Abstract
The mammalian mitochondrial translation machinery is responsible for the synthesis of 13 mitochondrially encoded proteins that are essential for energy production. These proteins are subunits of the oxidative phosphorylation complexes embedded in the inner membrane of mitochondria. Mitochondrial protein synthesis is highly similar to that of bacterial systems; however, there are subtle differences between these systems in terms of their mechanisms and components. In this review, we will discuss the elements of mitochondrial translation, including the stages of protein synthesis and the factors involved in these processes. Although much still waits to be learned about the regulation of this system, a summary of what is currently known about the regulation of its protein components by post-translational modifications, specifically concerning energy metabolism, will also be included in this chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Mitochondrial Protein Synthesis
- Bacterial Ribosome
- Mitochondrial Translation
- OXPHOS Complex
- Mammalian Mitochondrion
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
2.1 Background
2.1.1 The Role of Mitochondrial Translation in Energy Metabolism
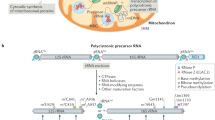
Mitochondria provide more than 90 % of the energy used by mammalian cells through the process of oxidative phosphorylation (OXPHOS). The 13 mitochondrially encoded proteins that are synthesized by the mitochondrial translation machinery are integral components of the electron transfer and ATP synthase complexes and are essential for energy metabolism. They are localized in the inner membrane (IM) of mitochondria and include seven subunits of complex I (NADH:ubiquinone oxidoreductase), one subunit of complex III (ubiquinone:cytochrome c oxidoreductase), three subunits of complex IV (cytochrome c:oxygen oxidoreductase), and two subunits of complex V (ATP synthase) (Fig. 2.1). The remaining subunits of the OXPHOS complexes, in addition to the ~1,500 proteins that support energy metabolism, are the products of nuclear genes. These proteins are synthesized by cytoplasmic ribosomes and imported into the mitochondria.
Oxidative phosphorylation and regulation of the mitochondrial translation machinery. Mammalian mitochondria contain a 16.5 kb circular genome (mtDNA) which encodes for 22 tRNAs, two rRNAs, and nine monocistronic and two dicistronic mRNAs. Mammalian mitochondrial ribosomes (28S and 39S subunits) are responsible for the synthesis of 13 mitochondrially encoded proteins that are subunits of complex I (blue), III (pink), IV (cyan), and ATP synthase, also known as complex V (orange). Metabolic levels of acetyl-CoA, NADH/NAD+, and ATP are important for the regulation of the mitochondrial translation machinery through reversible acetylation and phosphorylation
Mitochondrial energy metabolism, including the production of mitochondrial translation components, requires coordination of mitochondrial transcription and cytoplasmic translation. As described below, all the proteins supporting mitochondrial protein synthesis are synthesized by the cytoplasmic ribosomes. However, little is known about the retrograde regulation of mitochondrial energy metabolism and translation. The availability of the high energy molecules, Acetyl-CoA, NADH/NAD+, and ATP, could be the major regulator of energy metabolism and protein synthesis in mammalian mitochondria via post-translational modifications by reversible acetylation and phosphorylation (Fig. 2.1). A brief analysis of this hypothesis will be discussed at the end of this chapter.
2.1.2 The Mitochondrial Genome
In mammals, mitochondrial DNA (mtDNA) contains about 16.5 kilobase pairs. It is a circular genome that encodes for 13 of the proteins of the OXPHOS complexes, as well as 22 tRNAs and two rRNAs required for protein synthesis (Fig. 2.1). Nuclear DNA encodes for the majority of mitochondrial proteins and most subunits of the OXPHOS complexes. The 13 polypeptides encoded in the mammalian mitochondrial genome are synthesized from nine monocistronic mRNAs and two dicistronic mRNAs, which have overlapping reading frames (Jackson 1991; Anderson et al. 1982; Wolstenholme 1992). These mRNAs are quite unusual, almost entirely lacking 5′ and 3′ untranslated nucleotides. The translational start codon is generally located within three nucleotides of the 5′ end of the mRNA; therefore, mammalian mitochondrial mRNAs are defined as leaderless mRNAs (Anderson et al. 1982; Montoya et al. 1981). Analysis of the 5′ ends of human mitochondrial mRNAs reveals post-transcriptional processing of the 5′ ends in 8 of the 11 mRNAs (Montoya et al. 1981). Moreover, they have minimal secondary structures at their 5′ ends (Jones et al. 2008).
In animal mitochondria, the 22 tRNAs are encoded by the mitochondrial genome. They are shorter than other tRNAs and lack the conserved nucleotides that play important roles in tRNA-folding (Watanabe 2010). These tRNAs fold into the basic cloverleaf structure of canonical tRNAs. However, they lack a number of the tertiary interactions that are highly conserved in prokaryotic and eukaryotic cytoplasmic tRNA (Watanabe 2010; Helm et al. 2000). Although no crystal structure information is available for mitochondrial tRNAs, an L-shaped tRNA with a caved-in elbow region was found to be tightly bound to the mitochondrial ribosome at the P-site in cryo-EM reconstitution studies (Sharma et al. 2003).
Another interesting feature of the mitochondrial genome is the presence of only two rRNA species, 12S and 16S, for the small and large subunit rRNAs, respectively (Pietromonaco et al. 1991). A recent report suggests that a 5S rRNA is also imported into mitochondria; however, it has not been confirmed whether the imported 5S rRNA is incorporated into the mitochondrial ribosome (Smirnov et al. 2011). In addition to leaderless and short tRNAs, mitochondria contain rRNAs that are considerably smaller than their counterparts in bacterial and eukaryotic cytoplasmic ribosomes (Koc et al. 2010). The compactness of the mammalian mitochondrial genome suggests that this phenomenon is not coincidental and has possibly evolved to minimize damage from the oxidative environment of mammalian mitochondria.
2.1.3 Mitochondrial Translation Machinery
The mammalian mitochondrial translation machinery is composed of ribosomes, tRNAs, mRNAs, recycling factors, and the factors of translation initiation, elongation, and termination. Although emerging studies suggest RNA import into the mitochondria, all of the RNA components that are involved directly in the translation machinery, including rRNAs, tRNAs, and mRNAs, are encoded by the mitochondrial genome (Attardi 1985; Attardi et al. 1982). All the other components (translation factors and ribosomal proteins) are products of nuclear-encoded genes and are translated into proteins by cytoplasmic ribosomes before they are imported into the mitochondria.
The largest component of the translation machinery is the mitochondrial 55S ribosome, which is comprised of 28S and 39S subunits, called the small and large subunits, respectively (O’Brien 1971). Mitochondrial ribosomes have a molecular mass of about 2.7 × 106 Da, approximately the size of bacterial ribosomes. As mentioned above, mitochondrial rRNAs are shorter than their bacterial counterparts. They are truncated in specific locations, either in certain regions of their secondary structure or in entire domains (Koc et al. 2010). The missing regions in mammalian mitochondrial rRNAs are, for the most part, located on the periphery of bacterial rRNAs (Zweib et al. 1981; Glotz et al. 1981) and appear to be replaced by ribosomal proteins (Koc et al. 1999, 2000, 2001a, b, c). In fact, cryo-EM reconstitution studies have revealed that the truncated rRNA regions and domains are substituted with mitochondrial ribosomal proteins (Sharma et al. 2003). The combination of shorter rRNAs and a greater quantity of proteins has led us to describe mammalian mitochondrial ribosomes as “protein-rich” (Sharma et al. 2003; Koc et al. 2001b, c). For a more detailed and comprehensive analysis of the structural characteristics of mammalian mitochondrial ribosomes, readers should refer to Chap. 1.
The small subunit of the bovine mitochondrial ribosome has about 29 proteins, of which 14 have homologs in prokaryotic ribosomes. The remaining 15 proteins are specific to mitochondrial ribosomes. In contrast, the large subunit of bovine mitochondrial ribosomes contains about 48 proteins. Of these, 28 are homologs of bacterial ribosomal proteins, while the remaining 20 are unique to mitochondrial ribosomes. Only 15 of the mitochondria-specific proteins have homologs in the yeast mitochondrial ribosome (Koc et al. 2001b, c; Smits et al. 2007). This observation indicates that there is a significant divergence between the protein composition of mitochondrial ribosomes in higher and lower eukaryotes. Again, this high protein and shortened rRNA arrangement was probably favored during the evolution of mitochondria from endosymbiotic bacteria in order to protect rRNA, which is more prone to oxidative damage than proteins.
