Abstract
Mitochondrial ribosomes are known to be quite divergent from cytoplasmic ribosomes in both composition and structure even as their main functional cores, such as the mRNA decoding and peptidyl transferase sites, are highly conserved. The translational factors that interact with these ribosomes to facilitate the process of protein synthesis in mitochondria have also likewise acquired unique structural features, apparently to complement the structure and function of the mitochondrial ribosome. In this chapter, we describe the current state of structural knowledge of the mammalian mitochondrial ribosome, some of its component proteins, and key translational factors.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Bacterial Ribosome
- Mitochondrial Translation
- Peptidyltransferase Center
- Cytoplasmic Ribosome
- Mitochondrial Ribosome
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1.1 Introduction
Mitochondrial ribosomes (mitoribosomes) are responsible for synthesizing a limited number of polypeptide chains, which form essential components of the complexes involved in oxidative phosphorylation (OXPHOS). The OXPHOS complexes reside in the mitochondrial inner membrane (mtIM) and are responsible for generating about 90 % of the energy (ATP) required by the cell. All proteins required for mammalian mitochondrial translation, including the mitochondrial ribosomal proteins (MRPs), are encoded by the nuclear genome, translated in the cytoplasm and then imported into the mitochondria. The mammalian mitochondrial genome is relatively small (16.8 kb) and encodes for 37 genes, including two ribosomal RNAs (12S and 16S rRNAs), 22 mitochondrial transfer RNAs (tRNAmt), and 13 polypeptides of the OXPHOS complexes. Unlike cytoplasmic ribosomes, whose X-ray crystallographic structures are known for several bacterial (e.g., Schuwirth et al. 2005; Selmer et al. 2006), archaeal (Ban et al. 2000) and eukaryotic (e.g., Ben-Shem et al. 2011; Klinge et al. 2011) species, the three-dimensional (3D) structures of organellar ribosomes have been studied primarily using the single-particle cryo-electron microscopy (cryo-EM) and molecular modeling (Sharma et al. 2003, 2007, 2009; Mears et al. 2006; Manuell et al. 2007). This chapter concerns the mammalian mitoribosome, whose 3D cryo-EM structure is known for Bos taurus (Sharma et al. 2003). Several components of the mammalian mitochondrial translational machinery, including mito-rRNAs, MRPs, tRNAmts, tRNAmt synthetases and translational factors, have been associated with a number of human genetic diseases (see O’Brien 2002; O’Brien et al. 2005; Pearce et al. 2013; Watanabe 2010). Multiple review articles have been published recently on the structure of the mammalian mitoribosome (Agrawal and Sharma 2012; Agrawal et al. 2011; Christian and Spremulli 2012; Koc et al. 2010). In the present article, we first elucidate the unique characteristic of the cryo-EM structure of the mammalian mitoribosome in comparison to those of the cytoplasmic ribosomes. We then summarize some of the mammalian mitochondrial translation factors that interact with the mitoribosome during translation and discuss their singular features. Description of the mitochondrial translation machinery is mainly presented with reference to bacterial translation since the overall mechanism for translation in mitochondria is more similar to that in bacteria rather than to that in eukaryotic cytoplasm.
1.2 The Mammalian 55S Mitoribosome
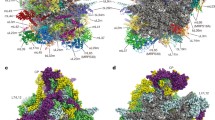
Mitochondria are thought to originate through an early endosymbiotic event (~1.8 billion years ago) between an α-protobacteria and a primitive host cell (Gray et al. 2001), and therefore, mitoribosomes were proposed to be structurally similar to their bacterial counterparts. The mammalian mitochondrial rRNAs are smaller than those in bacteria, and the MRPs are generally both larger (when they are homologous to bacteria) and greater in number. It was believed that the overall structural organization of the mitoribosome would still be very similar to its bacterial counterparts, except that the additional MRPs would structurally replace the missing bacterial rRNA segments that were deleted in the mito-rRNAs during the course of evolution. The first cryo-EM structure determined for the 55S mammalian mitoribosome revealed that this paradigm about the structural replacement of deleted bacterial rRNA segments by MRPs was only partially valid, as about 80 % of the missing bacterial rRNA segments were not found to be replaced by the MRPs (Sharma et al. 2003). In general, the loss of rRNA segments correspond to the loss of interacting bacterial proteins, suggesting a complementary evolution of the binding ribonucleoprotein (RNP) partners (Mears et al. 2006). Furthermore, the overall 3D structure of the mammalian mitoribosome was found to be significantly altered and highly porous as compared to its bacterial counterpart (Sharma et al. 2003; for recent reviews also see Agrawal et al. 2011; Agrawal and Sharma 2012), primarily due to the occupation of new spatial positions by mito-specific MRPs and extensions and insertions within several of MRPs that are homologous to their bacterial counterparts. However, the conserved mito-rRNA segments that form the functional core of a ribosome are intact in their spatial positions in the mitoribosome structure, except for few conserved rRNA segments that are present in the peripheral regions of the structure (Mears et al. 2006; Agrawal et al. 2011). Even though the molecular mass of the mammalian mitoribosome (~2.71 MDa) is very similar to that of a bacterial ribosome (~2.3 MDa) its overall dimension is much larger by about 60 Å in diameter. Its dimension is similar to that of a eukaryotic (yeast) cytoplasmic ribosome (Fig. 1.1) with a much higher molecular mass (~3.3 MDa). These observations are consistent with the observed porousness of the mammalian mitoribosome structure (Sharma et al. 2003).
A side-by-side comparison of structures of the mammalian mitochondrial ribosome with cytoplasmic (bacterial and eukaryotic) ribosomes. RNA–protein segmented structures of ribosomes from a mammalian mitochondria (55S, Bos taurus), b bacteria (70S, Escherichia coli), and c yeast cytoplasm (80S, Sachharomyces cerevisiae) displayed with SSU on the left and LSU on the right side, as viewed from the SSU shoulder (sh) and LSU stalk base (Sb) sides. In b and c, atomic structures of the 70S (PDB ID codes 1VOX-Y) and 80S (PDB ID codes 3U5B-E and 3IZS) ribosomes have been low-pass filtered to roughly match the resolution of the cryo-EM map of the mammalian mitoribosome shown in a. d–f Structures of SSUs from the corresponding top panel, but shown from the SSU-LSU interface side. g–i Structures of LSUs from corresponding top panel, but shown from the LSU-SSU interface side. Mito-specific MRPs of SSU and LSU are colored yellow and blue, respectively; conserved ribosomal proteins [here “conserved” refers to bacterial homologues present in the mitoribosome] of SSU and LSU are colored green and aquamarine, respectively; and rRNAs of SSU and LSU are colored orange and purple, respectively. Landmarks of SSU: b body, h head, m mRNA gate, pt platform, sh shoulder, S12 protein S12, Asterisks in d–f point to the location of mRNA decoding site. Landmarks of LSU: CP central protuberance, L1 protein L1 stalk (note the change in conformation of the L1 stalk, which is known to be a highly dynamic structure), L9 protein L9, Sb Stalk base or MRP L11 region, St L7/L12 stalk (this region was disordered in the crystallographic structures of the 70S and 80S ribosomes, and hence absent in panels H and I), SRL α-sarcin-ricin stem loop, H69 LSU rRNA helix 69, A, P and E in h indicate the positions of three canonical tRNA-binding sites (note the difference in the region analogous to E site in panel g), Red asterisks in g–i indicate the general location of the peptidyltransferase center. Spider (Frank et al. 2000) and Chimera (Pettersen et al. 2004) software were used respectively for surface representation and visualization of the ribosome maps
Like any other known ribosome, the 55S mitoribosomes are made up of two unequally sized subunits: the small 28S subunit (SSU) composed of a 12S rRNAmolecule and about 30 nuclear genome encoded MRPs, and the large 39S subunit (LSU) composed of a 16S rRNA molecule and about 50 nuclear genome encoded MRPs. Since all MRPs are imported into the mitochondria, most possess a mitochondrial targeting sequence (MTS) at their N-terminus (Claros and Vincens 1996). Recent studies suggest that many MRPs and mitochondrial translational factors undergo acetyl-CoA-, NAD+-, and ATP-dependent post-translational modifications in mitochondria, primarily acetylation and phosphorylation (see, Koc and Koc 2012, and Chap. 2), implying that these modifications play regulatory roles in mitochondrial translation. MRPs also participate in the formation of several mito-specific protein–protein bridges between the SSU and LSU (Sharma et al. 2003). A side-by-side comparison with the structures of the cytoplasmic ribosomes (Fig. 1.1) reveals that the exterior of the mammalian mitoribosome is predominantly shielded by MRPs, with fewer exposed rRNA regions. The inter-subunit space, which includes the mRNA decoding (Ogle et al. 2001) and peptidyl transferase (Nissen et al. 2000) sites, and tRNAs and translation factors binding sites, has a relatively conserved composition and architecture with more exposed rRNA regions (Fig. 1.1d–i). The first average structure of a mitoribosome-bound tRNAmt revealed by the cryo-EM study (Sharma et al. 2003) showed a canonical L-shape, but with a “caved-in” elbow region, consistent with generally reduced size of D- and/or T-stem loops in tRNAmts (Watanabe 2010). The corridor of the inter-subunit space, i.e., the space between the shoulder of SSU and stalk base of LSU (marked with ‘sh’ and ‘Sb’, respectively, in Fig. 1.1a–c), where most large translation factors are known to interact with the cytoplasmic ribosomes (Agrawal et al. 1998, 2000; Datta et al. 2005; Stark et al. 1997; Valle et al. 2002; Gomez-Lorenzo et al. 2000; Spahn et al. 2004; Gao et al. 2009; Schmeing et al. 2009; Yassin et al. 2011b; Yokoyama et al. 2012), has a slightly wider opening in the mitoribosome primarily due to the deletion of bacterial rRNA segments from the shoulder region of its SSU.
