were amplified by PCR using the primers listed in Table 6.1, and the four fragments were digested with BamHI and EcoRI and inserted into the pGEX-4T-2 (GE Healthcare) vector. The TMEPAI gene was amplified by PCR with the forward primer 5′-ATGAGCCACTACAAGCTGTCTG-3′ and reverse primer 5′-CTAGAGAGGGTGTCCTTTC-3′, and the PCR product was purified and digested with BamHI and EcoRI, and inserted into the BamHI and EcoRI sites of the pET21b vector.
2.2 Protein Expression and Purification
Plasmids encoding GST-fusion proteins of Nedd4 were transformed into E. coli BL21-CodonPlus (DE3)-RIL (Stratagene). Cells were induced with 0.1 mM IPTG overnight at 16 °C, and lysed in lysis buffer (20 mM Tris–Cl pH 7.4, 100 mM NaCl, 2 mM DTT) containing protease inhibitors. The recombinant protein was purified on Glutathione–Sepharose beads (GE Healthcare). After sonication, the extracts were clarified by centrifugation. The recovered supernatants were allowed to bind to Glutathione Sepharose beads for 3 h at 4 °C. The matrix was washed three times with lysis buffer, then the beads were eluted with elution buffer (100 mM Tris–Cl pH 7.4, 100 mM NaCl, 20 mM reduced glutathione). His-HA-TMEPAI was expressed in E. coli BL21-CodonPlus (DE3)-RIL (Stratagene) cells that transformed with pET21b-His-HA-TMEPAI plasmid, and induced with 0.5 mM IPTG overnight at 16 °C. The His-HA-tagged protein was purified on Ni-Sepharose beads after lysis of the bacteria in Na–P lysis buffer (50 mM Na–P pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol) plus protease inhibitors. After sonication, the extracts were clarified by centrifugation. The recovered supernatants were allowed to bind to Ni-Sepharose beads for 3 h at 4 °C. The matrix was washed three times with Na–P lysis buffer containing 50 mM imidazole, the beads were eluted with elution buffer (50 mM Na–P pH 8.0, 300 mM NaCl, 5 mM β-mercaptoethanol, and 250 mM imidazole).
2.3 GST Pull-Down Assays and Western Blot
For the GST pull-down assays, 5 μg GST-fusion proteins were first immobilized on the Glutathione–Sepharose beads. The matrix was incubated for 2 h in rotation at 4 °C, then washed three times with cold GST pull-down buffer (20 mM Tris–Cl pH 7.4, 100 mM NaCl, 1 % Triton-100, 2 mM DTT). At the same time, 0.5 μg protein of His-HA-TMEPAI was resuspended in the GST pull-down buffer and clarified by centrifugation at 14,000 rpm for 10 min. The supernatant was added into the GST-beads and incubated for 2 h in rotation at 4 °C, then washed three times with cold GST pull-down buffer, and washed once with 1 × PBS. Finally, the beads were resuspended and boiled in 40 μL 2 × SDS loading buffer; after boiled for 5 min, the supernatants were collected and separated by 10 % SDS-PAGE, electroblotted onto a polyvinylidene difluoride membrane (Millipore) and analyzed by Western blot using anti-HA-tag antibody.
3 Result
3.1 The Schematic Structure of Nedd4
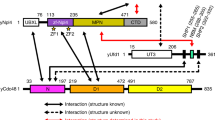
Nedd4 (NP-006145) contains 900 amino acids, including a N-terminal C2 domain for membrane binding, a central region containing four WW domains for protein–protein interaction, and a C-terminal HECT domain for ubiquitin-protein ligation (Fig. 6.1).
3.2 Nedd4 Interacts with TMEPAI Directly
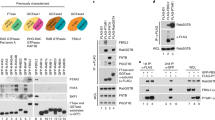
To verify that Nedd4 directly interacts with TMEPAI, GST-Nedd4, and His-HA-TMEPAI proteins were expressed in E. coli BL21-CodonPlus (DE3)-RIL and purified. GST pull-down experiment showed that GST-Nedd4, but not GST, pulled down purified His-HA-TMEPAI (Fig. 6.2), which indicates that GST-Nedd4 interacts with His-HA-TMEPAI directly.
3.3 Expression and Purification of the GST-Tagged WW Domains of Nedd4
To investigate which domains of Nedd4 are responsible for binding with TMEPAI, four plasmids encoding GST-tagged WW1, WW2, WW3, and WW4 domains of Nedd4 were constructed (Fig. 6.1). These four GST-fusion proteins were expressed and purified. The purified fusion proteins were subjected to SDS-PAGE and stained with Coomassie blue (Fig. 6.3).