Mammalian mitochondrial ribosomes resemble bacterial ribosomes more closely than eukaryotic cytoplasmic ribosomes, as shown in the homology of bacterial and mammalian mitochondrial protein components. About half of the mitochondrial ribosomal proteins (MRPs) have homologs in bacterial ribosomes, while the remaining proteins represent a new class of ribosomal proteins that is specific to mitochondria (Koc et al. 2001b, c, 2010). Conversely, bacterial ribosomes contain certain proteins not preserved in mitochondrial ribosomes. The distribution of mitochondria-specific proteins on the exterior surface of the ribosome is visible in cryo-EM studies; however, additional high-resolution structural information is still needed to determine the exact function of these proteins in translation (Sharma et al. 2003). Our studies suggest that mitochondria-specific proteins are replacements of the bacterial 70S ribosomal proteins that do not have clear homologs in mitochondrial ribosomes (Koc et al. 2001b, c; Suzuki et al. 2001). In addition to their primary functions in protein synthesis, several mitochondria-specific MRPs, including MRPS29 (also known as DAP3) and MRPS30, are reported to be involved in apoptosis and various diseases (Henning 1993; Kissil et al. 1995; Mariani et al. 2001; Takeda et al. 2007; Bhatti et al. 2010; Stacey et al. 2008; Woolcott et al. 2009). The disease-causing mutants and defects of these MRPs and other mitochondrial translation components has been elegantly summarized in several recent reviews (Rotig 2011; Smits et al. 2010; Christian and Spremulli 2011).
2.1.4 Interactions of Mammalian Mitochondrial Ribosomes with the Inner Membrane
Along with mtDNA and transcription machinery, mitochondrial ribosomes are associated with the IM, and mitochondrially encoded subunits are co-translationally inserted into the OXPHOS complexes (Bogenhagen 2009; Wang and Bogenhagen 2006; Liu and Spremulli 2000; Mick et al. 2012; Ott and Herrmann 2010). These 13 proteins are located at the hydrophobic cores of the OXPHOS complexes, possibly to prevent proton leakage and provide tightly coupled mitochondria. Studies in yeast provided most of our knowledge of the interactions of mitochondrial ribosomes with the IM and the assembly of mitochondrially encoded subunits of OXPHOS (Ott and Herrmann 2010; Bonnefoy et al. 2001; Naithani et al. 2003; Hell et al. 1998). The majority of the proteins involved in these processes are either absent or not highly conserved in mammals. In mammals, only three IM proteins Oxa1L, Cox18, and LetM, have been found to interact with mitochondrial ribosomes, which are homologous to yeast IM proteins Oxa1p (a homolog of bacterial Yidc, Cox18p or Oxa2, and Mdm38p, respectively (Bonnefoy et al. 2009; Frazier et al. 2006; Gaisne and Bonnefoy 2006). It was recently proposed that the components conserved in both systems regulate mitochondrial translation in response to the assembly state of the OXPHOS complexes (Mick et al. 2012). It is possible that this response mechanism, which depends on the availability of its components, is the most efficient way to regulate mitochondrial biogenesis at multiple stages. Interaction of mitochondrial ribosomes with the IM and co-translational insertion of mitochondrially encoded components can be found in several recent reviews (Christian and Spremulli 2011; Ott and Herrmann 2010; Fox 2012).
2.2 Protein Synthesis in Mammalian Mitochondria
Protein synthesis occurs in four stages, designated as initiation, elongation, termination, and ribosome recycling. Here, we will briefly describe the protein factors and mechanisms involved in these processes. The majority of factors involved in protein synthesis in mammalian mitochondria were initially discovered and characterized in the Spremulli laboratory (see recent reviews by Spremulli et al. (2010, 2011, 2011) for the detailed structural and mechanistic aspects of these proteins).
2.2.1 Translation Initiation
Translation initiation starts with the dissociation of ribosomes into their subunits. The formylated Met-tRNA (fMet-tRNA) is base-paired to the start codon on the mRNA in the P-site of the small subunit. This process is stimulated by three translation initiation factors in bacteria: IF1, IF2, and IF3. Although IF1 has been viewed as a universal translational initiation factor in bacteria, only two mammalian mitochondrial initiation factors, mitochondrial initiation factor 2 (IF2mt) and mitochondrial initiation factor 3 (IF3mt), have been identified to date (Koc and Spremulli 2002; Liao and Spremulli 1990, 1991). We have shown that these two factors are sufficient to assemble an initiation complex on 55S ribosomes with fMet-tRNA in vitro (Koc and Spremulli 2002; Grasso et al. 2007).
In the current model (Fig. 2.2), the first step is the dissociation of the 55S ribosome into its subunits by IF3mt and formation of the 28S-IF3mt complex. It is proposed that mRNA could enter via a protein-rich mRNA entrance and bind to the 28S subunit first (Sharma et al. 2003); however, the exact mechanism of mRNA binding to the ribosome is not known. Currently, the order of mRNA, fMet-tRNA, and IF2mt:GTP binding to the 28S-IF3mt complex is not clear (Christian and Spremulli 2011; Grasso et al. 2007). For simplicity, these processes are shown together in Step 3. Toeprinting analysis suggests that mRNA movement is paused on the 28S subunit to inspect the codon at the 5′ end of the mRNA (Christian and Spremulli 2010). During this pause, IF2mt:GTP can promote the binding of fMet-tRNA to the 5′ AUG start codon of leaderless mitochondrial mRNAs on the 28S the ribosome (Step 3). Next, the large subunit joins the 28S initiation complex, IF2mt hydrolyzes GTP to GDP, and the initiation factors are released, resulting in a complete 55S initiation complex (Step 4).
Model for the initiation stage mitochondrial protein synthesis. The process of initiation begins with the 39S and 28S subunits associated as a 55S ribosome. Mitochondrial initiation factor 3 (IF3) binds and dissociates the 55S ribosomes into its subunits (Step 1). The IF3mt remains bound to the 28S subunit (Step 2). Thereafter, fMet-tRNA, mRNA, and mitochondrial initiation factor 2 (IF2), which is bound to GTP (red circle), bind to the 28S subunit (Step 3). The presence of the proper start codon (AUG) allows the mRNA to be locked into place by codon–anticodon interactions, and this setup forms the 28S initiation complex. The 39S subunit joins the initiation complex, and GTP is hydrolyzed to GDP (orange circle) to allow IF2mt and IF3mt to dissociate (Step 4)
As mentioned above, mammalian mitochondrial mRNAs lack the 5′ methylguanylate cap structure and the canonical Shine-Dalgarno helix found in eukaryotic and bacterial mRNAs, respectively, to position the start codon at the P-site of the ribosome. However, the location of the start codon within one or two nucleotides of the 5′-end of leaderless mRNAs has been shown to be critical in initiation complex formation in vitro (Christian and Spremulli 2010). These unusual features of leaderless mRNAs suggest the presence of a novel mRNA recognition and binding mechanism to ribosomes, possibly provided by mitochondria-specific ribosomal proteins or additional protein factors in mammalian mitochondria. It is possible that this is one of the most highly regulated steps in mitochondrial translation, but this remains to be investigated.
2.2.1.1 Mitochondrial Translation Initiation Factor 2
Mammalian mitochondrial IF2mt, which is homologous to bacterial IF2, was initially characterized and purified from bovine liver (Schwartzbach et al. 1996). The full-length human, bovine, and mouse IF2mt are all 727 amino acids (aa) long. The predicted mature form, which includes residues 78-727, can be stably expressed in E. coli (Claros and Vincens 1996). This recombinant protein is capable of promoting the binding of the initiator tRNA (fMet-tRNA) to mitochondrial 28S subunits or 55S ribosomes in the presence of GTP and synthetic mRNAs. Mammalian IF2mt can also stimulate the binding of fMet-tRNA to bacterial ribosomes; however, bacterial IF2 cannot stimulate formation of the initiation complex on mitochondrial ribosomes (Ma and Spremulli 1995).
