Abstract
Work over the last years has uncovered that during the highly integrative process of polar auxin transport, dynamic interactions of membrane proteins with other membrane or soluble proteins or modulatory drugs are providing a high degree of flexibility. This overall concept is supported by the recent release of a first, partial Arabidopsis interactome by the Arabidopsis Interactome Mapping Consortium. In this context, we have summarized the current knowledge of posttranscriptional regulation of auxin transport with an emphasis on protein–protein interaction and protein phosphorylation. We suggest a novel protein–protein interaction feedback loop of auxin transport. Further, we summarize evidence that this interaction loop is tightly interconnected with a previously described PIN polarity loop via AGC3 kinases represented by PINOID. These data are compatible with the view of a putative multi-protein auxin efflux complex that is building the basis for a plastic and economic control of auxin streams during PAT.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Gradients of the intercellular signaling compound auxin are the primary determinant for the establishment and maintenance of plant polarity in respect to their development, growth, and physiology (Robert and Friml 2009; Vanneste and Friml 2009). Although the exact mechanism how indol-3-acetic acid (IAA), the most common auxin, is transported from one plasma membrane domain to the next is still controversial (Schlicht et al. 2006), the intercellular distribution of IAA provided by polar auxin transport (PAT) is a unique process so far not found for other signaling molecules or in non-plant species (Vieten et al. 2007).
The chemiosmotic model has predicted the existence of secondary active auxin transporters that are thought to be directly or indirectly energized by the proton gradient established by the H+-ATPase (Vanneste and Friml 2009). Despite the fact that also novel auxin transporters have been recently found (Barbez et al. 2012), until now, members of three distinct transporters families have been convincingly implicated to be directly involved in polar auxin transport: AUXIN RESISTANT 1/LIKE AUX1 (AUX1/LAX) uptake symporters, PIN-FORMED (PIN) efflux carriers, and P-GLYCOPROTEIN/MULTIDRUG RESISTANCE (PGP/MDR/ABCB) efflux pumps. A subgroup of ER-localized members of so-called short PINs, such as PIN5 and PIN8, have recently been shown to be implicated in cellular homeostasis and not in PAT (Bosco et al. 2012; Mravec et al. 2009; Ding et al. 2012; see Chap. 2 for details).
Importantly, due to the chemical features of IAA allowing partial diffusion into cells, exporters are the primary control units for PAT (Geisler and Murphy 2006). Accordingly, their activity has been shown to be tightly controlled on both the transcriptional and posttranscriptional levels (Titapiwatanakun and Murphy 2009). In the last years, members of the described PIN and ABCB family apparently independently—but also coordinately—function as the molecular machines that establish and maintain these gradients according to a revised version of the chemiosmotic model (Bandyopadhyay et al. 2007; Geisler and Murphy 2006).
In this chapter, we will focus mainly on the posttranscriptional regulation of PINs and ABCBs with a special focus on protein–protein interaction in respect to its impact on individual transport capacities. We suggest a novel regulatory, interaction feedback loop. However, we will also integrate very recent findings indicating that ABCB activity is directly controlled by protein phosphorylation, indicating that regulatory phosphorylation/polarity and interaction loops are interconnected.
2 Posttranscriptional Regulation of Auxin Catalysts
Based on the “canalization concept” originally hypothesized in order to explain vascular patterning of the leaf, a positive feedback loop between auxin flux and the cell’s auxin transport capacity has been proposed (Sachs 1969; Stoma et al. 2008). Many of the involved molecular components have been identified, including auxin transport catalysts and mechanisms by which auxin itself feeds back on their polar locations (reviewed in detail in Vieten et al. 2007). However, it should be stressed that until now the proposed auxin flux sensor has not yet been identified in plants (Merks et al. 2007).