3.4 The WW2 and WW3 Domains of Nedd4 Interact with TMEPAI
To measure the binding affinity of the Nedd4 WW domains with TMEPAI, the GST pull-down assay was performed. 5 μg GST or GST-WW domain fusion proteins were first bound to Glutathione–Sepharose beads, then incubated with 0.5 μg His-HA-TMEPAI protein. GST pull-down assay showed that the WW2 and WW3 domain had a higher binding affinity with His-HA-TMEPAI, with a much less binding affinity for WW4 domain, but no binding affinity for WW1 domain (Fig. 6.4). These data indicate that the WW2 and WW3 domains of Nedd4 are mainly responsible for the interaction with TMEPAI.
4 Discussion
Previous studies have demonstrated that TMEPAI is a substrate of Nedd4 E3 ubiquitin ligase [5, 9, 11]. The mouse Nedd4 contains three WW domains and it has been shown that the WW2 domain plays an important role in protein interaction [12, 13]. In this study, we have identified that the WW2 and WW3 domains of Nedd4 play an important role in interacting with TMEPAI. This may represent a general mechanism for the interaction between Nedd4 and its substates. However, we need to make a series of mutants of these proteins for further study.
Protein ubiquitination is a posttranslational modification in which a ubiquitin, or a multiubiquitin chain, is attached onto proteins by the formation of an isopeptide bond between the C-terminal Gly76 carboxyl group of ubiquitin and the 3-amino group of an internal Lys residue of the substrates, so as to control its stability or as a signal of protein transportation in the cells [14–17]. Given lacking of the specificity of proteasome inhibitors, the enzymatic nature, abundance, and specific substrate recognition properties of E3 ubiquitin ligase indicate that E3 might be used to as even more specific and effective therapeutic targets. Targeting individual E3 would selectively stabilize a particular or limit a number of protein substrates, thus possibly limiting untoward side effects. Hence, drugs targeting E3 promise a better therapeutic ratio than proteasome inhibitors, and the potential E3 inhibitors have a stronger chance to inhibit the active catalytic site.
References
Rotin D, Kumar S (2009) Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol 10(6):398–409
Chen C, Matesic LE (2007) The Nedd4-like family of E3 ubiquitin ligases and cancer. Cancer Metastasis Rev 26(3–4):587–604
Bernassola F, Karin M, Ciechanover A et al (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14(1):10–21
Lu PJ, Zhou XZ, Lu KP et al (1999) Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 283(5406):1325–1328
Scheffner M, Staub O (2007) HECT E3s and human disease. BMC Biochem 8:S1–S6
Magnifico A, Ettenberg S, Lipkowitz S (2003) WW domain HECT E3s target Cbl RING finger E3s for proteasomal degradation. J Biol Chem 278(44):43169–43177
Sudol M, Hunter T (2000) NeW wrinkles for an old domain. Cell 103(7):1001–1004
Xu LL, Shanmugam N, Srivastava S et al (2000) A novel androgen-regulated gene, PMEPA1, located on chromosome 20q13 exhibits high level expression in prostate. Genomics 66(3):257–263
Xu LL, Shi Y, Srivastava S et al (2003) PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Res 63(15):4299–4304
Macias MJ, Wiesner S, Sudol M (2002) WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett 513(1):30–37
Li H, Xu LL, Srivastava S et al (2008) A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J Biol Chem 283(43):28988–28995
Yang B, Kumar S (2010) Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ 17(1):68–77
Giuseppe G, Lucia DM, Alberto G et al (2003) EGF- and cell-cycle-regulated STAG1/PMEPA1/ERG1.2 belongs to a conserved gene family and is overexpressed and amplified in breast and ovarian cancer. Mol Carcinog 38(4):188–200
Ciechanover A (1998) The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J 17(24):7151–7160
Hershko A, Ciechanover A (1998) The ubiquitin system. Ann Rev Biochem 67:425–479
Ciechanover A (2005) Intracellular protein degradation: from a vague idea thru the lysosome and the ubiquitin–proteasome system and onto human diseases and drug targeting. Cell Death Differ 12(9):1178–1190
Ing B, Kotler AS, Rotin D et al (2007) Regulation of commissureless by the ubiquitin ligase DNedd4 is required for neuromuscular synaptogenesis in drosophila melanogaster. Mol Cell Biol 27(2):481–496
Acknowledgments
This research is supported by the program for Changjiang Scholars and Innovative Research Team in University (IRT1166).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this paper
Cite this paper
Jing, L., Huo, X., Li, Y., Li, Y., Diao, A. (2015). Identification of the Binding Domains of Nedd4 E3 Ubiquitin Ligase with Its Substrate Protein TMEPAI. In: Zhang, TC., Nakajima, M. (eds) Advances in Applied Biotechnology. Lecture Notes in Electrical Engineering, vol 332. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-662-45657-6_6
Download citation
DOI: https://doi.org/10.1007/978-3-662-45657-6_6
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-45656-9
Online ISBN: 978-3-662-45657-6
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)