When compared to the six-domain model of E. coli IF2, mammalian IF2mt covers domains III–VI, with an additional small insertion domain that is only found in animal mitochondria (Christian and Spremulli 2011; Atkinson et al. 2012; Yassin et al. 2011; Spremulli et al. 2004). The electron density map of the initiation complex formed with bovine IF2mt and 70S ribosomes at 10.8 Å reveals a three-dimensional model for the structure of this factor (Fig. 2.3a) (Yassin et al. 2011). Although domain III could not be modeled in this structure, the deletion mutations of IF2mt suggest that this domain interacts with the ribosome and makes important contacts with the 28S subunit (Spencer and Spremulli 2005). Domain IV, which is called the G-domain and is the most highly conserved domain, is also responsible for effective binding to 28S subunits. The presence of the G-domain is required for the proper arrangement of IF2mt domains, specifically domain V and its interactions with the large subunit near the ribosomal L7/L12 stalk and the sarcin-ricin loop (Allen et al. 2005; La Teana et al. 2001). The small insertion domain is located between domains V and VIC1, and its structural aspects are discussed in Chap. 1 by Agrawal et al. The region of IF2mt responsible for interacting with fMet-tRNA has been mapped to the VIC2 subdomain (Fig. 2.3a) (Spencer and Spremulli 2004). The mammalian mitochondrial genome encodes only a single tRNAMet gene that is partitioned between initiation and elongation after aminoacylation. This is also a unique feature that is only found in the animal mitochondrial translation system. Translation initiation starts with a formylated Met-tRNA (fMet-tRNA); therefore, only a fraction of Met-tRNA is formylated by a mitochondrial Met-tRNA formylase (Takeuchi et al. 1998). The formylated form interacts with IF2mt and is used in translation initiation, while the unformylated Met-tRNA interacts with EF-Tumt and is channeled into elongation. It is postulated that this partitioning begins with the competition between transformylase and EF-Tumt (Spencer and Spremulli 2004).
Structural models of mammalian mitochondrial initiation factors. a Model for the 3-D structure of IF2mt, based on the cryo-EM map of IF2mt:70S initiation complex (PDB# 3IZY). Domain III has been omitted from the structure due to the resolution obtained with the cryo-EM images. Domain IV is shown in blue, domain V is shown in purple, the insertion domain is shown in orange, domain VI-C1 is shown in green, and domain VI-C2 is shown in pink. b Structural model of IF3mt based on the crystal structure of the N-terminal domain of G. stearothermophilus IF3 and the NMR structure of the murine IF3mt C-terminal domain (PDB# 1TIF and 2CRQ)
2.2.1.2 Mitochondrial Translation Initiation Factor 3
We discovered mammalian IF3mt in homology searches of human and mouse ESTs using IF3 sequences from various species as queries (Koc and Spremulli 2002). The coding region of human IF3mt contains 278 amino acid residues. IF3mt is highly conserved in mammals, while the sequence homology is 35–40 % in vertebrates. Recently, a clear homolog in Saccharomyces cerevisiae was reported (Atkinson et al. 2012). Recombinant human IF3mt was generated after removing the 31-residue signal peptide predicted by MitoProtII (Koc and Spremulli 2002). Bacterial factors contain two globular domains separated by a linker region (Biou et al. 1995). The structural model of IF3mt based on the crystal structure of the N-terminal and linker domains of Geobacillus stearothermophilus IF3 and the NMR structure of the C-terminal domain of mouse IF3mt indicates that the mammalian factor is also organized in a dumbbell shape (Fig. 2.3b) (Moreau et al. 1997). The N-terminal homology domain of IF3mt contains an α-helix folded against four β-sheets (Fig. 2.3b). The C-terminal domain of IF3mt is folded into a similar structure, except with two α-helices (Fig. 2.3b). Although the linker region contains a helical segment in bacteria, the mitochondrial linker is predicted to be a partial α-helix. Deletion mutations in IF3mt biochemical studies suggest that this linker is highly flexible prior to binding to the ribosome (Christian and Spremulli 2009).
IF3mt promotes the dissociation of 55S ribosomes into their subunits and stimulates the binding of fMet-tRNA to mitochondrial 28S subunits in the presence of IF2mt and mRNA (Koc and Spremulli 2002). It also prevents premature binding of fMet-tRNA to ribosomes prior to mRNA binding (Bhargava and Spremulli 2005). However, the proofreading activity found in bacterial IF3 is not conserved in IF3mt (Petrelli et al. 2001). This lack of proofreading activity is postulated to be due to the recognition of both AUG and AUA as start codons and to the partitioning of tRNAMet between translation initiation and elongation (Christian and Spremulli 2011). The deletion and point mutations of IF3mt and their roles in translation initiation have been studied extensively (Christian and Spremulli 2009; Bhargava and Spremulli 2005; Haque and Spremulli 2008). The removal of the N-terminal domain and the linker region only slightly affects IF3mt activity, whereas truncation of the C-terminal domain completely inactivates IF3mt function and its binding to the 28S subunit. It has also been shown that the C-terminal domain of IF3mt is sufficient by itself to promote initiation complex formation with the 55S ribosome (Christian and Spremulli 2009). In fact, Ala mutations of several critical residues at the C-terminal domain of human IF3mt, located at positions 170–171 and 175, result in loss of its dissociation function, and therefore, initiation complex formation (Christian and Spremulli 2009). This observation clearly supports the IF3mt mechanism of action proposed in Fig. 2.2. The N- and C-terminal extensions of IF3mt also play roles in monitoring the sequence of initiation complex formation by reducing the affinity of the factor for the 39S subunit and preventing the premature binding of fMet-tRNA, respectively (Fig. 2.4) (Christian and Spremulli 2009, 2011).
Model for the elongation and termination steps of mitochondrial protein synthesis. During elongation, the tRNA with the growing polypeptide chain or fMet-tRNA is located in the P-site of the ribosome. A second aa-tRNA is brought to the A-site of the ribosome by EF-Tumt (Tu), which is bound to GTP (shown as a red circle) (Step 1). As GTP is hydrolyzed to GDP (depicted as an orange circle), EF-Tumt is released from the ribosome, and at this step, EF-Tsmt (Ts) is required for GDP-GTP exchange (Step 2). In the next step, peptide bond synthesis is catalyzed and the growing polypeptide chain is transferred to the aa-tRNA in the A-site (Step 3). Next, EF-G1mt:GTP (G1 with a red circle) binds to the ribosome’s A-site and catalyzes its translocation (Step 4). This moves the deacetylated tRNA out of the P-site and shifts the newly acetylated tRNA to the P-site (Step 5). Elongation proceeds until the ribosome recognizes a termination codon. The beginning of termination is signaled by the termination codon (shown here as UAG), which enters the ribosome’s A-site (Step 1). Both mtRF1a (F1a) and GTP bind to the A-site, and the polypeptide chain is released with hydrolysis of GTP (Step 2). The mechanism for the release of mtRF1a and GDP from the ribosome is still unknown. RRF1mt (RRF) then binds to the A-site, followed by EF-G2mt (G2, also known as RRF2mt) (Step 3). These two release factors promote the dissociation of the ribosomal subunits as well as the release of the mRNA and deacetylated tRNA (Step 4). The ribosome is free to perform further cycles of translation after RRF1mt and EF-G2mt are released
In bacteria, IF3 binds to the platform region of the small subunit, and this region is one of the most highly conserved regions of the mitochondrial ribosome (Sharma et al. 2003; Koc et al. 2001b; McCutcheon et al. 1999; Pioletti et al. 2001). We have identified the contacts between IF3mt and the 28S subunit using cross-linking assays in combination with identification of cross-linked ribosomal proteins by mass spectrometry (Haque et al. 2011). In this analysis, MRPs with bacterial homologs (such as MRPS5, MRPS9, MRPS10, and MRPS18-2) and proteins with no homologs in bacterial ribosomes (such as MRPS29, MRPS32, MRPS36, and PTCD3) are identified as the 28S subunit proteins that interact with IF3mt. The interaction between IF3mt and MRPS29 (DAP3) is unexpected, because IF3mt was found to be located at the lower part of the 28S subunit by immunoelectron microscopy (O’Brien et al. 2005). The locations of MRPS32, MRPS36, and PTCD3 in the small subunit are not known; however, we have shown that PTCD3 is one of the proteins that interacts with mRNA on the 28S subunit (Koc and Spremulli 2003). The group of bacterial homologs of MRPs cross-linked to IF3mt is different from the ribosomal proteins that interact with the bacterial IF3 in the platform of the 30S subunit. This difference is possibly due to the mitochondria-specific ribosomal proteins surrounding the platform region in the 28S subunit (Koc and Spremulli 2003).