Recent work on the posttranscriptional regulation of auxin catalysts has mainly focused on the trafficking pathways of PIN proteins (and also of AUX1) that have been studied intensively, and endosomal cycling is thought to play an essential role in PIN localizations (Feraru and Friml 2008; Jurgens and Geldner 2007; Kleine-Vehn and Friml 2008). Compared to PINs, the trafficking routes of ABCBs have not yet been analyzed in detail. However, keeping the functional ABCB–PIN interactions on the plasma membrane (see below) and the BFA sensitivity of the ABCB1 plasma membrane location (Titapiwatanakun et al. 2009; Wu et al. 2010) in mind, one might speculate that ABCBs follow known PIN routes: ABCB1, like PIN1, is internalized into endosomal compartments upon BFA treatments, unlike ABCB19 that is widely BFA insensitive and follows apparently a trafficking pathway that is distinct from PIN1/ABCB1 (Titapiwatanakun et al. 2009). Finally, plasma membrane locations of ABCB1, ABCB19, and ABCB4 but not PIN1 or PIN2 (Bouchard et al. 2006) have been shown to be dependent on the action of the FKBP42, TWISTED DWARF1 (TWD1), although the underlying mechanism is not yet clear (Wu et al. 2010).
The current picture that emerges is that PIN (and most probably also AUX1/LAX) transporters both obviously providing the majority of bulk PAT streams are mainly regulated by influencing their subcellular location and polarity, while ABCBs that show widely nonpolar locations and are more stable on the plasma membrane are obviously controlled by regulation of their catalytic activity. The underlying mechanisms for regulatory modules, protein–protein interaction/modulatory drugs, and protein phosphorylation are summarized in more detail in the following.
3 Regulation of Auxin Catalysts by Protein–Protein Interaction
Work over the last years has revealed that during the highly integrative process of PAT, dynamic interactions of membrane proteins with other membrane or soluble components (hereafter referred to as PAT modulators) are thought to provide a high degree of flexibility that usually characterizes higher plants. This overall concept is supported by the recent release of a first, partial Arabidopsis interactome by the Arabidopsis Interactome Mapping Consortium (2011). Apparently, the Arabidopsis interactome reveals a strong enrichment of a few network communities, including those for transmembrane transport and vesicle trafficking. Strikingly, the largest transmembrane transport community shares a high amount of proteins with the vesicle-trafficking community, suggesting a strong physical and functional overlap and interaction. While the Arabidopsis interactome, in contrast to those of yeast, human, C. elegans, and Drosophila, awaits its completion also for Arabidopsis protein–protein interaction (PPI) tools and experimental and theoretical large-scale protein interaction maps have been developed (Chen et al. 2012; De Bodt et al. 2012; Geisler-Lee et al. 2007; Lee et al. 2010; Li et al. 2011). In agreement with the overrepresentation of Arabidopsis transporter proteins (over 1,200 proteins) have been assigned as transporters although only 267 transporters have been as such characterized (Lalonde et al. 2010). A focus was laid on the elucidation of membrane proteins and their signaling networks (Chen et al. 2012; Lalonde et al. 2010). However, previous experimental work on PAT in respect to protein–protein interaction has focused on functional interactions between ABCBs and the immunophilin-like FKBP42, TWISTED DWARF1 (TWD1) on the one hand and on the interaction between ABCBs and PINs on the other.
3.1 TWD1–ABCB Interaction
Work over the last decade has established the physical and functional interaction of the immunophilin-like protein FKBP42, called TWISTED DWARF1 (TWD1)/ULTRACURVATA 2 (UCU2) (Perez-Perez et al. 2004), with ABCBs, ABCB1, and ABCB19 (Geisler and Bailly 2007). TWD1/FKBP42 belongs to the FK506-binding protein (FKBP) subfamily of PPIases (cis–trans peptidyl-prolyl isomerases), which are thought to catalyze the cis–trans isomerization of cis-prolyl bonds (Geisler and Bailly 2007). Many FKBPs have a PPIase (rotamase) activity, creating the impression that FKBPs function primarily in protein folding. However, extensive research during the past decade has elucidated two independent functions for FKBPs (1) a PPIase activity classically inhibited by binding of clinically relevant immunosuppressant drugs, such as FK506 (tacrolimus) or rapamycin (sirolimus), and (2) a chaperone function that is independent of the PPIase activity and unaffected by immunosuppressant drugs (Barik 2006; Blecher et al. 1996; Harrar et al. 2003).