The unique features of two mitoribosomal subunits and some of their MRPs are described in the next two sections. Our laboratory has been modeling 3D structures of MRPs and docking them into our most recent 7.0 Å resolution cryo-EM map to develop a pseudo-atomic model of the 55S mitoribosome. Here our focus is primarily on some of the homologous MRPs for which atomic structures and relative positions within the bacterial ribosome are known and which possess mito-specific insertion and/or extension segments.
1.2.1 The 28S Small Subunit
The overall shape of the mitoribosome small subunit (SSU) is dramatically altered compared to bacterial ribosomes (Fig. 1.1d–f) due to the deletion of a significant portion of bacterial rRNA segments (Fig. 1.2a–c). These deletions amount to almost 40 % of the SSU rRNA as there are 1,542 nucleotides (nts) in E. coli versus 950 nts in mammalian mito-SSU. The decoding-site region, which is the central functional core of the SSU (marked with asterisks in Fig. 1.1d–f) is conserved but the peripheral architecture of the mito-SSU is significantly altered. The intactness of all three characteristic domains of a SSU, the body (b), head (h) and platform (pt), suggests that the basic architecture of SSU is important for its function, despite the fact that the diameter of the mito-SSU body is reduced and its shoulder region is significantly diminished due to the absence of the bacterial rRNA helices 16 and 17.
Structural components of the mammalian mito-SSU. a Secondary structures of small ribosomal subunit RNAs from bacteria (16S, grey) and mammalian mitochondria (12S, green). Portions of rRNA that form components of the SSU head, body (shoulder and bottom), and platform are indicated. The rRNA helices are numbered according to bacterial 16S rRNA. b–c Three dimensionally folded structures of the bacterial (b) and mammalian mitochondrial (c) SSU rRNAs, as derived from X-ray crystallography (PDB ID code 1VOX) and cryo-EM (based on Sharma et al. 2003, and Sharma et al., manuscript in preparation) studies, respectively. The head (hd), platform (pt), shoulder (sh), and spur (sp) regions are labeled. Note the diminished sh region in the mito-rRNA fold due to the absence of bacterial rRNA helices 16 and 17, and a channel created in the main body due to absence of bacterial rRNA helices 12 and 21. d Ab initio models of the homologous SSU MRPs as generated using I-TASSER (Roy et al. 2010), but not fitted into the cryo-EM of the mito-SSU. The MRP segments homologous to bacterial ribosomal proteins are depicted in blue and the mito-specific insertions/extensions are shown in red. N- and C-termini are identified in each case
The bovine mitoribosome contains 31 MRPs (named S1 to S31), of which 14 are bacterial homologues and 17 MRPs are specific to mitoribosome (Koc et al. 2000; Koc et al. 2001a; Suzuki et al. 2001a; Emine Koc, personal communication). One of the homologous SSU MRPs, S18 is present in three-isoforms, referred to as MRP S18A, S18B and S18C (Koc et al. 2001a). There is no homologue for the six bacterial ribosomal SSU proteins S3, S4, S8, S13, S19 and S20 in the mitoribosome. The mito-specific MRPs significantly contribute to the unique architecture of the mito-SSU by primarily occupying its head, lower body and solvent side (Sharma et al. 2003) (Fig. 1.1a–f). Addition of significantly large mass of mito-specific MRPs to the lower body is primarily responsible for an overall elongate shape and an unusually large dimension of the mito-SSU along its long axis.
The homologous SSU MRPs possess a varying size range of amino acid extension segments (8–215 amino acid) at their N- and/or C-terminus, except for MRPs S6 and S12, which are shorter in the mitoribosome (Table 1.1; Fig. 1.2d). For, example, The MRP S9, located in the head of SSU, carries the longest N-terminus extension (NTE) of 215 amino acid residues, which can be accounted by the large extra mass of mito-specific protein cryo-EM density present in the immediate vicinity of the protein’s homologous segment (Sharma et al. 2003). Among all homologous SSU MRPs, S7 is the only MRP that has a 20 amino acid insertion within its conserved domain (Fig. 1.2d) that is tightly accommodated in an additional mass of density observed within the cryo-EM map (unpublished results from our laboratory).
One of the unique features of the mammalian mitochondrial translation is that most of its mRNAs lack a 5′ untranslated region (5′-UTR) (Temperley et al. 2010). In the cryo-EM structure of the mito-SSU the mRNA entrance has a unique gate-like feature, which is made up of mito-specific MRPs (Sharma et al. 2003; marked by “m” in Fig. 1.1a), which may be directly involved in the recruitment of mitochondrial mRNAs to initiate the translation. In the bacterial ribosome, proteins S3, S4 and S5 occupy the mRNA entrance (Yusupova et al. 2001). Of these, proteins homologous to S3 and S4 are absent in the mitoribosome, but are replaced by certain other mito-specific MRPs or the extensions of homologous MRPs. For example, MRP S5 has 120 and 52 amino acid NTE and C-terminal extension (CTE), respectively, that may partially compensate for the loss of bacterial S4 and S8 proteins that reside on opposite sides of S5 in the bacterial ribosome (Wimberly et al. 2000). However, these extensions would not account for the SSU’s gate-like feature, which can only be explained by the presence of additional mito-specific MRPs that are yet to be identified.
1.2.2 The 39S Large Subunit
About 50 % of bacterial rRNA segments are also deleted in the mito-large subunit (LSU) (3,024 nts in E. coli versus 1,560 nts in mammalian mito-LSU) (Fig. 1.3a–c), but the overall shape of the mito-LSU is not altered as dramatically as that of the mito-SSU (Fig. 1.1). The presence of all three characteristic features of a bacterial LSU, such as the L1 protuberance (L1), the central protuberance (CP) and the L7/L12 stalk (St), within the cryo-EM structure of the mito-LSU suggests that the basic architectures of both ribosomal subunits are important for their protein synthesis function. Like in mito-SSU, rRNA deletions have not affected the structure of the central functional core (the peptidyltransferase center region) of the mito-LSU, but they have significantly altered the composition of its tRNA exit site (E site), nascent polypeptide exit site, and several other peripheral regions of LSU (Fig. 1.1g–h). For example, 11 of the 12 interaction sites of tRNA involving rRNA segments at the bacterial ribosomal E site are absent in the mito-LSU (Mears et al. 2006), strongly suggesting that binding of tRNAmt at the putative mitoribosomal E site is either very weak or such a site does not exist on the mammalian mitoribosome. Similarly, the lower two-thirds of the nascent polypeptide exit tunnel (Nissen et al. 2000) is almost completely remodeled in the mito-LSU and is occupied primarily by the mito-specific MRPs (Sharma et al. 2003; also see, Agrawal et al. 2011).