2.2.2 Translation Elongation in Mammalian Mitochondria
During elongation, mRNA is decoded sequentially as amino acids are incorporated into the growing polypeptide chain (Fig. 2.4). In contrast to the differences observed between the mitochondrial and bacterial translation initiation processes, elongation is highly conserved in these systems (Spremulli et al. 2004). The basic mechanism of mitochondrial translation elongation is summarized in Fig. 2.4. In the first step, a ternary complex formed with GTP, mitochondrial elongation factor Tu (EF-Tumt), and aminoacyl-tRNA (aatRNA) enters the A-site of the 55S ribosome, which is occupied with either fMet-tRNA or aa-tRNA at the P-site (Step 1). Formation of the correct cognate ternary complex hydrolyzes GTP and causes the release of EF-Tu:GDP in Step 2. Elongation Factor Ts (EF-Tsmt) supports the GDP/GTP exchange by forming an EF-Tumt:EF-Tsmt complex. The peptidyl-transferase activity of the ribosome catalyzes peptide bond formation and extends the peptide chain by one residue on the peptidyl-tRNA at the A-site, leaving a deacylated tRNA in the P-site (Step 3). In the next step, deacetylated tRNA is removed (Step 4) and peptidyl-tRNA at the A-site is translocated into the P-site (Step 5) by mitochondrial Elongation Factor G1 (EF-G1mt). In bacteria, the deacylated tRNA is progressively released from the E-site; however, a corresponding E-site tRNA-binding site was not found in the cryo-EM studies of the mitochondrial ribosome (Sharma et al. 2003). At the final stage of elongation, ribosome with peptidyl-tRNA at the P-site is ready for either a subsequent elongation cycle or for termination. Mammalian mitochondrial elongation factors, specifically EF-Tumt and EF-Tsmt, have been extensively studied, either using their native forms from bovine mitochondria or recombinant proteins from E. coli (Schwartzbach and Spremulli 1989; Woriax et al. 1995, 1996; Xin et al. 1995). Here, we will briefly discuss the properties of the mammalian mitochondrial elongation factors. For more comprehensive information on mitochondrial elongation factors, readers are referred to previous reviews (Christian and Spremulli 2011; Spremulli et al. 2004).
2.2.2.1 Mitochondrial Elongation Factors Tu and Ts
EF-Tumt and EF-Tsmt were the first mitochondrial translation factors identified and characterized from bovine mitochondria (Schwartzbach and Spremulli 1989). EF-Tumt is a 452 aa long protein in mammalian mitochondria. The mature form of the bovine protein is 409 residues long and is 55–60 % identical to bacterial EF-Tu. Crystal structures of the bacterial EF-Tu ternary complexes from several different species have been solved (Nissen et al. 1995, 1999). The crystal structure of bovine EF-Tumt has similarities to bacterial EF-Tu and is folded into three domains (Jeppesen et al. 2005) (Fig. 2.5). Domain I binds guanine nucleotides and forms the 3′-end of the aa-tRNA-binding site along with domain II. These domains also interact with the small subunit during the tRNA delivery to the peptidyl-transferase active center. The extended acceptor-TΨC helix of the aa-tRNA interacts with domain III. A binding pocket for the 5′-end and the acceptor stem of the aa-tRNA is provided by all three domains (Fig. 2.5).
Model of the bovine EF-Tumt:EF-Tsmt complex with tRNA. In this model of the bovine EF-Tumt:EF-Tsmt complex (PDB# 1XB2) with E. coli Cys-tRNACys (PDB# IB23), EF-Tumt, EF-Tsmt, and tRNA are shown in violet, blue, and orange, respectively. Individual domains observed in the structure are indicated. The positions of two disease-causing mutations found in EF-Tumt (R336) and EF-Tsmt (R325) are labeled in red. EF-Tumt is phosphorylated at highly conserved Thr273 and Ser312 residues (shown in cyan) near the tRNA-binding site and acetylated at Lys88 and Lys 256 (shown in yellow)
Studies performed with chimeric bacterial EF-Tu and EF-Tumt proteins reveal that domains I and II of the mitochondrial factor are primarily responsible for aa-tRNA delivery to the ribosome (Bullard et al. 1999; Hunter and Spremulli 2004a). It is also suggested that the formation of codon–anticodon interactions possibly causes conformational changes in the body of the tRNA and sends a signal to domain I, triggering GTP hydrolysis. This observation is extremely valuable in explaining mitochondrial myopathies caused by mutations in mitochondrial tRNAs, specifically those causing conformational changes, rendering them inefficient in triggering the GTPase activity of EF-Tu (Hunter and Spremulli 2004b, c, d; Shi et al. 2012; Hao and Moraes 1997; Kelley et al. 2000; Ling et al. 2007) (Fig. 2.5).
In mammals, the most common form of EF-Tsmt is 325 amino acid residues in length, and the mature protein is about 31 kDa (Xin et al. 1995). Although bacterial EF-Ts is found as a free protein, EF-Tsmt has been identified to be complexed with EF-Tumt (Schwartzbach and Spremulli 1989). EF-Tsmt is only 25–30 % identical to its bacterial counterpart. In agreement with this low sequence homology, the structure of EF-Tsmt is also less conserved in comparison to the structural homology between mitochondrial and bacterial EF-Tus. One of the most remarkable differences is the complete loss of the coiled-coil domain structure of the C-terminal domain, in contrast to the similar folds found in the N-terminal domain (Fig. 2.5). Likewise, the β-strand folds located in the central domain of EF-Tsmt are organized differently in the bacterial protein. The central domain of EF-Tsmt interacts with domains I and II in EF-Tumt, and this region undergoes substantial conformational changes during the nucleotide exchange process between EF-Tumt and EF-Tsmt (Fig. 2.5) (Jeppesen et al. 2005).
The mechanistic and structural aspects of mammalian mitochondrial translation elongation factors described above have been extensively studied and reviewed by the Spremulli laboratory (Christian and Spremulli 2011; Spremulli et al. 2004). The in vitro studies performed by this laboratory have shed light on the role of these factors in metabolic diseases and cancers, as reported recently (Shi et al. 2012; Smeitink et al. 2006; Skrtic et al. 2011; Akama et al. 2010). For a comprehensive representation of the roles of these factors in human diseases, readers should refer to the previous reviews (Rotig 2011; Akama et al. 2010).
2.2.2.2 Mitochondrial Elongation Factor G1
There are two forms of EF-Gmt: EF-G1mt and EF-G2mt (Tsuboi et al. 2009; Hammarsund et al. 2001). EF-G1mt catalyzes the translocation of peptidyl-tRNA from the A-site to the P-site while dislocating P-site tRNA from the ribosome (Fig. 2.4). EF-G2mt is involved in termination. Full-length EF-G1mt is 751 aa residues, and both native and recombinant forms of mammalian EF-G1mt are active in catalyzing translocation in both mitochondrial and bacterial translation elongation in vitro (Tsuboi et al. 2009; Chung and Spremulli 1990; Bhargava et al. 2004). On the other hand, bacterial EF-G does not support mitochondrial translocation (Chung and Spremulli 1990). Another unusual feature of mammalian EF-G1mt is the resistance of this factor to fusidic acid (Bhargava et al. 2004). The specificity of EF-G1mt can be attributed to the structural differences between the EF-G sequence and the L7/L12 stalk region of mitochondrial and bacterial ribosomes (Bhargava et al. 2004; Terasaki et al. 2004).
2.2.3 Termination of Mitochondrial Translation and Ribosome Recycling
The final stage of protein synthesis is termination and the dissociation of the ribosome complex for recycling. In the proposed model, UAA and UAG serve as stop codons in mammalian mitochondria (Fig. 2.4a). In the first step of termination, these stop codons move into the A-site and are recognized by mtRF1a (referred to as F1a in Fig. 2.4). In humans, two arginine codons, AGA and AGG, also function as terminal codons; however, these codons promote -1 frameshifting and reposition the standard UAG codon at the A-site for termination (Temperley et al. 2010). The binding of mtRF1a to the ribosome triggers GTP hydrolysis and the release of the completed peptide during Step 2. In the following step, ribosomes are ready to be recycled and to release mtRF1a. This step requires RF3 in bacteria. However, no factor homologous to RF3 has been found in mitochondria; thus, the mechanism of mtRF1a dissociation is not known. In Step 3, mitochondrial ribosome recycling factor (RRF1mt, abbreviated as RRF in Fig. 2.4) and EFG2mt (also known as RRF2mt and abbreviated as G2 in Fig. 2.4) act together to release mRNA and P-site tRNA and to dissociate 55S ribosomes into the subunits. The release of these factors from the ribosomes, accompanied by GTP hydrolysis, prepares them for the next round of protein synthesis in Step 4 (Fig. 2.4) (Chrzanowska-Lightowlers et al. 2011).