The twisted dwarf1 T-DNA insertional mutant (twd1-1) has been isolated in a mutant screen for developmental phenotypes. In order to understand the pleiotropic developmental growth phenotype of twd1, characterized by a dwarf plant size, reduced cell elongation, disoriented growth of all organs, and misshapen epidermal cells (causing a twist), a yeast two-hybrid screen for putative interactors using the soluble portion of TWD1 as bait was performed (Geisler et al. 2003). The rationale to do so was based on the fact that TWD1 owns no detectable PPIase activity using standard test substrates but contains three repetitions of a so-called tetratricopeptide repeat (TPR). This qualified TWD1 as a multi-domain (high-molecular weight) FKBP, containing typically up to three N-terminal putative FK506-binding domains (FKBDs), typically followed by a TPR domain and a calmodulin-binding domain (CaM-BD; see Fig. 1), both known to mediate protein–protein interactions to calmodulin and heat-shock proteins, respectively (Geisler and Bailly 2007). Both calmodulin and HSP90 in vitro binding to TWD1 was demonstrated although the physiological relevance of these interactions is entirely unclear (see Sect. 6).
Model summarizing the effect of PIN1–ABCB and TWD1–ABCB interactions on auxin export. ABCB and PIN proteins are able to function as independent auxin export catalysts; however, tissue-specific ABCB–PIN pairings alter positively and negatively (+/−) IAA export, NPA sensitivity, substrate specificity, and eventually also transport directionality of ABCBs. Interaction is provided between the cytoplasmic loop of plasma membrane-bound PINs interfering with the NBD2 of ABCBs. TWD1 functions as a positive (+) modulator of ABCB activity by interaction between the TWD1 FKBD (blue) and the ABCB NBD2. Interaction is thought to be either disrupted by NPA binding to the TWD1 FKBD (blue) and the ABCB interface between NBD and intracellular loops (Bailly and Geisler, unpublished) or by flavonoid binding to the ABCB NBDs (Conseil et al. 1998), resulting in ABCB inhibition
The two-hybrid screen for TWD1 interactors resulted, besides HSP90, in the identification of C-terminal nucleotide-binding domains (NBDs) of ABCB1 and members of the ABCC/MRP family. TWD1–ABCB1 interaction was verified by using in vitro pull-down assays, NPA- and TWD1-HA affinity chromatography (Geisler et al. 2003) and bioluminescence resonance energy transfer in yeast (BRET; Bailly et al. 2008). Mapping of interacting domains demonstrated unexpectedly that not the C-terminal TPR domain but the N-terminal putative PPIase domain (FKBD) provided this interaction (Geisler et al. 2003). Interaction was not affected by immunosuppressant drugs, suggesting, together with the absence of a detectable PPIase activity, an evolutionary shift of function toward protein–protein interaction (Geisler and Bailly 2007; Geisler et al. 2003). Both interaction and ABCB regulation are provided by the N-terminal FKBD, as the soluble FKBD upon co-expression in yeast can functionally replace the full-length TWD1 (Bailly et al. 2008).
Phenotypic twd1 analysis revealed reduced hypocotyl lengths in the dark and under light, elongated root length (in the dark), and an obvious overlap between twd1 and abcb1abcb19 loss-of-function alleles, especially at early stages, suggesting a regulatory impact of TWD1 on ABCB1/B19-mediated auxin transport capacities (Geisler and Bailly 2007; Geisler et al. 2003). Co-expression of ABCB1 with TWD1 (but not with Arabidopsis FKBP12) in yeast reduced ABCB1 IAA export activity to vector control levels as shown by transport and growth assays (Bouchard et al. 2006). This was surprising as it is the opposite of what one would have expected from previous in planta data. TWD1 has also a negative impact on ABCB1 activity when co-expressed in the heterologous plant system N. benthamiana (Henrichs et al. 2012). However, in mammalian HeLa cells, TWD1 has an activating effect on ABCB1, suggesting that a third factor might be absent in yeast and tobacco (Bouchard et al. 2006). In this respect, TWD1 might compete for ABCB1 activation by high levels of yeast (or tobacco) FKBP12, the first shown to activate ABCB1 and mouse ABCB3/MDR3 (Hemenway and Heitman 1996). Assuming higher affinity and/or abundance of heterologous FKBP12 compared to TWD1, this would result in a net reduction of ABCB1-mediated auxin transport (Bailly et al. 2008; Henrichs et al. 2012). This concept is supported by the recent finding that yeast FKBP12 is able to widely complement twd1 (Henrichs et al. 2012). Moreover, TWD1 does apparently not compete for endogenous FKBP12 activation in Arabidopsis as AtFKBP12 has no significant effect on ABCB1 activity (Bouchard et al. 2006).