Structural components of the mammalian mito-LSU. a Secondary structures of large ribosomal subunit RNAs from bacteria (23S, grey) and mammalian mitochondria (16S, green), Dashed lines indicate unassigned segments of the mito-rRNA. The rRNA helices are numbered according to the bacterial 23S rRNA. b–c Three dimensionally folded structures of the bacterial (b) and mammalian mitochondrial (c) LSU rRNAs, as derived from X-ray crystallography (PDB ID code 1VOY) and cryo-EM (based on Sharma et al. 2003; Mears et al. 2006; and Sharma et al., manuscript in preparation) studies, respectively. d Unfitted ab initio models of the homologous LSU MRPs. The MRP segments homologous to bacterial ribosomal proteins are depicted in blue and the mito-specific insertions/extensions are shown in red. MRP L47 is tentatively included in this list, as the major portion of its NTD domain appears to be a structural homolog of the bacterial L29 (see text). N- and C-termini are identified in each case
The mito-LSU contains 50 MRPs, out of which 29 are homologous to bacterial ribosomal proteins (Table 1.2), and 21 MRPs are mito-specific (Koc et al. 2001a, b, 2010; Suzuki et al. 2001b). Homologues of the bacterial ribosomal proteins L5, L6, L25, L26 and L31 are absent in the mito-LSU. Similar to SSU MRPs, most homologous MRPs of LSU possess NTE and/or CTE, ranging in size from 8 to 120 amino acid segments, except for MRPs L2, L14, L33, L34 and L36 (Fig. 1.3d). L5, which is present within the central protuberance (CP) of the bacterial ribosome and forms the B1 group bridges with the SSU is replaced by MRPs specific to the mito-LSU (Sharma et al. 2003). The immediate binding partners of L5, the 5S rRNA and the LSU rRNA helix 84 are also absent in the mito-LSU. However, a recent report suggests that 5S rRNA is transported into mitochondria along with MRP L18 and it could become part of the mitoribosome LSU (Smirnov et al. 2011). It is possible that the 5S rRNA associates with only a small fraction of the mitoribosome, and therefore has gone undetected in the averaged 3D cryo-EM map (Sharma et al. 2003).
In mitochondria, all synthesized polypeptides are inserted into mtIM, and therefore polypeptide conducting exit tunnel in their ribosomes are tailor-made (Sharma et al. 2003, 2009; Agrawal et al. 2011), apparently to facilitate the co-translational release and incorporation of nascent hydrophobic polypeptides into the mtIM (Gruschke and Ott 2010). Unlike the situation in the structures of cytoplasmic ribosomes (e.g. Nissen et al. 2000), the polypeptide exit tunnel in mito-LSU has two solvent-accessible openings, referred to as the conventional polypeptide exit site (PES), which is located ~90 Å away from the peptidyltransferase center, and a mito-specific polypeptide-accessible site (PAS), which prematurely opens up in the middle, at only ~65 Å away from the peptidyltransferase center. In the mammalian mitoribosome, the openings at both PES and PAS are primarily occupied by MRPs. Based on overall topology of the polypeptide-exit tunnel in the mammalian mitoribosome, it is conceivable that the nascent polypeptide chain could use either the conventional PES or the mito-specific PAS route, depending upon its folding requirements. In the bacterial ribosome, PES is encircled by four proteins L22, L23, L24 and L29 and inner lining of the lower two-thirds of the polypeptide exit tunnel is primarily made by domains I and III of the 23S rRNA. In case of mitoribosome the analogous section of the tunnel is either surrounded by MRPs or is left unoccupied, as the size of domains I and III are dramatically reduced in the mito-LSU 16S rRNA (Fig. 1.3a), giving rise to PAS (see Sharma et al. 2003; also see Agrawal et al. 2011). The conventional PES is also primarily surrounded with mito-specific MRPs. The homologous MRPs L22, L23, and L24, each with mito-specific extensions, are present in the mito-LSU (Fig. 1.3d). These extensions may account for the part of the large extra mass of mito-specific MRP density present at the mito-PES. However, there is still an ambiguity to whether or not the mito-specific MRP L47 is a homologue of bacterial L29. Bioinformatics analysis such as profile–profile search for distinct homologue has shown that MRP L47 is related to bacterial L29, whereas BLAST and Pfam analysis do not reveal any similarity among these two proteins (Smits et al. 2007). The ab initio model of MRP L47 shows a conserved structural domain similar to that in the bacterial L29, in addition to the large NTE and CTE (Fig. 1.3d). Based on distinct similarity between the central domain of MRP L47 3D model and structure of the bacterial L29, it is tempting to categorize MRP L47 as a bacterial L29 homologue.
In mitochondria the process of polypeptide integration into the mtIM is facilitated by the Oxa1 proteins, homologues of bacterial YidC. Oxa1L is the human homologue of the yeast Oxa1p, which is known to be involved in the biogenesis of membrane proteins and which assists in the insertion of proteins from mitochondrial matrix to the mtIM (Luirink et al. 2001; Ott and Herrmann 2010). The yeast homologue of MRP L23 (mrp20 in yeast) is known to interact with the C-terminus tail (CTT) of Oxa1p in order to recruit the mitoribosome for co-translational insertion of the mitochondrially-encoded polypeptides into mtIM (Jia et al. 2003; Szyrach et al. 2003; Keil et al. 2012). Interestingly, the cross-linking studies of Oxa1L-CTT with the bovine mito-LSU did not produce any crosslinks to the conventional PES proteins, such as the MRPs L22, L23, L24 and L29. However, the Oxa1L-CTT does crosslink to some other MRPs, including MRPs L13, L20, L28, L48, L49, and L51 (Haque et al. 2010). Of these, L13, L20, and L28 are known to be situated at distant locations from the polypeptide exit tunnel in the bacterial ribosome (Schuwirth et al. 2005). Of these, MRPs L13 and L20 possess only small CTEs, and therefore, are less likely to reach the polypeptide exit tunnel to interact with the mitoribosome-bound Oxa1L-CTT. However, MRP L28 has relatively large extensions on both its N-and C-termini (Fig. 1.3d), which could potentially reach the PAS but less likely to reach all the way to PES. Among remaining MRPs that are cross-linked to Oxa1L-CTT are mito-specific MRPs L48, L49 and L51. The exact locations of these MRPs on the mito-LSU are not presently known. As described in the previous paragraph, large mass of unidentified mito-specific MRP densities are present at both PAS and PES that could be accounted for by all three cross-linked mito-specific MRPs. Since no crosslink to MRP L47, the putative bacterial L29 homologue, was detected with the Oxa1L-CTT, it is possible that MRP L47 is shielded by mito-specific MRPs L48, L49 or L51 on the mitoribosome.
1.3 Mitochondrial Translation Factors
Like cytoplasmic ribosomes, the mitoribosome requires well-coordinated interactions with a number of mitochondrial translational factors (henceforth referred to as mito-translational factor) to conduct protein synthesis. Mito-translational factors bear greater similarity to their bacterial rather than to their eukaryotic counterparts. However, most mammalian mito-translational factors have acquired special structural features (Fig. 1.4) that may be required for effective and accurate translation on the significantly modified mitoribosomes. Like MRPs, all mito-translational factors are also encoded in the nucleus, synthesized in the cytoplasm with N-terminus MTSs (Table 1.3), and then imported into the mitochondrial matrix. In the following sections, we describe the structural organization of various mammalian mito-translational factors that directly interact with the mitoribosome and possess mito-specific amino-acid sequence insertions and/or extensions, based on sequence alignment of the mature (i.e., after the removal of MTSs) mito-translational factors (Fig. 1.4a) with those of their bacterial counterparts, and their 3D ab initio models (Fig. 1.4b). We describe them in a general order of their involvement in the process of protein synthesis, using bacterial translation system as the model (e.g., see, Schmeing and Ramakrishnan 2009). Accordingly, the mitochondrial translation can be divided into four stages of protein synthesis, i.e., initiation, elongation, termination, and recycling.