2.2.3.1 Mitochondrial Release Factors
There are three mitochondrial proteins with significant homology to bacterial RF1: mtRF1a, c12orf65, and ICT1. All of these factors contain the GGQ motif, which is critical for the termination of protein synthesis because they all hydrolyze the peptidyl-tRNA bond (Chrzanowska-Lightowlers et al. 2011). Full-length human mtRF1a contains 445 amino acid residues and has been shown to participate in the termination of 11 human mitochondrial mRNAs at UAA and UAG codons (Zhang and Spremulli 1998; Soleimanpour-Lichaei et al. 2007). Therefore, it is described as the mitochondrial release factor used in the decoding process (Christian and Spremulli 2011; Soleimanpour-Lichaei et al. 2007). Currently, the function of c12orf65 is not known; however, ICT1 functions as a ribosome-dependent peptidyl-tRNA hydrolase because it is tightly associated with the large subunit of the mitochondrial ribosome (Richter et al. 2010). It is also proposed to have a role in the recycling of stalled ribosomes on damaged mRNAs or mRNAs lacking stop codons (Chrzanowska-Lightowlers et al. 2011). This is a possible replacement for the tmRNA mechanism found in bacteria (Christian and Spremulli 2011; Haque and Spremulli 2010).
2.2.3.2 Ribosome Recycling in Mammalian Mitochondria
The coding region of the mitochondrial ribosome recycling factor (mtRRF) is a 262 aa residue-long protein in mammals and is 25–30 % identical to bacterial RRF. In human cell lines, mtRRF has been shown to be essential (Rorbach et al. 2008). There are several alternatively spliced isoforms of mtRRF listed in protein databases such as NCBI and UniProt; however, it is not known whether these shorter isoforms function in the termination of protein synthesis. The bacterial form of mtRRF works with EF-G, the only translocation factor found in bacteria. In mammals, however, mtRRF and EF-G2mt cooperate to disassemble the ribosome:mRNA:tRNA complex in the termination steps (Steps 3 and 4) of mitochondrial translation (Fig. 2.4).
As described above, two proteins homologous to bacterial EF-G (EF-G1mt and EF-G2mt) are present in mammalian mitochondria. While EF-G1mt is responsible for translocation during chain elongation, EF-G2mt, also known as RRF2mt, functions in combination with mtRRF at the termination step (Tsuboi et al. 2009). In many organisms, there is only a single EF-G involved in both elongation and termination, as occurs in bacteria. However, mammalian EF-G2mt only serves in the termination process, and GTP hydrolysis is required for its release, along with mtRRF, from the ribosome (Tsuboi et al. 2009). In bacterial termination, GTP hydrolysis is necessary for ribosome recycling.
2.3 Regulation of Mitochondrial Protein Synthesis
Most of our knowledge of mammalian mitochondrial protein synthesis comes from either in vitro studies or from studies of disease-causing or lethal mutations in humans. Yet, the regulation of the mitochondrial translation machinery, which is essential for cellular homeostasis and survival, has not been explored in great detail. The regulation of mitochondrial protein synthesis requires coordinated expression of nuclear and mitochondrial genes at the transcriptional and translational levels. This retrograde signaling is proposed to be regulated by molecules derived from mitochondrial metabolism, such as reactive oxygen species (ROS), NAD+, Acetyl-coA, and ATP (Wallace 2012; Finley and Haigis 2009; Schieke and Finkel 2006). In addition, it has been suggested that organelle biogenesis in plants and animals is regulated by dual or multiple localizations of proteins and small RNA species, such as 5S rRNA and microRNAs, into the nucleus, cytosol, and mitochondria (Smirnov et al. 2011; Duchene and Giege 2012; Vedrenne et al. 2012; Ernoult-Lange et al. 2012). Regulation of protein synthesis by these small RNAs in yeast and mammalian mitochondria will be discussed in Chap. 4.
Recent technological progress in system biological approaches has demonstrated that these approaches have the potential to solve metabolite-regulated signaling processes in mammalian mitochondria. As indicated above, Acetyl-coA, NAD+, and ATP are the most essential metabolites of oxidative energy metabolism and are used for a variety of cellular functions in health and disease (Fig. 2.1). More importantly, these metabolites are involved in post-translational modifications and the modulation of protein activities by reversible acetylation and phosphorylation in mammalian mitochondria (O’Rourke et al. 2011; Hebert et al. 2013; Zhao et al. 2010). High-throughput proteomic surveys of mitochondrial proteins employed by many different laboratories have confirmed that these post-translational modifications are very common and are regulated by the activities of mitochondrially localized kinases and NAD+-dependent deacetylases (sirtuins), including SIRT3, SIRT4, and SIRT5 (Hebert et al. 2013; Zhao et al. 2010; Choudhary et al. 2009a; Kim et al. 2006; Michishita et al. 2005; Schwer et al. 2006; Onyango et al. 2002; Bell and Guarente 2011).
Protein synthesis in eukaryotic cytoplasm by 80S ribosomes is predominantly regulated by the phosphorylation of translation factors, specifically eIF2a (Kimball et al. 1998; Hershey 1989). Phosphorylation of eIF2a at Ser 51 results in the differential expression of mRNAs and directs many critical cellular events, such as cell growth and apoptosis. However, the regulatory role of phosphorylation on bacterial and mammalian mitochondrial translation systems has been overlooked until recently. Our recent studies have revealed that the regulation of mitochondrial translation by post-translational modifications, specifically by phosphorylation and acetylation, is far more widespread than originally thought (Miller et al. 2008, 2009; Yang et al. 2010). Additionally, the phosphorylation and acetylation of mammalian mitochondrial ribosomal proteins and translation factors have been mapped in high-throughput proteomics approaches using tandem mass spectrometry (Zhao et al. 2010; Choudhary et al. 2009a; He et al. 2001; Zhang et al. 2009; Sajid et al. 2011; Koc and Koc 2012). However, the regulation mechanism of mitochondrial translation factors by reversible acetylation and phosphorylation remains largely to be discovered. A comprehensive list of post-translationally modified bacterial and mammalian mitochondrial ribosomal proteins and translation factors can be found in a recent review by our group (Koc and Koc 2012).
Surprisingly, we observe that post-translationally modified aa residues are found in the functionally critical regions of translation factors and MRPs, supporting the hypothesis that these modifications play a role in the regulation of mitochondrial translation and, consequently, OXPHOS. For example, inhibition of bacterial protein synthesis by phosphorylation of EF-Tu has long been known; however, phosphorylation of Thr 118 has been recently demonstrated to inhibit tRNA binding in Mycoplasma tuberculosis EF-Tu (Sajid et al. 2011; Alexander et al. 1995). In two recent large-scale mouse phosphoproteome analyses, phosphorylation at Thr273 and Ser312 residues was mapped by mass spectrometry, and, interestingly, these two residues are located in the tRNA-binding region (Fig. 2.5) (Choudhary et al. 2009b; Huttlin et al. 2010). Moreover, most post-translationally modified regions of MRPs that are exposed to solvent are typically rich in Lys, Ser, Thr, and Tyr residues to support acetylation and phosphorylation. One of the unexpected observations included in our review is the conservation of modifications in both bacterial and mitochondrial ribosomes (Miller et al. 2009; Yang et al. 2010; Koc and Koc 2012; Soung et al. 2009). It is clear that these reversible modifications in the exposed parts of ribosomal proteins are likely to be involved in the recruitment of translation factors, rRNA and mRNA interactions, and subunit association.
One of the most active regions of mitochondrial and bacterial ribosomes during translation is the L7-L12 stalk. It is involved in the recruitment of translation factors IF2, EF-Tu, and EF-G in both bacteria and mammalian mitochondrial ribosomes (Uchiumi et al. 1999, 2002). This flexible region of the large subunit is highly conserved and is composed of L11, L10, and multiple copies of L7/L12, the exact number depending on the species. There is ample evidence that phosphorylation and acetylation in this region are involved in regulatory roles in both mitochondrial and bacterial ribosomes (Ilag et al. 2005; Traugh and Traut 1972; Mikulik et al. 2011; Gordiyenko et al. 2008). We have demonstrated that MRPL10 is the major acetylated protein in the mammalian mitochondrial ribosome and that an NAD+-dependent deacetylase, SIRT3, is involved in the deacetylation of MRPL10 (Yang et al. 2010). In SIRT3 knock-out mice mitochondria, we have shown that the acetylation of MRPL10 results in the enhancement of mitochondrial translation. MRPL12 was also found to be acetylated in unpublished studies from our laboratory and in a recent high-throughput mapping of acetylated proteins in SIRT3 knock-out mice mitochondria (Hebert et al. 2013; Yang et al. 2010; Han et al. 2013). This observation suggests that the role of L12 acetylation in bacteria is also conserved in mammalian mitochondrial ribosomes (Gordiyenko et al. 2008; Ramagopal and Subramanian 1974). Due to the fact that the stalk itself is very flexible and that mitochondria contain free pools of MRPL12 copies, it is likely that the components of the stalk transiently associate with the ribosome to regulate protein synthesis in mitochondria. In fact, a free pool of MRPL12 has been reported to interact with mitochondrial RNA polymerase (Wang et al. 2007; Surovtseva et al. 2011).