However, a positive regulatory in planta role on ABCBs and in PAT was indicated by reduced reflux capacities in abcb and twd1 roots measured by employing an IAA-specific microelectrode (Bouchard et al. 2006) and by demonstrating that locations of PIN1 and PIN2 were unchanged (Wu et al. 2010). This concept was underlined by the finding that twd1 roots, similarly to those of abcb1 abcb19 (but not abcb1 and abcb19 single) mutants, reveal elevated levels of free IAA (especially in mature parts) and altered gravitropic responses (Bailly et al. 2008). Together with the major predicted roles of ABCB1 and ABCB19 in basipetal (ABCB1) and acropetal (ABCB19) auxin transport in roots (Bandyopadhyay et al. 2007; Blakeslee et al. 2007; Geisler and Murphy 2006), this suggests TWD1 to function as a central regulator of ABCB-mediated long-range auxin transport controlling plant physiology and development (Bailly et al. 2006; Geisler and Bailly 2007).
The crystal structures of the TWD1 FKBD and the full-length protein without membrane anchor have been determined (Granzin et al. 2006; Weiergraber et al. 2006). Modeling of FK506 docking indicates that, consistent with experimental data, TWD1 (like HsFKBP38) sterically excludes immunosuppressant drugs like FK506 (Bailly et al. 2006). Even more informative is that FK506 binding positively correlates with the presence of a PPIase activity. Therefore, the current picture that emerges is that stress-related FKBPs have apparently maintained a conserved PPIase activity to fulfill their proposed chaperone function, whereas others (such as human FKBP38, TWD1, or PAS1) have lost (or only retained low) PPIase activity (Geisler and Bailly 2007). A structural shift toward functionality in protein–protein interaction was provided by recent NMR assignments of the FKBP-type PPIase domain of FKBP42. Signal intensities revealed an additional structure element that is atypical for such FKBP domains (Burgardt et al. 2012).
In silico modeling of the protein–protein interaction with key interacting partners, HSP90, ABCB1, and ABCC1, has facilitated the prediction of docking sites at the molecular level. Although the docking domains of TWD1 that interact with the nucleotide-binding fold of ABCB- and ABCC-like ABC transporters are different (FKB and TPR domains, respectively), both transporters use overlapping surface areas on the transporters, suggesting a new paradigm for the regulation of ABC transporter activity (Granzin et al. 2006). Co-crystallization of FKBPs with interacting partners will be the method of choice to understand TWD1-ABC transporter at the molecular level.
TWD1 is surprisingly difficult to solubilize by detergents, which has initially supported the prediction of a glycophosphatidyl inositol (GPI) anchoring (Geisler et al. 2003). However, no GPI moiety has been detected biochemically (Murphy et al. 2002), and TWD1 has not been identified in any of the proteomic approaches on raft-like structures (Morel et al. 2006). Recently, based on NMR analysis of a C-terminal TWD1 peptide, a perpendicular orientation of the TWD1 anchor forming a so-called amphipathic in-plane membrane (IPM) anchor has been predicted (Scheidt et al. 2007).
Using electron microscopy on HA-TWD1-OX plants, TWD1 has, besides on the plasma membrane, additionally been localized to the tonoplast (Kamphausen et al. 2002). This is of relevance because, as mentioned above, TWD1 has been shown to functionally interact with ABCC1/MRP1 and ABCC2/MRP2 (Geisler et al. 2004). Moreover, recently employing a C-terminal CFP tag fused to a genomic construct expressing TWD1 under its own promoter, a convincing location that was restricted solely to the ER was found (Wu et al. 2010). Like the previous HA-tagged cDNA construct expressing TWD1 under the constitutive, also this genomic construct fully complemented the twisted syndrome. This conflict is even more puzzling as expression of TWD1 from the TWD1:TWD1–CFP construct resulted in a clear plasma membrane location in tobacco leaves but no obvious ER signal (Henrichs et al. 2012). In summary, it appears that TWD1 resides on multiple subcellular compartments in analogy to its mammalian ortholog, FKBP38 (Edlich and Lucke 2011; Shirane and Nakayama 2003).