Mammalian mitochondrial translation factors. a Bar diagrams showing mammalian mito-specifc segments (red) as compared to their bacterial counterparts. Numbers on top of each bar diagram refer to amino acids. Missing numbers at the start of each bar diagram would correspond to MTSs, except for RRFmt, where MTS has been proposed to be a functional component of the factor. RF1amt is not shown, as it does not contain a contiguous mito-specific segment. The GGQ domain of ICT1 posseses a unique 10 amino acid insertion segment (depicted in pink). b Ab initio models of the mitochondrial translational factors as generated using I-TASSER, except for IF2mt, which is based on cryo-EM study (Yassin et al. 2011b), and EF-Tumt, which is known from X-ray-crystallography (Jeppesen et al. 2005). The nomenclature of structural domains of various factors is based on general consensus in the field, and domains are identified mostly by roman numerals, except for the GTP-binding domains, which are labeled as G-Domain. NTD and CTD refer to IF3mt’s N- and C-terminal domains, respectively; in b, N- and C-termini are also identified in each case. The color codes used for various domains of corresponding factors are matched between a and b
1.3.1 Translation Initiation
The initiation stage concludes with the docking of the fMet-tRNA Metmt at the start codon of the mRNA onto the ribosomal peptidyl site (P site) on the 55S mitoribosome. Bacterial translation involves three initiation factors, IF1, IF2 and IF3. However, homologues of only two bacterial factors, IF2mt and IF3mt, were identified in the mammalian mitochondria. It was proposed that IF2mt performs the task of both IF1 and IF2 in mitochondria (Spencer and Spremulli 2005; Gaur et al. 2008). Like bacterial IF3, IF3mt functions to actively dissociate the 55S mitoribosome into its two subunits, while it remains bound to the mito-SSU to prevent the re-association of the latter with the mito-LSU (Spencer and Spremulli 2005); whereas IF2mt forms a ternary complex with the initiator fMet-tRNA Meti and GTP to deliver the fMet-tRNA Meti into the initiator P site (e.g., see Allen et al. 2005 for IF2; and Yassin et al. 2011b for IF2mt) of the mito-SSU. During this process, the CCA end of the initiator tRNA interacts with the conserved IF2mt’s domain VI-C2 (Spencer and Spremulli 2004; Yassin et al. 2011b). Once the mito-SSU initiation complex is formed, IF2mt promotes its association with the mito-LSU via the latter’s interaction with IF2mt domains IV (the GTPase domain) and VI. The activation of IF2mt’s GTPase leads to the dissociation of IF2mt and of IF3mt, resulting in the formation of a 55S initiation complex, which then enters the elongation phase of protein synthesis.
1.3.1.1 Mitochondrial Initiation Factor 2
The mitochondrial initiation factor 2 (IF2mt) comprises of six structural domains (Fig. 1.4), which are homologous to domains III–VI of bacterial IF2 (Spencer and Spremulli 2005; Gaur et al. 2008; Yassin et al. 2011b), except for a mito-specific 37 amino-acid residues insertion domain. The insertion domain is present in the inter-domain linker between domains V and VI-C1. Mutation of several conserved basic residues of the insertion domain reduces the ability of the Bos taurus IF2mt to bind mito-SSU, implying that the insertion domain makes direct contacts with the SSU (Spencer and Spremulli 2005). Among all mitochondrial translation factors, the ribosome-bound structure is known only for the IF2mt, which was obtained by cryo-EM. The cryo-EM study of the IF2mt•GDPNP•fMet-tRNA Meti in complex with the bacterial 70S ribosome (Yassin et al. 2011b) strongly supported the previous biochemical (Spencer and Spremulli 2005) and genetic (Gaur et al. 2008) studies. It showed that the insertion domain protrudes from rest of the IF2mt mass onto the ribosomal-SSU’s aminoacyl-tRNA binding site (A site), where it interacts with conserved ribosomal elements (the SSU rRNA helices 18 and 44, and protein S12) that are also known to interact with the bacterial IF1 (Fig. 1.5) as well as the A-site tRNA (Carter et al. 2001). Thus, the study suggested that the mito-specific insertion domain mimics the function of the bacterial IF1 by sterically precluding the binding of initiator tRNA to the ribosomal A site (Yassin et al. 2011b). However, some questions remain unresolved as (i) the insertion domain had to be linked to the rest of IF2mt in the homology model through very long unstructured regions on both its ends (Yassin et al. 2011a), and (ii) there is neither a sequence homology nor any structural similarity between the insertion domain and the bacterial IF1 (Fig. 1.5c, d). However, both structures bear similar surface charge distributions, which might play an important role in recognition of the same binding pocket on the SSU. Owing to the long unstructured linker region, it is conceivable that the insertion domain is a highly dynamic structure that remains folded onto the rest of the IF2mt molecule in its unbound state and the extended conformation is attained only upon its interaction with the ribosome, as has been observed for bacterial class I release factors (e.g., Rawat et al. 2003). Moreover, the ribosome-bound IF2mt structure presents a compelling evidence of the integration of function of bacterial IF1 onto IF2mt that might have occurred during the course of evolution so that one less protein had to be transported into the mitochondrial matrix. Integration of relatively small IF1 feature to IF2mt would also ensure its efficient transport to the protein synthesis site within highly dense mitochondrial matrix.
Structure of the ribosome-bound mammalian IF2mt, as derived by cryo-EM. a The cryo-EM map of the E. coli 70S ribosome (30S subunit, yellow; 50S subunit, blue) in complex with IF2mt (red), initiator tRNA (green) at the P/I position. b Fitting of atomic models of the IF2mt and initiator tRNA (P/I stands for P-site tRNA at initiator position) into the corresponding cryo-EM densities (meshwork) extracted from the map of the 70S•IF2mt•GMPPNP•fMet-tRNA complex shown in a. The color codes used for various domains of IF2mt are the same as in Fig. 1.4. Asterisk (*) point to the region that would correspond to domain III of IF2mt. Binding positions of the insertion domain (red, c) and IF1 (green, d) onto a common binding pocket of SSU of the ribosome. Landmarks of the ribosome are as in Figs. 1.1 and 1.2 (adopted from Yassin et al. 2011b)
1.3.1.2 Mitochondrial Initiation Factor 3
Mitochondrial initiation factor 3 (IF3mt) shares the basic domain organization of the eubacterial IF3, with an N-terminal domain (NTD) and a C-terminal domain (CTD) connected by a flexible helical linker (Biou et al. 1995; Christian and Spremulli 2009; Petrelli et al. 2001). In addition, both its conserved domains harbor mito-specific NTE and CTE of ~30 amino acid residues each (Fig. 1.4). IF3mt stimulates initiation complex formation on the mito-specific leaderless mRNAs (Christian and Spremulli 2010). Both its mito-specific NTE and CTE have been implicated in optimizing the binding of IF3mt to enable its own dissociation from the 55S mitoribosome (Bhargava and Spremulli 2005; Christian and Spremulli 2009; Haque and Spremulli 2008). The CTE and the linker region of IF3mt have also been implicated in dissociation of the fMet-tRNA•IF2mt from the mito-SSU in the absence of mRNA, suggesting their role in preventing the premature occupation of the P site.
The binding position of bacterial IF3 on the bacterial SSU is known from cryo-EM (McCutcheon et al. 1999; Julian et al. 2011) and by hydroxyl radical probing (Dallas and Noller 2001) studies. Interestingly, each of these studies places IF3 on the rim of the platform of the ribosomal SSU, but with different assignments of orientation for the two IF3 domains. While the structure of a mito-SSU•IF3mt is not yet published, our preliminary cryo-EM reconstructions suggest that the overall configuration of IF3mt binding on the mito-SSU would be similar to what was proposed earlier for the bacterial ribosome. The cross-linking studies of IF3mt with the mito-SSU (Haque et al. 2011) identified several MRPs that may be present in immediate vicinity of the platform rim of the mito-SSU. This includes homologous MRPs S5, S9, S10 and S18 and several mito-specific MRPs, S29, S32, S36, and PTCD3. Among the homologous MRPs, S18 is situated on the platform and therefore, its direct interaction with IF3mt can be readily explained. The presence of the globular portions of S5, S9, and S10 on the solvent side of the bacterial SSU (Wimberly et al. 2000) suggests that the interaction of these MRPs with IF3mt, which sits on the rim of the SSU platform, would be unlikely. However, C-terminus of S9 is exposed on the SSU-LSU interface side and MRPs S5 and S10 both possess long mito-specific extensions (Fig. 1.2d), which have potential to reach to the inter-subunit face to produce crosslinks with mito-SSU bound IF3mt. In addition, there is a large mass of unidentified cryo-EM density within the mito-SSU head region that could account for the cross-linked mito-specific MRPs. Of these, docking of the homology model of PTCD3 into the cryo-EM map nicely explains the bulk of unassigned density in the SSU head at the interface of the mRNA channel (unpublished results in our laboratory), from where it could potentially crosslink to IF3mt.
1.3.2 Translation Elongation
Translation elongation is a cyclic process (see for example, Agrawal et al. 2000; Schmeing and Ramakrishnan 2009) where an amino acid residue, as specified by the mRNA codon, is added to the growing nascent peptide, followed by progression of the ribosome along the mRNA by a codon step. The elongation stage alternates between aminoacyl-tRNA (aa-tRNA) delivery and translocation of the mRNA•(tRNA)2 complex on the ribosome, and ends when the ribosome encounters a stop codon. Like in bacterial translation system, mitochondrial translation also involves two canonical translation elongation factors (EFs) that interact directly with the mitoribosome. (i) EF-Tumt, which promotes the accurate binding of aa-tRNAmt, in form of a ternary complex aa-tRNAmt•EF-Tumt•GTP, to the vacant aa-tRNA binding site (A site) of the ribosome; and (ii) EF-G1mt, which, after the peptide-bond formation, binds to the ribosome as EF-G1mt•GTP and promotes translocation of the mRNA•peptidyl-tRNA complex to free up the ribosomal A site, or the mRNA decoding site, for the next round of elongation. Two forms of EF-Gmt are present in most organisms (Hammarsund et al. 2001). The second isoform, EF-G2mt, is exclusively involved at the recycling stage, and is described later under the Sect. 1.3.4.2.