In addition to the MRPs with bacterial homologs, we observed extensive modifications of mitochondria-specific proteins found in the small subunit of the 55S ribosome (Miller et al. 2009). One of the most extensively studied pro-apoptotic proteins of the 28S subunit, MRPS29 (DAP3), is both acetylated and phosphorylated. Its phosphorylation near the GTP-binding domain has been shown to be crucial to its pro-apoptotic function in various cell lines (Takeda et al. 2007; Miller et al. 2008; Miyazaki et al. 2004). Aberrant expression of DAP3 has been detected in thyroid oncocytic, brain, and breast cancer tissues (Mariani et al. 2001; Jacques et al. 2009; Wazir et al. 2012). Clearly, further studies need to be done with mitochondrial translation factors and ribosomal proteins to shed light on their roles and the mechanisms by which they are regulated, including and starting with PTMs, to support cellular homeostasis.
2.4 Future Directions
Major milestones have been reached toward the complete characterization and identification of the components of mammalian mitochondrial translation. However, some mechanistic and structural aspects of mitochondrial translation have yet to be understood. One of the major unanswered questions that remains concerning the mechanism of mitochondrial protein synthesis is the recognition of leaderless mRNAs. The evidence so far points to a protein-dominated mechanism for mRNA recognition and binding to the mitochondrial ribosome. In addition, the components that facilitate co-translational insertion of mitochondrially encoded subunits of OXPHOS complexes into the IM need to be discovered and studied in greater detail in mammals. Retrograde signaling pathway(s) providing communication links between the mitochondrial and cytoplasmic translation machineries remain to be uncovered. Finally, the modulation of the translation machinery by reversible acetylation and phosphorylation and the enzymes involved in these reversible modifications are urgently waiting to be investigated.
References
Agrawal RK, Sharma MR, Yassin A, Lahiri I, Spremulli LL (2011) Structure and function of organaller ribosomes as revealed by cryo-EM in ribosomes: structure, functions and dynamics. Springer, New York
Akama K, Christian BE, Jones CN, Ueda T, Takeuchi N, Spremulli LL (2010) Analysis of the functional consequences of lethal mutations in mitochondrial translational elongation factors. Biochim Biophys Acta 1802:692–698
Alexander C, Bilgin R, Lindschau C, Mestes J, Kraal B, Hilgenfeld R, Erdmann V, Lippmann C (1995) Phosphorylation of elongation factor Tu prevents ternary complex formation. J Biol Chem 270:14541–14547
Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121:703–712
Anderson S, de Brujin M, Coulson A, Eperon I, Sanger F, Young I (1982) Complete sequence of bovine mitochondrial DNA: conserved features of the mammalian mitochondrial genome. J Mol Biol 156:683–717
Atkinson GC, Kuzmenko A, Kamenski P, Vysokikh MY, Lakunina V, Tankov S, Smirnova E, Soosaar A, Tenson T, Hauryliuk V (2012) Evolutionary and genetic analyses of mitochondrial translation initiation factors identify the missing mitochondrial IF3 in S. cerevisiae. Nucleic Acids Res 40:6122–6134
Attardi G (1985) Animal mitochondrial DNA: an extreme example of genetic economy. Int Rev Cytology 93:93–145
Attardi G, Chomyn A, Montoya J, Ojala D (1982) Identifaction and mapping of human mitochondrial genes. Cytogenet Cell Genet 32:85–98
Bell EL, Guarente L (2011) The SirT3 divining rod points to oxidative stress. Mol Cell 42:561–568
Bhargava K, Spremulli LL (2005) Role of the N- and C-terminal extensions on the activity of mammalian mitochondrial translational initiation factor 3. Nucleic Acids Res 33:7011–7018
Bhargava K, Templeton P, Spremulli LL (2004) Expression and characterization of isoform 1 of human mitochondrial elongation factor G. Protein Expr Purif 37:368–376
Bhatti P, Doody MM, Rajaraman P, Alexander BH, Yeager M, Hutchinson A, Burdette L, Thomas G, Hunter DJ, Simon SL (2010) Novel breast cancer risk alleles and interaction with ionizing radiation among U.S. radiologic technologists. Radiat Res 173:214–224
Biou V, Shu F, Ramakrishnan V (1995) X-ray crystallography shows that translational initiation factor IF3 consists of two compact α/β domains linked by an α-helix. EMBO J 14:4056–4064
Bogenhagen DF (2009) Biochemical isolation of mtDNA nucleoids from animal cells. Methods Mol Biol 554:3–14
Bonnefoy N, Bsat N, Fox TD (2001) Mitochondrial translation of Saccharomyces cerevisiae COX2 mRNA is controlled by the nucleotide sequence specifying the pre-Cox2p leader peptide. Mol Cell Biol 21:2359–2372
Bonnefoy N, Fiumera HL, Dujardin G, Fox TD (2009) Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta 1793:60–70
Bullard JM, Cai Y-C, Zhang Y, Spremulli LL (1999) Effects of domain exchanges between Escherichia coli and mammalian mitochondrial EF-Tu on interactions with guanine nucleotides, aminoacyl-tRNA and ribosomes. Biochim Biophys Acta 1446:102–114
Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009a) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840
Choudhary C, Olsen JV, Brandts C, Cox J, Reddy PN, Bohmer FD, Gerke V, Schmidt-Arras DE, Berdel WE, Muller-Tidow C et al (2009b) Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell 36:326–339
Christian BE, Spremulli LL (2009) Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry 48:3269–3278
Christian BE, Spremulli LL (2010) Preferential selection of the 5′-terminal start codon on leaderless mRNAs by mammalian mitochondrial ribosomes. J Biol Chem 285:28379–28386
Christian BE, Spremulli LL (2011) Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 1819:1035–1054
Chrzanowska-Lightowlers ZM, Pajak A, Lightowlers RN (2011) Termination of protein synthesis in mammalian mitochondria. J Biol Chem 286:34479–34485
Chung HK, Spremulli LL (1990) Purification and characterization of elongation factor G from bovine liver mitochondria. J Biol Chem 265:21000–21004
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:770–786
Duchene AM, Giege P (2012) Dual localized mitochondrial and nuclear proteins as gene expression regulators in plants? Front Plan Sci 3:221
Ernoult-Lange M, Benard M, Kress M, Weil D (2012) P-bodies and mitochondria: which place in RNA interference? Biochimie 94:1572–1577
Finley LW, Haigis MC (2009) The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res Rev 8:173–188
Fox TD (2012) Mitochondrial protein synthesis, import, and assembly. Genetics 192:1203–1234
Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P (2006) Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol 172:553–564
Gaisne M, Bonnefoy N (2006) The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res 6:869–882
Glotz C, Zweib C, Brimacombe R (1981) Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplasts and human and mouse mitochondrial ribosomes. Nuc Acids Res 9:3287–3306
Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV (2008) Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J Mol Biol 380:404–414
Grasso DG, Christian BE, Spencer A, Spremulli LL (2007) Overexpression and purification of mammalian mitochondrial translational initiation factor 2 and initiation factor 3. Methods Enzymol 430:59–78
Hammarsund M, Wilson W, Corcoran M, Merup M, Einhorn S, Grander D, Sangfelt O (2001) Identification and characterization of two novel human mitochondrial elongation factor genes, hEFG2 and hEFG1, phylogenetically conserved through evolution. Hum Genet 109:542–550
Han M-JC, H.; Koc H, Yang Y, Tong Q, Koc EC (2013) SIRT3 regulates mitochondrial translation by modulating MRPL12 binding to the ribosome (in preparation)
Hao H, Moraes CT (1997) A disease-associated G5703A mutation in human mitochondrial DNA causes a conformational change and a marked decrease in steady-state levels of mitochondrial tRNAASN. Mol Cell Biochem 17:6831–6837
Haque ME, Spremulli LL (2008) Roles of the N- and C-terminal domains of mammalian mitochondrial initiation factor 3 in protein biosynthesis. J Mol Biol 384:929–940
Haque ME, Spremulli LL (2010) ICT1 comes to the rescue of mitochondrial ribosomes. EMBO J 29:1019–1020
Haque ME, Koc H, Cimen H, Koc EC, Spremulli LL (2011) Contacts between mammalian mitochondrial translational initiation factor 3 and ribosomal proteins in the small subunit. Biochim Biophys Acta 1814:1779–1784
He H, Chen M, Scheffler NK, Gibson BW, Spremulli LL, Gottlieb RA (2001) Phosphorylation of mitochondrial elongation factor Tu in ischemic myocardium: basis for chloramphenicol-mediated cardioprotection. Circ Res 89:461–467
Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ et al (2013) Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49:186–199
Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA (1998) Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci U S A 95:2250–2255
Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6:1356–1379
Henning KA (1993) The molecular genetics of human diseases with defective DNA damage processing. PhD thesis, Stanford University, Stanford, USA
Hershey JW (1989) Protein phosphorylation controls translation rates. J Biol Chem 264:20823–20826
Hunter SE, Spremulli LL (2004a) Interaction of mitochondrial elongation factor Tu with aminoacyl-tRNAs. Mitochondrion 4:21–29
Hunter SE, Spremulli LL (2004b) Mutagenesis of Arg335 in bovine mitochondrial elongation factor Tu and the corresponding residue in the Escherichia coli factor affects interactions with mitochondrial aminoacyl-tRNAs. RNA Biol 1:95–102
Hunter SE, Spremulli LL (2004c) Mutagenesis of glutamine 290 in Escherichia coli and mitochondrial elongation factor Tu affects interactions with mitochondrial aminoacyl-tRNAs and GTPase activity. Biochemistry 43:6917–6927
Hunter SE, Spremulli LL (2004d) Effects of mutagenesis of residue 221 on the properties of bacterial and mitochondrial elongation factor EF-Tu. Biochim Biophys Acta 1699:173–182
Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143:1174–1189
Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, Robinson CV (2005) Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc Natl Acad Sci U S A 102:8192–8197
Jackson RJ (1991) The ATP requirement for initiation of eukaryotic translation varies according to mRNA species. Eur J Biochem 200:285–294
Jacques C, Fontaine JF, Franc B, Mirebeau-Prunier D, Triau S, Savagner F, Malthiery Y (2009) Death-associated protein 3 is overexpressed in human thyroid oncocytic tumours. Br J Cancer 101:132–138
Jeppesen MG, Navratil T, Spremulli LL, Nyborg J (2005) Crystal structure of the bovine mitochondrial elongation factor Tu·Ts complex. J Biol Chem 280:5071–5081
Jones CN, Wilkinson KA, Hung KT, Weeks KM, Spremulli LL (2008) Lack of secondary structure characterizes the 5′ ends of mammalian mitochondrial mRNAs. RNA 14:862–871
Kelley SO, Steinberg SV, Schimmel P (2000) Functional defects of pathogenic human mitochondrial tRNAs related to structural fragility. Nat Struct Biol 7:862–865
Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L et al (2006) Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23:607–618
Kimball S, Fabian J, Pavitt G, Hinnebusch A, Jefferson L (1998) Regulation of Guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2α; Role of the α- and σ-subunits of eIF2B. J Biol Chem 273:12841–12845
Kissil JL, Deiss LP, Bayewitch M, Raveh T, Khaspekov G, Kimchi A (1995) Isolation of DAP3, a novel mediator of interferon-gamma-induced cell death. J Biol Chem 270:27932–27936
Koc EC, Koc H (2012) Regulation of mammalian mitochondrial translation by post-translational modifications. Biochim Biophys Acta 1819:1055–1066
Koc EC, Spremulli LL (2002) Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J Biol Chem 277:35541–35549
Koc EC, Spremulli LL (2003) RNA-binding proteins of mammalian mitochondria. Mitochondrion 2:277–291
Koc EC, Blackburn K, Burkhart W, Spremulli LL (1999) Identification of a mammalian mitochondrial homolog of ribosomal protein S7. Biochem Biophys Res Comm 266:141–146
Koc EC, Burkhart W, Blackburn K, Moseley A, Koc H, Spremulli LL (2000) A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J Biol Chem 275:32585–32591
Koc EC, Burkhart W, Blackburn K, Koc H, Moseley A, Spremulli LL (2001a) Identification of four proteins from the small subunit of the mammalian mitochondrial ribosome using a proteomics approach. Prot Sci 10:471–481
Koc EC, Burkhart W, Blackburn K, Moseley A, Spremulli LL (2001b) The small subunit of the mammalian mitochondrial ribosome: identification of the full complement of ribosomal proteins present. J Biol Chem 276:19363–19374
Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL (2001c) The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276:43958–43969
Koc EC, Haque ME, Spremulli LL (2010) Current views of the structure of the mammalian mitochondrial ribosome. Israel J Chem 50:45–59
La Teana A, Gualerzi CO, Dahlberg AE (2001) Initiation factor IF 2 binds to the alpha-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA 7:1173–1179
Liao H-X, Spremulli LL (1990) Identification and initial characterization of translational initiation factor 2 from bovine mitochondria. J Biol Chem 265:13618–13622
Liao H-X, Spremulli LL (1991) Initiation of protein synthesis in animal mitochondria: purification and characterization of translational initiation factor 2. J Biol Chem 266:20714–20719
Ling J, Roy H, Qin D, Rubio MA, Alfonzo JD, Fredrick K, Ibba M (2007) Pathogenic mechanism of a human mitochondrial tRNAPhe mutation associated with myoclonic epilepsy with ragged red fibers syndrome. Proc Natl Acad Sci U S A 104:15299–15304
Liu M, Spremulli LL (2000) Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Biol Chem 275:29400–29406
Ma L, Spremulli LL (1995) Cloning and sequence analysis of the human mitochondrial translational initiation factor 2 cDNA. J Biol Chem 270:1859–1865
Mariani L, Beaudry C, McDonough WS, Hoelzinger DB, Kaczmarek E, Ponce F, Coons SW, Giese A, Seiler RW, Berens ME (2001) Death-associated protein 3 (Dap-3) is overexpressed in invasive glioblastoma cells in vivo and in glioma cell lines with induced motility phenotype in vitro. Clin Cancer Res 7:2480–2489
McCutcheon J, Agrawal R, Philips SM, Grassucci R, Gerchman S, Clemons WM, Ramakrishnan V, Frank J (1999) Location of translational initiation factor IF3 on the small ribosomal subunit. Proc Natl Acad Sci U S A 96:4301–4306
Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16:4623–4635
Mick DU, Dennerlein S, Wiese H, Reinhold R, Pacheu-Grau D, Lorenzi I, Sasarman F, Weraarpachai W, Shoubridge EA, Warscheid B et al (2012) MITRAC links mitochondrial protein translocation to respiratory-chain assembly and translational regulation. Cell 151:1528–1541
Mikulik K, Bobek J, Zikova A, Smetakova M, Bezouskova S (2011) Phosphorylation of ribosomal proteins influences subunit association and translation of poly (U) in Streptomyces coelicolor. Mol Biosystems 7:817–823
Miller JL, Koc H, Koc EC (2008) Identification of phosphorylation sites in mammalian mitochondrial ribosomal protein DAP3. Protein Sci 17:251–260
Miller JL, Cimen H, Koc H, Koc EC (2009) Phosphorylated proteins of the mammalian mitochondrial ribosome: implications in protein synthesis. J Proteome Res 8:4789–4798
Miyazaki T, Shen M, Fujikura D, Tosa N, Kon S, Uede T, Reed JC (2004) Functional role of death associated protein 3 (DAP3) in anoikis. J Biol Chem 279:44667–44672
Montoya J, Ojala D, Attardi G (1981) Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 290:465–470
Moreau M, de Cock E, Fortier P-L, Garcia C, Albaret C, Blanquet S, Lallemand J-Y, Dardel F (1997) Heteronuclear NMR studies of E. coli translation initiation factor IF3. Evidence that the inter-domain region is disordered in solution. J Mol Biol 266:15–22
Naithani S, Saracco SA, Butler CA, Fox TD (2003) Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell 14:324–333
Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark B, Nyborg J (1995) Crystal structure of the ternary complex of Phe-tRNAphe, EF-Tu and a GTP analog. Science 270:1464–1472
Nissen P, Thirup S, Kjeldgaard M, Nyborg J (1999) The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure 7:143–156
O’Brien TW (1971) The general occurrence of 55S ribosomes in mammalian liver mitochondria. J Biol Chem 246:3409–3417
O’Brien TW, O’Brien BJ, Norman RA (2005) Nuclear MRP genes and mitochondrial disease. Gene 354:147–151
Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP (2002) SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A 99:13653–13658
O’Rourke B, Van Eyk JE, Foster DB (2011) Mitochondrial protein phosphorylation as a regulatory modality: implications for mitochondrial dysfunction in heart failure. Congest Heart Fail 17:269–282
Ott M, Herrmann JM (2010) Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta 1803:767–775
Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO (2001) Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J 20:4560–4569
Pietromonaco SF, Denslow ND, O’Brien TW (1991) Proteins of mammalian mitochondrial ribosomes. Biochimie 73:827–835