Interestingly, based on the finding that ABCB1-, B4-, and B19-GFP are retained on the ER in twd1, a chaperone function independent of a PPIase activity for TWD1 in plasma membrane secretion of ABCBs was re-proposed (Wu et al. 2010). These data are in conflict with a previous work showing that ABCB1 resides on plasma membrane fractions after sucrose density gradient centrifugation (Bouchard et al. 2006). However, ER retention of ABCB1/B4/B19 suggests an alternative TWD1 functionality that implies that the twisted dwarf phenotype is caused not by a lack of ABCB activation but their delocalization. Obviously, these two scenarios are not exclusive. Moreover, one might also imagine that in the absence of TWD1, inactive ABCBs are removed from the plasma membrane and that ER locations represent degradation locations. In any case, a comparison of twd1 with abcb1,b4,b19 plants as well as an analysis of TWD1–ABCB4 interaction is highly desirable.
Regardless of the mode of membrane anchoring and the intracellular compartment of interaction, a relevant question is why the need for membrane anchoring of TWD1 at all? It has been suggested that FKBP38 acts as a mitochondrial docking molecule that concentrates two anti-apoptotic membrane proteins at the mitochondria (instead of the ER), thus preventing apoptosis (Edlich and Lucke 2011). A similar mechanism might be involved in TWD1 regulation of ABC transporters both on the tonoplast and plasma membrane: Membrane anchoring might thus increase the probability of contacts by reducing the spatiality of TWD1 diffusion. Furthermore, restraining the mobility of TWD1 by membrane anchoring might serve as a means to decouple the regulation of transporters located on different membranes (Scheidt et al. 2007).
3.2 ABCB–PIN Interaction
Previous work established that ABCB and PIN proteins are able to function as independent auxin export catalysts (Petrasek et al. 2006). However, both transporter classes lack the degree of substrate specificity seen in planta when expressed in heterologous systems (Blakeslee et al. 2007). Moreover, abcb1 and, to an even higher degree, abcb19 and pin1 mutant roots show a high degree of unspecific basipetal BA transport not found for wild type (Blakeslee et al. 2007). This, together with the fact that ABCB1, B19, and PIN1/P2 show widely overlapping root expression profiles, suggested functional interaction between these auxin efflux catalysts (Blakeslee et al. 2007).
Subcellular co-localization, yeast two-hybrid interaction, and co-immunoprecipitation analyses provided clear evidence for distinct ABCB1,19-PIN1,2 pairings (Blakeslee et al. 2007; Rojas-Pierce et al. 2007). However, it should be mentioned that not all possible ABCB1/B19–PIN1/P2 combinations have been clearly proven and that the hardest set of evidence exists for ABCB19–PIN1 and ABCB1–PIN2 interactions (Blakeslee et al. 2007). Not surprisingly, these interactions correlate with their proposed overlapping functions in apical and basipetal auxin transport, respectively. As was the case for ABCB/TWD1 pairs, interaction of ABCBs and PINs employs the C-terminal NBDs of the ABCBs binding the central cytoplasmic PIN loops (Blakeslee et al. 2007).
Co-expression of ABCBs in HeLa cells with PIN1 increased export, NPA sensitivity, and substrate specificity of ABCB1/B19, while PIN2 had only significant effects on ABCB specificity. Similar results were found with ABCB1 co-expressed with PIN1 and PIN2 (Blakeslee et al. 2007). Functional interaction was supported by synergistic abcb19 pin1 plant phenotypes and abcb1 abcb19 pin2 root agravitropism (Blakeslee et al. 2007). Interestingly, dynamics of PIN1 cycling are reduced in ABC19 locations, and PIN1 plasma membrane location was more easily perturbed in abcb19 roots, suggesting that ABCB19 stabilizes PIN1 in plasma membrane microdomains (Titapiwatanakun et al. 2009).
As mentioned already, based on interaction and transport studies, there was no indication for ABCB–AUX1 interaction (Blakeslee et al. 2007). However, functional co-expression of PIN1 reversed the import direction of ABCB4 in HeLa cells, while PIN2 enhanced ABCB4 activity (Blakeslee et al. 2007). However, trials to verify PIN–ABCB4 interaction failed so far (Titapiwatanakun et al. 2009).
In summary, these data provide evidence for independent ABCB and PIN transport mechanisms but also tissue-specific ABCB–PIN pairings that function interactively. In these, PIN proteins seem to add a vectorial dimension to ABCB-mediated nonpolar cellular auxin export required for PAT.