1.3.2.1 Mitochondrial Elongation Factor Tu
Mitochondrial elongation factor Tu (EF-Tumt) is the most highly conserved among all translation factors that associate with the mitoribosome. EF-Tumt has an overall 55 and 75 % sequence identity and similarity, respectively, with bacterial EF-Tu. It is also the only mitochondrial translational factor for which an atomic structure is currently available, in complex with its GTP exchange factor, EF-Tsmt (Jeppesen et al. 2005), and as expected, it shows an overall structural similarity to its bacterial counterpart (Kjeldgaard et al. 1993; Nissen et al. 1995). It comprises of three domains, the G domain (the GTP-binding GTPase domain), and domains II and III. The 3′ end of the aa-tRNA resides in the crevice between the G domain and domain II as its CCA arm interacts with domain III (Schmeing et al. 2009; Akama et al. 2010). Although the factor-binding region on the mitoribosome is significantly open due to deletion of the bacterial SSU rRNA helices h16 and h17 in the mito-SSU rRNA (Sharma et al. 2003) (Figs. 1.1a, d, and 1.2a–c), most of the ribosomal components that are known to interact with the functional sites of bacterial EF-Tu (G domain and domain II) and bound tRNAs (anticodon stem-loop ASL region) as part of the ternary complex (Valle et al. 2002; Schmeing et al. 2009; Schuette et al. 2009; Agirrezabala et al. 2011) are conserved in the mitoribosome. These include components such as α-sarcin-ricin stem loop (SRL), GTPase-associated center (comprised of LSU rRNA helices 43, 44 and protein L11), SSU protein S12, and the decoding site (comprised of portions of SSU rRNA helices 18 and 44). Thus, all essential structural elements that are involved in the proofreading step in bacterial ribosome are conserved in the mitoribosome. However, EF-Tumt possesses a CTE of 11 amino acid residues (Fig. 1.4), whose functional significance is unknown.
The E. coli ternary complex can deliver the aa-tRNA to the mitoribosome when constituted with canonical aa-tRNAs, but not when in complex with aa-tRNAmts (Bullard et al. 1999), suggesting that the nature of the interaction of the aa-tRNA with EF-Tumt must also account for the differences in the shape and stability of tRNAmts, several of which are smaller in size and lack their D- and/or T-arms (Hanada et al. 2000; Watanabe 2010). Furthermore, the tRNA itself participates in the signal transduction process of decoding. On cognate codon-anticodon interaction, the anticodon stem-loop is pulled into the A site and gets distorted. Part of that signal is transmitted via the tRNA scaffold to the G domain triggering the GTPase activity of EF-Tumt (Valle et al. 2002; Schmeing et al. 2009; Schuette et al. 2009). The coupling of tRNAmt structure to that of the EF-Tumt suggests that the mito-specific CTE may play a role in positioning the shorter aa-tRNAmts effectively in the decoding site for proofreading. It may compensate for the non-canonical shape and size of mammalian tRNAmts, in a mechanism that may be similar to that of the much longer CTE of C. elegans EF-Tu1mt. The CTE in C. elegans EF-Tu1mt has been proposed to compensate for the lack of the T-arm in the C. elegans tRNAmt (Ohtsuki and Watanabe 2007). In the bacterial ribosome•aa-tRNA•EF-Tu complex, domain III interacts with the T stem-loop of the tRNA such that its C-terminus points towards the shoulder of the SSU (Schmeing et al. 2009). Accordingly, the CTE in EF-Tumt may also interact with the structurally diminished shoulder of the mito-SSU. Such an interaction would help stabilizing the aa-tRNAmt•EF-Tumt•GTP ternary complex interaction with the mitoribosome. A structural characterization of the mitoribosome•aa-tRNAmt•EF-Tumt•GTP complex could help in delineating the function of the CTE in EF-Tumt.
1.3.2.2 Mitochondrial Elongation Factor G1
Mitochondrial elongation factor G1 (EF-G1mt) catalyzes the translocation of the mRNA•(tRNAmt)2 complex on the mitoribosome. It carries a mito-specific CTE besides the well-defined five structurally conserved domains, which are homologous to the bacterial EF-G (Ӕvarsson et al. 1994; Czworkowski et al. 1994) (Fig. 1.4). Unlike its bacterial counterpart, EF-G1mt is inactive in ribosome-recycling (Tsuboi et al. 2009) and is highly resistant to fusidic acid (Chung and Spremulli 1990; Bhargava et al. 2004). Bacterial ribosomal elements that are known to interact with EF-G domains are mostly conserved in the mitoribosome (Bhargava et al. 2004), except for certain rRNA components in the mito-SSU shoulder (Sharma et al. 2003). Phylogenetic analysis indicates that the EF-G1mt like proteins have evolved to facilitate the translocation process on the ribosome (Atkinson and Baldauf 2011). In bacterial ribosomes, EF-G binds and stabilizes the ratcheted state of ribosome to catalyze the translocation step (Agrawal et al. 1999; Frank and Agrawal 2001; Agirrezabala and Frank 2009). Cryo-EM data from our laboratory suggests that the inter-subunit ratcheting is less pronounced in the mitoribosome (Sharma et al., manuscript in preparation), as compared to that observed for the bacterial ribosome (Agrawal et al. 1999; Frank and Agrawal 2000). It is possible that the remodeling of EF-G1mt enables it to function on the mitoribosome without the requirement of substantial ratchet-like reorganization. In addition to the mito-specifc CTE, which appears to be directly involved in facilitating translocation on the mitoribosome (Sharma et al., manuscript in preparation), the G domain of EF-G1mt has also been extensively remodeled, including an insertion of GEV in the highly conserved switch I loop (Atkinson and Baldauf 2011). Such a remodeling of the G domain has been implicated in conferring fusidic acid resistance to EF-G1mt. In addition to the mito-specific features in EF-G1mt, the central protuberance of the mito-LSU bears a unique and dynamic structural feature that may also contribute to the tRNAmt translocation process. This feature was identified as the P-site finger (Sharma et al. 2003), as it interacts with the T-stem loop side of the P-site tRNAmt on the mitoribosome.
1.3.3 Translation Termination
A stop codon (UAA/UAG) at the A site marks the end of the open-reading frame (ORF) and it is recognized by the class I release factors (RFs) that catalyse the hydrolysis of the ester bond of peptidyl-tRNA, leading to release of the nascent polypeptide chain form the ribosome and translation termination. In mitochondria, at least four proteins possessing the peptide-hydrolyzing domain of a typical RF, the GGQ domain, have been identified, which include RF1mt, RF1amt, immature colon carcinoma transcript-1 (ICT1), and C12orf65 (Fig. 1.4, note that the figure includes only those factors that possess mito-specific insertions or extensions, suggesting that RF1amt does not carry a mito-specific segment). Of these, so far only RF1amt has been characterized as a canonical RF in mammalian mitochondria (see Richter et al. 2010). However, in the following sections, we briefly describe each of these four factors.
1.3.3.1 Mitochondrial Release Factor 1a
Mitochondrial release factor 1a (RF1amt) is a mitochondrial class 1 release factor that recognizes both UAA/UAG stop codons and terminates translation of all 13 mitochondrially-encoded polypeptides (Chrzanowska-Lightowlers et al. 2011). The domain organization of RF1amt is similar to that of bacterial class I factors (Vestergaard et al. 2001). Domain II, the codon-recognition domain of the factor, contains structural elements that identify the stop codon; the codon recognition loop with the conserved tri-peptide motif PXT and the tip of helix α5. Domain III bears the universal GGQ motif, which interacts with the acceptor end of the P-site tRNA. The glutamine residue of the GGQ motif catalyzes the hydrolysis of the peptidyl-tRNA by coordinating a water molecule, as shown by structural studies of analogous bacterial complexes (Rawat et al. 2003; Laurberg et al. 2008; Weixlbaumer et al. 2008). Correct stop codon recognition by domain II is required for proper placement of the GGQ motif of domain III into the peptidyltransferase center. The codon recognition signal is transmitted via LSU rRNA helix 69 and the inter-domain switch loop within class I RF (Laurberg et al. 2008; Korostelev 2011). Since there is no expected difference between the structural organizations of the bacterial RF1/2 and RF1amt, it is likely that RF1amt interacts with the mitoribosome in a similar fashion as its bacterial counterpart (Laurberg et al. 2008; Weixlbaumer et al. 2008).