Pioletti M, Schlunzen F, Harms J, Zarivach R, Gluhmann M, Avila H, Bashan A, Bartels H, Auerbach T, Jacobi C et al (2001) Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3. EMBO J 20:1829–1839
Ramagopal S, Subramanian AR (1974) Alteration in the acetylation level of ribosomal protein L12 during growth cycle of Escherichia coli. Proc Natl Acad Sci U S A 71:2136–2140
Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM (2010) A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J 29:1116–1125
Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA et al (2008) The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res 36:5787–5799
Rotig A (2011) Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta 1807:1198–1205
Sajid A, Arora G, Gupta M, Singhal A, Chakraborty K, Nandicoori VK, Singh Y (2011) Interaction of Mycobacterium tuberculosis elongation factor Tu with GTP is regulated by phosphorylation. J Bacteriol 193:5347–5358
Schieke SM, Finkel T (2006) Mitochondrial signaling, TOR, and life span. Biol Chem 387:1357–1361
Schwartzbach C, Spremulli LL (1989) Bovine mitochondrial protein synthesis elongation factors: identification and initial characterization of an elongation factor Tu-elongation factor Ts complex. J Biol Chem 264:19125–19131
Schwartzbach C, Farwell M, Liao H-X, Spremulli LL (1996) Bovine mitochondrial initiation and elongation factors. Methods Enzymol 264:248–261
Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E (2006) Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A 103:10224–10229
Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK (2003) Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115:97–108
Shi H, Hayes M, Kirana C, Miller R, Keating J, Macartney-Coxson D, Stubbs R (2012) TUFM is a potential new prognostic indicator for colorectal carcinoma. Pathology 44:506–512
Skrtic M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N et al (2011) Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20:674–688
Smeitink JA, Elpeleg O, Antonicka H, Diepstra H, Saada A, Smits P, Sasarman F, Vriend G, Jacob-Hirsch J, Shaag A et al (2006) Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet 79:869–877
Smirnov A, Entelis N, Martin RP, Tarassov I (2011) Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev 25:1289–1305
Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ (2007) Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res 35:4686–4703
Smits P, Smeitink J, van den Heuvel L (2010) Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotech 2010:737385
Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R et al (2007) mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol Cell 27:745–757
Soung GY, Miller JL, Koc H, Koc EC (2009) Comprehensive analysis of phosphorylated proteins of Escherichia coli ribosomes. J Proteome Res 8:3390–3402
Spencer AC, Spremulli LL (2004) Interaction of mitochondrial initiation factor 2 with mitochondrial fMet-tRNA. Nucleic Acids Res 32:5464–5470
Spencer AC, Spremulli LL (2005) The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim Biophys Acta 1750:69–81
Spremulli LL, Coursey A, Navratil T, Hunter SE (2004) Initiation and elongation factors in mammalian mitochondrial protein biosynthesis. Prog Nucleic Acid Res Mol Biol 77:211–261
Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, Jonsson GF, Jakobsdottir M, Bergthorsson JT, Gudmundsson J, Aben KK (2008) Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet 40:703–706
Surovtseva YV, Shutt TE, Cotney J, Cimen H, Chen SY, Koc EC, Shadel GS (2011) Mitochondrial Ribosomal Protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc Natl Acad Sci U S A 108:17921–17926
Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K (2001) Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J Biol Chem 276:21724–21736
Takeda S, Iwai A, Nakashima M, Fujikura D, Chiba S, Li HM, Uehara J, Kawaguchi S, Kaya M, Nagoya S et al (2007) LKB1 is crucial for TRAIL-mediated apoptosis induction in osteosarcoma. Anticancer Res 27:761–768
Takeuchi N, Kawakami M, Omori A, Ueda T, Spremulli LL, Watanabe K (1998) Mammalian mitochondrial methionyl-tRNA transformylase from bovine liver: purification, characterization and gene structure. J Biol Chem 273:15085–15090
Temperley R, Richter R, Dennerlein S, Lightowlers RN, Chrzanowska-Lightowlers ZM (2010) Hungry codons promote frameshifting in human mitochondrial ribosomes. Science 327:301
Terasaki M, Suzuki T, Hanada T, Watanabe K (2004) Functional compatibility of elongation factors between mammalian mitochondrial and bacterial ribosomes: characterization of GTPase activity and translation elongation by hybrid ribosomes bearing heterologous L7/12 proteins. J Mol Biol 336:331–342
Traugh JA, Traut RR (1972) Phosphorylation of ribosomal proteins of Escherichia coli by protein kinase from rabbit skeletal muscle. Biochemistry 11:2503–2509
Tsuboi M, Morita H, Nozaki Y, Akama K, Ueda T, Ito K, Nierhaus KH, Takeuchi N (2009) EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol Cell 35:502–510
Uchiumi T, Hori K, Nomura T, Hachimori A (1999) Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J Biol Chem 274:27578–27582
Uchiumi T, Honma S, Nomura T, Dabbs ER, Hachimori A (2002) Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J Biol Chem 277:3857–3862
Vedrenne V, Gowher A, De Lonlay P, Nitschke P, Serre V, Boddaert N, Altuzarra C, Mager-Heckel AM, Chretien F, Entelis N et al (2012) Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am J Hum Genet e 91:912–918
Wallace DC (2012) Mitochondria and cancer. Nat Rev Cancer 12:685–698
Wang Y, Bogenhagen DF (2006) Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem 281:25791–25802
Wang Z, Cotney J, Shadel GS (2007) Human mitochondrial ribosomal protein MRPL12 interacts directly with mitochondrial RNA polymerase to modulate mitochondrial gene expression. J Biol Chem 282:12610–12618
Watanabe K (2010) Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc Jpn Acad Ser B Phys Biol Sci 86:11–39
Wazir U, Jiang WG, Sharma AK, Mokbel K (2012) The mRNA expression of DAP3 in human breast cancer: correlation with clinicopathological parameters. Anticancer Res 32:671–674
Wolstenholme D (1992) Animal mitochondrial DNA. In: Wolstenholme D, Jeon K (eds) Mitochondrial genomes. Academic Press, New York, pp 173–216
Woolcott CG, Maskarinec G, Haiman CA, Verheus M, Pagano IS, Le Marchand L, Henderson BE, Kolonel LN (2009) Association between breast cancer susceptibility loci and mammographic density: the Multiethnic Cohort. Breast Cancer Res 11:R10
Woriax V, Burkhart W, Spremulli LL (1995) Cloning, sequence analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim Biophys Acta 1264:347–356
Woriax V, Spremulli G, Spremulli LL (1996) Nucleotide and aminoacyl-tRNA specificity of the mammalian mitochondrial elongation factor EF-Tu:Ts complex. Biochim Biophys Acta 1307:66–72
Xin H, Woriax VL, Burkhart W, Spremulli LL (1995) Cloning and expression of mitochondrial translational elongation factor Ts from bovine and human liver. J Biol Chem 270:17243–17249
Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q et al (2010) NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem 285:7417–7429
Yassin AS, Haque ME, Datta PP, Elmore K, Banavali NK, Spremulli LL, Agrawal RK (2011) Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc Natl Acad Sci U S A 108:3918–3923
Zhang Y, Spremulli LL (1998) Identification and cloning of human mitochondrial translational release factor 1 and the ribosome recycling factor. Biochim Biophys Acta 1443:245–250
Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Prot 8:215–225
Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H et al (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327:1000–1004
Zweib C, Glotz C, Brimacombe R (1981) Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nuc Acids Res 9:3621–3640
Acknowledgments
We thank Ms. Tamara B. Trout for critical reading of this chapter and for technical assistance in the generation of figures.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Koc, E.C., Koc, H. (2013). Mechanism and Regulation of Protein Synthesis in Mammalian Mitochondria. In: Duchêne, AM. (eds) Translation in Mitochondria and Other Organelles. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-39426-3_2
Download citation
DOI: https://doi.org/10.1007/978-3-642-39426-3_2
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-39425-6
Online ISBN: 978-3-642-39426-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)