3.3 PAT Modulators
The auxin efflux complex is thought to consist of at least two proteins: a membrane integral transporter and regulatory subunit binding the noncompetitive, synthetic auxin efflux inhibitor, 1-N-naphtylphtalamic acid (NPA), qualifying it as NPA-binding protein (NBP) (Bernasconi et al. 1996; Cox and Muday 1994; Luschnig 2001; Michalke et al. 1992). Until today the identity, number, and affinity of putative NBPs are still controversial (Cox and Muday 1994; Michalke et al. 1992; Sussman and Gardner 1980), but there is apparently a consensus that PIN proteins do not itself act as NBPs (Lomax et al. 1995).
The regulatory impact of flavonoids, a class of plant-derived secondary compounds, on PAT was initially based on their ability to compete with NPA for transporter binding sites (Jacobs and Rubery 1988; Lomax et al. 1995; Morris 2000). This concept is further supported by auxin-related phenotypes of Arabidopsis mutants with altered flavonoid levels (Buer et al. 2007; Peer and Murphy 2006, 2007; Taylor and Grotewold 2005), although fundamental physiological processes occur in the absence of flavonoids. Aglycone molecules, such as quercetin and kaempferol, have been shown to inhibit PAT and consequently to enhance localized auxin accumulation (Brown et al. 2001). Currently they are seen as transport modulators (Peer and Murphy 2007); nevertheless, the mechanisms by which flavonoids physically interfere with auxin efflux components are not yet clear.
ABCB1 and ABCB19 have been identified—together with TWD1—in high-affinity fractions as NBPs (Geisler et al. 2003; Murphy et al. 2002; Noh et al. 2001), which obviously does not directly prove that all three are high-affinity NBPs (see below). However, it was shown that high micromolar NPA concentrations cause inhibition of auxin efflux catalyzed by ABCB1 and ABCB19 (Bouchard et al. 2006; Terasaka et al. 2005). NPA binding studies using microsomes prepared from abcb mutants and yeast or HeLa cells expressing ABCB1 and ABCB19 verified this assumption (Benjamins et al. 2001; Kim et al. 2010; Noh et al. 2001; Rojas-Pierce et al. 2007).
ABCB1 and ABCB19 both bind NPA but own apparently different NPA affinities. This has led to the idea that ABCB19 represents the major target of NPA (Rojas-Pierce et al. 2007). Using plasma membrane-enriched microsomes from loss-of-function plants, it was shown that abcb19 but not abcb1 (or pin1) showed significantly reduced NPA binding, although the synthetic ATI, gravacin, removed NPA to similar wild-type levels (Rojas-Pierce et al. 2007).
By means of chemical genomics, another auxin transport inhibitor, called BUM (2-[4-(diethylamino)-2-hydroxybenzoyl]benzoic acid), was identified by its potential to efficiently block auxin-regulated plant physiology and development (Kim et al. 2010). In many respects, BUM resembles the functionality of NPA but has an IC50 that is roughly a factor 30 lower. Physiological analysis and binding assays identified ABCBs, primarily ABCB1, as key targets of BUM and NPA, while PIN proteins were shown not to be directly affected. BUM is complementary to NPA by having distinct ABCB target spectra and impacts on basipetal polar auxin transport in the shoot and root. In comparison to gravacin, it lacks interference with ABCB membrane trafficking.
Two findings were suggesting that TWD1–ABCB interaction was disrupted by NPA: First, concentrations needed to block ABCB transport activity expressed in heterologous systems were far higher than what was needed in planta. And, second, excess washing with NPA excluded TWD1 but not ABCBs from NPA chromatographies (Geisler et al. 2003). Using a yeast-based BRET (bioluminescence resonance energy transfer) system, NPA and BUM, but not competitive ATIs, TIBA, or CPD, were shown to disrupt TWD1–ABCB1 interaction (Bailly et al. 2008; Kim et al. 2010). Further, all flavonoids tested disrupted the interaction (Bailly et al. 2008), while gravacin (Rojas-Pierce et al. 2007) had no significant effect on the interaction (Kim et al. 2010) verifying the idea that TWD1 is probably not a target of it. The flavonol quercetin (IC50 ≈ 200 nM) was the most efficient and, surprisingly, was also active as glucoside (Bailly et al. 2008). Yeast IAA export assays in the presence of ATIs verified the BRET measurements on the transport levels. Mutant analysis indicated that the TWD1 FKBD is responsible for both interaction and drug regulation of ABCB1. This assumption was confirmed by specific NPA binding studies using whole yeast, highly pure FKBD, and plant microsomes prepared from TWD1 loss- and gain-of-function mutants (Bailly et al. 2008). Using gravitropism analysis, imaging of auxin fluxes upon gravistimulation, and measuring root auxin fluxes, this concept was further substantiated by the finding that twd1 and to lesser amount also abcb1 abcb19 (but not the single abcb loss-of-function alleles) were NPA insensitive (Bailly et al. 2008).