1.3.3.2 Mitochondrial Release Factor 1
Mitochondrial release factor 1 (RF1mt) has been shown to be inactive as a peptidyl hydrolyase on the bacterial ribosome (Soleimanpour-Lichaei et al. 2007), apparently because the codon recognition elements of RF1mt are significantly different from those of canonical RFs. The conserved tripeptide of the codon recognition loop of RF1 is PXV instead of the conserved PXT, which is followed by a GXS insertion. Another codon recognition element on the tip of α-helix 5 has an RT insertion in its preceding loop (Soleimanpour-Lichaei et al. 2007; Chrzanowska-Lightowlers et al. 2011; Huynen et al. 2012). Besides the conserved domain organization of a class I RF, RF1mt has a 48 amino acid long mito-specific NTE. The function of this RF, including its mito-specific extension (Fig. 1.4a), is yet unknown. An analysis of a ribosome-bound homology model of RF1mt alludes to RF1mt as having a role analogous to that of the tmRNA in bacterial ribosome (Huynen et al. 2012). The study suggests that bulkier RF1mt codon recognition motif is unlikely to be accommodated in an mRNA occupied A site and that it probably recognizes the empty A site of a stalled mitoribosome.
1.3.3.3 Immature Colon Carcinoma Transcript-1
Immature colon carcinoma transcript-1 (ICT1) is a component of the mito-LSU and its activity is essential for cell viability. Having the universal GGQ motif but lacking the codon recognition domain, it is active as a ribosome-dependent peptidyl-tRNA hydrolyase in a non-codon dependent manner. Its activity has been implicated in the hydrolysis of prematurely terminated peptidyl-tRNA in the stalled mitoribosome (Richter et al. 2010). This functionality is similar to that of the bacterial YaeJ (Antonicka et al. 2010; Gagnon et al. 2012). However, its position on the mitoribosome is yet to be mapped, which is required to understand how this factor, being a component of the mitoribosome LSU, disengages itself from the nascent polypeptide hydrolysis site when not required.
1.3.3.4 C12orf65
C12orf65 is another essential mitochondrial RF lacking the stop codon recognition domain. However, despite the presence of the universal GGQ domain, its peptidyl-hydrolyase activity has not been demonstrated in vitro. The fact that the C12orf65 deficiency can be suppressed by over expression of ICT1 suggests that the protein is catalytically active in vivo (Antonicka et al. 2010). Since it is present in the mitochondrial matrix, C12orf65 has been suggested to play a role in recycling the abortive peptidyl-tRNA species, released from the mitoribosome during the elongation phase of translation. Phylogenetic analysis suggests that though C12orf65 is of eukaryotic origins, it shares a C-terminal helix rich in basic residues with YaeJ and ICT1 (Duarte et al. 2012). In YaeJ this helix is responsible for sensing the empty mRNA channel on a stalled ribosome (Gagnon et al. 2012).
1.3.4 Ribosome Recycling
After the translation termination, the post-termination ribosome complex (PoTC) remains occupied by the translated mRNA and a deacylated tRNA at the P/E position. As in bacteria (see Yokoyama et al. 2012), two mito-translational factors, RRFmt and EF-G2mt, work in conjunction to facilitate the recycling step in mitochondria (Tsuboi et al. 2009) to release the deacylated tRNAmt and mRNA from the PoTC, and perhaps, to dissociate the 55S mitoribosome into its two subunits with the involvement of the third factor, IF3mt. The cryo-EM studies of the bacterial PoTC•RRF and PoTC•RRF•EF-G complexes suggest that alternate inter-ribosomal subunit ratcheting (upon RRF binding; Barat et al. 2007; Yokoyama et al. 2012) and unratcheting (upon subsequent EF-G binding; Yokoyama et al. 2012) in conjunction with a steric clash between domain II of RRFmt and SSU (Barat et al. 2007) facilitates disassembly of the PoTC.
1.3.4.1 Mitochondrial Ribosome Recyling Factor
The conserved fold of the mitochondrial ribosome recyling factor (RRFmt) is similar to that of a bacterial RRF that contains two well-defined structural domains (Selmer et al. 1999). Domain I is a three helix bundle connected by flexible elbow linkers to domain II, a βαβ sandwich. In addition, RRFmt has a mito-specific 78 amino acid residues long NTE (Rorbach et al. 2008). The homology model (Fig. 1.4b) suggests that its NTE is mainly α-helical. Most components of the ribosome (LSU rRNA helices, 69, 71, 80 and 93, and SSU protein S12) that are known to interact with the bacterial RRF (Agrawal et al. 2004; Gao et al. 2005; Barat et al. 2007; Weixlbaumer et al. 2007; Pai et al. 2008; Dunkle et al. 2011; Yokoyama et al. 2012) or the chloroplast ribosome (Sharma et al. 2007, 2010) are conserved in the mitoribosome. Therefore, it is likely that the conserved domains I and II of RRFmt make similar contacts on the mitoribosome.
In bacteria, RRF stabilizes the ribosome in its ratcheted state, which causes the destabilization of several inter-subunit bridges (B1 group bridges, and bridges B2a and B3; see Gabashvili et al. 2000; Yusupov et al. 2001; Sharma et al. 2003, for the bridge positions and nomenclature), and apparently primes the ribosome for the subsequent EF-G binding (Barat et al. 2007; Yokoyama et al. 2012), which catalyzes the final disassembly step. As indicated earlier (Sect. 1.3.2.2), the mitoribosome does not undergo ratcheting to the same degree as the bacterial ribosome does. It is possible that the long mito-specific NTE of RRFmt is involved in disruption of the mitoribosome’s B1 group bridges, which are also made of mito-specific MRPs, to offset the requirement of a pronounced ratcheted state.
1.3.4.2 Mitochondrial Elongation Factor G2
Mitochondrial elongation factor G2 (EF-G2mt) is a mito-specific paralog of EF-G that interacts with the 55S mitoribosome•RRFmt complex to catalyze the disassembly of the PoTC, therefore the factor has been also referred to as RRF2mt (Tsuboi et al. 2009). Unlike the canonical EF-G, EF-G2mt is unable to catalyze translocation and does not require hydrolysis of GTP to accomplish the ribosomal subunit splitting. The ability of EF-G2mt to functionally interact with RRFmt and the ribosome, to bring about ribosome splitting, mainly lies within its domains III and IV (Tsuboi et al. 2009). There is a mito-specific 25 amino acid insertion within EF-G2mt’s domain II. The cryo-EM study of the bacterial PoTC•RRF•EF-G complex shows that domains III, IV and V (but not domain II) of the structurally homologous bacterial EF-G interact with the domain II of RRF (Yokoyama et al. 2012). Thus, the function of the mito-specific insertion in domain II of EF-G2mt remains elusive. However, based on its location on the 3D model of EF-G2mt (Fig. 1.4b) it is conceivable and as suggested earlier for the mito-specific extension in EF-Tumt (Sect. 1.3.2.1), that the insertion may be involved in facilitating the factor’s interaction with the structurally depleted shoulder of the mito-SSU, rather than directly interacting with the mito-specific NTE of RRFmt. Further structural studies in context of the mitoribosome would be needed to resolve this intriguing interplay between the two recycling factors of the mitochondrial translation.
1.4 Concluding Remarks
An overall comparison of the previously determined cryo-EM structure of the mammalian mitoribosome with atomic structures of the cytoplasmic ribosomes is presented in this article, highlighting some of the unique features of the mitoribosome. The retention of key architectural elements in the mitoribosome underpins a notably conserved basic functioning despite compositional changes during its long structural evolution. The cryo-EM structure suggests that the mammalian mitoribosome has acquired several novel features related to mitochondrial protein synthesis. We have come a long way in improving the resolution of the cryo-EM structures of the mitoribosome, which is currently at 7 Å resolution in our laboratory. In the absence of any atomic structures of MRPs or mitochondrial translational factors, with the exception of EF-Tumt, molecular interpretation of the cryo-EM structures of the mitoribosome and its functional complexes is currently based on experimentally supported docking of homology models into the cryo-EM maps. However, identification and modeling of 38 non-homologous mito-specific MRPs in the cryo-EM map, especially those with undefined secondary structure motifs, continues to be a challenging task. The main barrier in achieving a high resolution structure appears to be an inherently heterogeneous composition of the mitoribosome, primarily due to a dramatic reduction in size of mito-rRNAs and significant increase in the number of MRPs. Many of these MRPs may be loosely attached to the rest of the mitoribosome in the absence of a direct interaction with the main rRNA scaffolds. This situation is dramatically different from the cytoplasmic ribosomes, which possess large rRNA scaffolds for interaction with their mostly basic ribosomal proteins, to produce relatively stable complex amenable to X-ray crystallographic structure determination. While the compositional fragility of the mitoribosome poses a challenge for X-ray crystallographic technique, it is highly suited to the single-particle cryo-EM method. This technique can provide structures for the mitoribosome and its functional complexes at ever increasing resolution to understand the functions of various insertion and extension sequences in both MRPs and mitochondrial translation factors, and to unravel mechanistic and molecular details of mitochondrial protein synthesis.