These data are in agreement with the current concept that the efflux complex consists of at least two proteins: a transporter and an NPA-binding regulatory subunit (Luschnig 2001; Morris 2000; Petrasek et al. 2003). ABCBs represent apparently integral membrane-embedded NBPs identified by Bernasconi et al. (1996) and Ruegger et al. (1997). Therefore, TWD1 might be the peripheral NBP (Cox and Muday 1994), which is in line with the recently proposed perpendicular orientation of the TWD1 C-terminus. However, this perception is also supported by the fact that the NBP has been suggested to be required for auxin efflux transporter positioning (Gil et al. 2001). Interestingly, a low-affinity NPA binding site has been associated with the transporter because its block results in transport inhibition, while the high-affinity site does not interfere directly with auxin transport (Michalke et al. 1992). In this respect, it will be of high interest to quantify NPA binding affinities for TWD1 and ABCBs.
On the other hand, these data suggest a novel mode of drug-mediated regulation of ABCB activities via an interacting FKBP: The TWD1 FKBD owns a receptor-like function and is therefore capable of integrating negative (ATI) inputs on ABCB1 (see Fig. 1). This concept is supported by in silico docking analysis of ATIs on the crystal structure of the FKBD, providing indication for an ATI binding pockets (Bailly and Geisler, unpublished). Interestingly, this pocket is overlapping with surfaces thought to dock to the ABCB1 NBD2 (Granzin et al. 2006), providing a mechanistic ratio for disruption of TWD1–ABCB interaction. Based on computational binding, NPA docks to ABCB pockets flanked by coupling helices and Q loops of NBD1 and NBD2 at the NBD–ICL interface (Kim et al. 2010). One could easily imagine that NPA blocks efficiently the main mechanistic of the transporter during transition of conformational changes between the NBDs and the ICLs.
Recent data supported that TWD1 does not bind flavonols, like quercetin, itself (Henrichs et al. 2012); therefore, their potential to disrupt TW1–ABCB interaction suggests that they bind to plant ABCBs. In agreement, flavonoids function as inhibitors of plant (Geisler et al. 2005; Terasaka et al. 2005) and mammalian ABCBs (Morris and Zhang 2006), most probably by mimicking ATP and competing for ABCB nucleotide-binding domains (Conseil et al. 1998). Different targets and binding domains clearly indicate distinct modes of actions for NPA and flavonols and question the simplified view that flavonols act as plant-endogenous NPA homologs.
The eligible question that arises still is why nature invented second NPA binding affinities on ABCBs besides TWD1. The simplest answer to this might be that there is apparently a need for ABCB regulation by ATIs in the absence of TWD1.
4 Regulation of Auxin Transporter Activity by Protein Phosphorylation
4.1 PIN Phosphorylation
Several studies indicate that reversible protein phosphorylation is an important regulatory mechanism for PAT. As mentioned above, endocytotic cycling represents a highly regulated mechanism for polar PIN locations that among others has been shown to be regulated by protein phosphorylation events (Friml et al. 2004; Michniewicz et al. 2007). Flowering plants do not contain orthologs of animal protein kinase A (PKA), cyclic GMP-dependent protein kinase (PKG), or protein kinase C (PKC). However, a family of so-called AGCVIII Ser/Thr protein kinases, named after their mammalian homologs, is thought to own similar function in growth factor signal transduction (Galvan-Ampudia and Offringa 2007).