References
Ӕvarsson A, Brazhnikov E, Garber M, Zheltonosova J, Chirgadze Y, al-Karadaghi S, Svensson LA, Liljas A (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J 13:3669–3677
Agirrezabala X, Frank J (2009) Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q Rev Biophys 42:159–200
Agirrezabala X, Schreiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J (2011) Structural insights into cognate versus near-cognate discrimination during decoding. EMBO J 30:1497–1507
Agrawal RK, Sharma MR (2012) Structural aspects of mitochondrial translational apparatus. Curr Opin Struct Biol 22:797–803
Agrawal RK, Penczek P, Grassucci RA, Frank J (1998) Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Natl Acad Sci U S A 95:6134–6138
Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J (1999) EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol 6:643–647
Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J (2000) Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol 150:447–460
Agrawal RK, Sharma MR, Kiel MC, Hirokawa G, Booth TM, Spahn CM, Grassucci RA, Kaji A, Frank J (2004) Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: functional implications. Proc Natl Acad Sci U S A 101:8900–8905
Agrawal RK, Sharma MR, Yassin AS, Lahiri I, Spremulli L (2011) Structure and function of organellar ribosomes as revealed by cryo-EM. In: Rodnina M, Wintermeyer W, Green R (eds) Ribosomes: structure, function, and dynamics. SpringerWien, New York, pp 83–96
Akama K, Christian BE, Jones CN, Ueda T, Takeuchi N, Spremulli LL (2010) Analysis of the functional consequences of lethal mutations in mitochondrial translational elongation factors. Biochim Biophys Acta 1802:692–698
Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J (2005) The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121:703–712
Antonicka H, Ostergaard E, Sasarman F, Weraarpachai W, Wibrand F, Pedersen AM, Rodenburg RJ, van der Knaap MS, Smeitink JA, Chrzanowska-Lightowlers ZM et al (2010) Mutations in C12orf65 in patients with encephalomyopathy and a mitochondrial translation defect. Am J Hum Genet 87:115–122
Atkinson GC, Baldauf SL (2011) Evolution of elongation factor G and the origins of mitochondrial and chloroplast forms. Mol Biol Evol 28:1281–1292
Ban N, Nissen P, Hansen J, Moore PB, Steitz TA (2000) The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289:905–920
Barat C, Datta PP, Raj VS, Sharma MR, Kaji H, Kaji A, Agrawal RK (2007) Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol Cell 27:250–261
Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M (2011) The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334:1524–1529
Bhargava K, Spremulli LL (2005) Role of the N- and C-terminal extensions on the activity of mammalian mitochondrial translational initiation factor 3. Nucleic Acids Res 33:7011–7018
Bhargava K, Templeton P, Spremulli LL (2004) Expression and characterization of isoform 1 of human mitochondrial elongation factor G. Protein Expr Purif 37:368–376
Biou V, Shu F, Ramakrishnan V (1995) X-ray crystallography shows that translational initiation factor IF3 consists of two compact alpha/beta domains linked by an alpha-helix. EMBO J 14:4056–4064
Bullard JM, Cai YC, Zhang Y, Spremulli LL (1999) Effects of domain exchanges between Escherichia coli and mammalian mitochondrial EF-Tu on interactions with guanine nucleotides, aminoacyl-tRNA and ribosomes. Biochim Biophys Acta 1446:102–114
Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291:498–501
Christian BE, Spremulli LL (2009) Evidence for an active role of IF3mt in the initiation of translation in mammalian mitochondria. Biochemistry 48:3269–3278
Christian BE, Spremulli LL (2010) Preferential selection of the 5′-terminal start codon on leaderless mRNAs by mammalian mitochondrial ribosomes. J Biol Chem 285:28379–28386
Christian BE, Spremulli LL (2012) Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 1819:1035–1054
Chrzanowska-Lightowlers ZM, Pajak A, Lightowlers RN (2011) Termination of protein synthesis in mammalian mitochondria. J Biol Chem 286:34479–34485
Chung HK, Spremulli LL (1990) Purification and characterization of elongation factor G from bovine liver mitochondria. J Biol Chem 265:21000–21004
Claros MG, Vincens P (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786
Czworkowski J, Wang J, Steitz TA, Moore PB (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J 13:3661–3668
Dallas A, Noller HF (2001) Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 8:855–864
Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK (2005) Interaction of the G’ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell 20:723–731
Duarte I, Nabuurs SB, Magno R, Huynen M (2012) Evolution and diversification of the organellar release factor family. Mol Biol Evol 29:3497–3512
Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH (2011) Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332:981–984
Frank J, Agrawal RK (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406:318–322
Frank J, Agrawal RK (2001) Ratchet-like movements between the two ribosomal subunits: their implications in elongation factor recognition and tRNA translocation. Cold Spring Harb Symp Quant Biol 66:67–75
Frank J, Penczek P, Agrawal RK, Grassucci RA, Heagle AB (2000) Three-dimensional cryoelectron microscopy of ribosomes. Methods Enzymol 317:276–291
Gabashvili IS, Agrawal RK, Spahn CM, Grassucci R, Svergun D, Frank J, Penczek P (2000) Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100:537–549
Gagnon MG, Seetharaman SV, Bulkley D, Steitz TA (2012) Structural basis for the rescue of stalled ribosomes: structure of YaeJ bound to the ribosome. Science 335:1370–1372
Gao N, Zavialov AV, Li W, Sengupta J, Valle M, Gursky RP, Ehrenberg M, Frank J (2005) Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell 18:663–674
Gao YG, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V (2009) The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326:694–699
Gaur R, Grasso D, Datta PP, Krishna PD, Das G, Spencer A, Agrawal RK, Spremulli L, Varshney U (2008) A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell 29:180–190
Gomez-Lorenzo MG, Spahn CM, Agrawal RK, Grassucci RA, Penczek P, Chakraburtty K, Ballesta JP, Lavandera JL, Garcia-Bustos JF, Frank J (2000) Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution. EMBO J 19:2710–2718
Gray, M.W., Burger, G., and Lang, B.F. (2001). The origin and early evolution of mitochondria. Genome Biol 2, REVIEWS1018
Gruschke S, Ott M (2010) The polypeptide tunnel exit of the mitochondrial ribosome is tailored to meet the specific requirements of the organelle. BioEssays 32:1050–1057
Hammarsund M, Wilson W, Corcoran M, Merup M, Einhorn S, Grander D, Sangfelt O (2001) Identification and characterization of two novel human mitochondrial elongation factor genes, hEFG2 and hEFG1, phylogenetically conserved through evolution. Hum Genet 109:542–550
Hanada T, Suzuki T, Watanabe K (2000) Translation activity of mitochondrial tRNA with unusual secondary structure. Nucleic Acids Symp Ser 44:249–250
Haque ME, Spremulli LL (2008) Roles of the N- and C-terminal domains of mammalian mitochondrial initiation factor 3 in protein biosynthesis. J Mol Biol 384:929–940
Haque ME, Spremulli LL, Fecko CJ (2010) Identification of protein–protein and protein-ribosome interacting regions of the C-terminal tail of human mitochondrial inner membrane protein Oxa1L. J Biol Chem 285:34991–34998
Haque ME, Koc H, Cimen H, Koc EC, Spremulli LL (2011) Contacts between mammalian mitochondrial translational initiation factor 3 and ribosomal proteins in the small subunit. Biochim Biophys Acta 1814:1779–1784
Huynen MA, Duarte I, Chrzanowska-Lightowlers ZM, Nabuurs SB (2012) Structure based hypothesis of a mitochondrial ribosome rescue mechanism. Biol Direct 7:14
Jeppesen MG, Navratil T, Spremulli LL, Nyborg J (2005) Crystal structure of the bovine mitochondrial elongation factor Tu.Ts complex. J Biol Chem 280:5071–5081
Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA (2003) Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J 22:6438–6447
Julian P, Milon P, Agirrezabala X, Lasso G, Gil D, Rodnina MV, Valle M (2011) The Cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol 9:e1001095
Keil M, Bareth B, Woellhaf MW, Peleh V, Prestele M, Rehling P, Herrmann JM (2012) Oxa1-ribosome complexes coordinate the assembly of cytochrome C oxidase in mitochondria. J Biol Chem 287:34484–34493
Kjeldgaard M, Nissen P, Thirup S, Nyborg J (1993) The crystal structure of elongation factor EF-Tu from Thermus aquaticus in the GTP conformation. Structure 1:35–50
Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334:941–948
Koc EC, Koc H (2012) Regulation of mammalian mitochondrial translation by post-translational modifications. Biochim Biophys Acta 1819:1055–1066
Koc EC, Burkhart W, Blackburn K, Moseley A, Koc H, Spremulli LL (2000) A proteomics approach to the identification of mammalian mitochondrial small subunit ribosomal proteins. J Biol Chem 275:32585–32591
Koc EC, Burkhart W, Blackburn K, Moseley A, Spremulli LL (2001a) The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem 276:19363–19374
Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL (2001b) The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276:43958–43969
Koc EC, Haque ME, Spremulli LL (2010) Current views of the structure of the mammalian mitochondrial ribosome. Isr J Chem 50:45–59
Korostelev AA (2011) Structural aspects of translation termination on the ribosome. RNA 17:1409–1421
Laurberg M, Asahara H, Korostelev A, Zhu J, Trakhanov S, Noller HF (2008) Structural basis for translation termination on the 70S ribosome. Nature 454:852–857
Luirink J, Samuelsson T, de Gier JW (2001) YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett 501:1–5
Manuell AL, Quispe J, Mayfield SP (2007) Structure of the chloroplast ribosome: novel domains for translation regulation. PLoS Biol 5:e209
McCutcheon JP, Agrawal RK, Philips SM, Grassucci RA, Gerchman SE, Clemons WM Jr, Ramakrishnan V, Frank J (1999) Location of translational initiation factor IF3 on the small ribosomal subunit. Proc Natl Acad Sci U S A 96:4301–4306
Mears JA, Sharma MR, Gutell RR, McCook AS, Richardson PE, Caulfield TR, Agrawal RK, Harvey SC (2006) A structural model for the large subunit of the mammalian mitochondrial ribosome. J Mol Biol 358:193–212
Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J (1995) Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science 270:1464–1472
Nissen P, Hansen J, Ban N, Moore PB, Steitz TA (2000) The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–930
O’Brien TW (2002) Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286:73–79
O’Brien TW, O’Brien BJ, Norman RA (2005) Nuclear MRP genes and mitochondrial diseases. Gene 354:147–151
Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292:897–902
Ohtsuki T, Watanabe Y (2007) T-armless tRNAs and elongated elongation factor Tu. IUBMB Life 59:68–75
Ott M, Herrmann JM (2010) Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta 1803:767–775
Pai RD, Zhang W, Schuwirth BS, Hirokawa G, Kaji H, Kaji A, Cate JH (2008) Structural Insights into ribosome recycling factor interactions with the 70S ribosome. J Mol Biol 376:1334–1347
Pearce S, Nezich CL, Spinazzola A (2013) Mitochondrial diseases: translation matters. Mol Cell Neurosci 55:1–12
Petrelli D, LaTeana A, Garofalo C, Spurio R, Pon CL, Gualerzi CO (2001) Translation initiation factor IF3: two domains, five functions, one mechanism? EMBO J 20:4560–4569
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Rawat UB, Zavialov AV, Sengupta J, Valle M, Grassucci RA, Linde J, Vestergaard B, Ehrenberg M, Frank J (2003) A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature 421:87–90
Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM (2010) A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J 29:1116–1125
Rorbach J, Richter R, Wessels HJ, Wydro M, Pekalski M, Farhoud M, Kuhl I, Gaisne M, Bonnefoy N, Smeitink JA et al (2008) The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res 36:5787–5799
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738
Schmeing TM, Ramakrishnan V (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461:1234–1242
Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR JR, Ramakrishnan V (2009) The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326:688–694
Schuette JC, Murphy FVt, Kelley AC, Weir JR, Giesebrecht J, Connell SR, Loerke J, Mielke T, Zhang W, Penczek PA et al (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J 28:755–765
Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH (2005) Structures of the bacterial ribosome at 3.5 Å resolution. Science 310:827–834
Selmer M, Al-Karadaghi S, Hirokawa G, Kaji A, Liljas A (1999) Crystal structure of Thermotoga maritima ribosome recycling factor: a tRNA mimic. Science 286:2349–2352
Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942
Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK (2003) Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115:97–108
Sharma MR, Wilson DN, Datta PP, Barat C, Schluenzen F, Fucini P, Agrawal RK (2007) Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci U S A 104:19315–19320
Sharma MR, Booth TM, Simpson L, Maslov DA, Agrawal RK (2009) Structure of a mitochondrial ribosome with minimal RNA. Proc Natl Acad Sci U S A 106:9637–9642
Sharma MR, Dönhöfer A, Barat C, Marquez V, Datta PP, Fucini P, Wilson DN, Agrawal RK (2010) PSRP1 is not a ribosomal protein, but a ribosome-binding factor that is recycled by the ribosome-recycling factor (RRF) and elongation factor G (EF-G). J Biol Chem 285:4006–4014
Smirnov A, Entelis N, Martin RP, Tarassov I (2011) Biological significance of 5S rRNA import into human mitochondria: role of ribosomal protein MRP-L18. Genes Dev 25:1289–1305
Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ (2007) Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res 35:4686–4703
Soleimanpour-Lichaei HR, Kuhl I, Gaisne M, Passos JF, Wydro M, Rorbach J, Temperley R, Bonnefoy N, Tate W, Lightowlers R et al (2007) mtRF1a is a human mitochondrial translation release factor decoding the major termination codons UAA and UAG. Mol Cell 27:745–757
Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J (2004) Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23:1008–1019
Spencer AC, Spremulli LL (2004) Interaction of mitochondrial initiation factor 2 with mitochondrial fMet-tRNA. Nucleic Acids Res 32:5464–5470
Spencer AC, Spremulli LL (2005) The interaction of mitochondrial translational initiation factor 2 with the small ribosomal subunit. Biochim Biophys Acta 1750:69–81
Stark H, Rodnina MV, Rinke-Appel J, Brimacombe R, Wintermeyer W, van Heel M (1997) Visualization of elongation factor Tu on the Escherichia coli ribosome. Nature 389:403–406
Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K (2001a) Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28S small subunit. J Biol Chem 276:33181–33195
Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K (2001b) Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome. Systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J Biol Chem 276:21724–21736
Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM (2003) Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 22:6448–6457
Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM (2010) Human mitochondrial mRNAs-like members of all families, similar but different. Biochim Biophys Acta 1797:1081–1085
Tsuboi M, Morita H, Nozaki Y, Akama K, Ueda T, Ito K, Nierhaus KH, Takeuchi N (2009) EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol Cell 35:502–510
Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J (2002) Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21:3557–3567
Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M (2001) Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell 8:1375–1382
Watanabe K (2010) Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc Jpn Acad Ser B Phys Biol Sci 86:11–39
Weixlbaumer A, Petry S, Dunham CM, Selmer M, Kelley AC, Ramakrishnan V (2007) Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol 14:733–737
Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V (2008) Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322:953–956
Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407:327–339
Yassin AS, Agrawal RK, Banavali NK (2011a) Computational exploration of structural hypotheses for an additional sequence in a mammalian mitochondrial protein. PLoS ONE 6:e21871
Yassin AS, Haque ME, Datta PP, Elmore K, Banavali NK, Spremulli LL, Agrawal RK (2011b) Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc Natl Acad Sci U S A 108:3918–3923
Yokoyama T, Shaikh TR, Iwakura N, Kaji H, Kaji A, Agrawal RK (2012) Structural insights into initial and intermediate steps of the ribosome-recycling process. EMBO J 31:1836–1846
Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF (2001) Crystal structure of the ribosome at 5.5 Å resolution. Science 292:883–896
Yusupova GZ, Yusupov MM, Cate JH, Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106:233–241
Acknowledgments
This work was supported by the National Institutes of Health grant R01 GM61576 (to R.K.A.).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Sharma, M.R., Kaushal, P.S., Gupta, M., Banavali, N.K., Agrawal, R.K. (2013). Insights into Structural Basis of Mammalian Mitochondrial Translation. In: Duchêne, AM. (eds) Translation in Mitochondria and Other Organelles. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-39426-3_1
Download citation
DOI: https://doi.org/10.1007/978-3-642-39426-3_1
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-39425-6
Online ISBN: 978-3-642-39426-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)