The Ser/Thr protein kinase PINOID (PID), belonging—together with PID2, WAG1, and WAG2—to the AGC3 clade of AGCVIII kinases (Galvan-Ampudia and Offringa 2007), is an important regulator of this process and was shown to function as molecular switch of PIN locations (Friml et al. 2004; Kleine-Vehn et al. 2009; Michniewicz et al. 2007; Rakusova et al. 2011; Zhang et al. 2010). Loss-of-function alleles reveal a pinoid (pin1-like) plant phenotype (Benjamins et al. 2001), while plants overexpressing PID reveal defects in gravitropism and a loss of root meristem organization probably due to auxin depletion (Friml et al. 2004). In pid inflorescences, PIN1 has been shown to be shifted from the upper to the lower side, explaining similar phenotypes found for pin1 and pid, while in PID-OX plants, PIN2 and PIN4 were shown to be mistargeted (Friml et al. 2004). Moreover, polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response was shown to be as well controlled by PID (Rakusova et al. 2011).
Recently, PID, WAG1, and WAG2 were shown to phosphorylate PIN carriers at a conserved TPRXS(N/S) motif in the central hydrophilic loop, leading to PIN recruitment into the apical recycling pathway (Dhonukshe et al. 2010; Huang et al. 2010). Moreover, disruption of PID and its three closest homologues completely abolishes the formation of cotyledons (Cheng et al. 2008). These findings, together with the fact that WAG1 and WAG2 are apolar and plasma membrane-associated, suggest that AGC3 kinases act in the same or in a parallel regulatory pathway of PAT (Santner and Watson 2006).
The current model suggests that PID, together with the trimeric serine–threonine protein phosphatase 2A (PP2A), antagonistically determines the fate of PIN cargoes for trafficking to the appropriate membrane by (de)phosphorylating conserved motifs of the hydrophilic loop of PIN proteins (Dhonukshe et al. 2010; Ding et al. 2011; Huang et al. 2010; Kleine-Vehn et al. 2009) (see Fig. 4 and Chap. 5 of this series for more details). In summary, this suggests a posttranscriptional polarity loop via antagonistic action by PID/PP2A (Benjamins and Scheres 2008; see Fig. 3). However, it is an open question if altered PIN polarity is indeed directly caused by PIN phosphorylation or not simply the consequence of altered PIN activity. The latter has been suggested for D6 protein kinase (Zourelidou et al. 2009).
4.2 ABCB Phosphorylation
Several lines of clinical evidence suggest ABCBs as general targets for phosphorylation-dependent regulation in a so-called linker region. This linker region connects the N- and C-terminal NBDs of ABCBs and has been shown to regulate ABCB by multiple phosphorylation events catalyzed by PKC (Chambers et al. 1990; Conseil et al. 2001). Linker phosphorylation modifies ABCB transport and associated ATPase activity (Szabo et al. 1997). An accumulation of serine residues was identified to be phosphorylated by PKA and/or PKC (Chambers et al. 1990; Conseil et al. 2001; Orr et al. 1993) that regulate the drug transport properties (Szabo et al. 1997). Employing different phospho-proteomics approaches, plant ABCB proteins have recently (among other ABC transporters) shown to be phosphorylated (Benschop et al. 2007; de la Fuente van Bentem et al. 2006; Nuhse et al. 2004; Peck 2006).
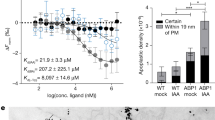
The first proof that also plant ABCBs are controlled by reversible protein phosphorylation came from the finding that the AGC4 kinase, PHOTROPIN1 (phot1), was shown to interact with both NBDs of ABCB19 but not with the NBD2 of ABCB1 (Christie et al. 2011). Interestingly, interaction was blocked by light irradiation as would be expected for a blue-light receptor kinase. In vitro phosphorylation experiments verified ABCB19 as phot1 kinase substrate, although phosphorylated domains and residues remain exclusive. Using co-expression in HeLa cells, auxin efflux activity of ABCB19 but not of B1 was specifically shown to be inhibited by phot1 co-expression, in a mode that is dependent on the phot1 kinase activity and that is accelerated by light irradiation (see Fig. 2b).
Model summarizing the analogous regulatory impact of AGC3 kinases, PID and phot1, on ABCB1 and ABCB19 activities. (a) In the absence of TWD1, PID has a positive (+) regulatory effect on ABCB1 activity, most probably by S634 linker phosphorylation. In its presence, PID phosphorylates an unknown residue (?) resulting in block of auxin efflux (−). Flavonols (Flav), such as quercetin, bind to PID resulting in PID inhibition and block of ABCB regulation by PID. (b) Phosphorylation of the NBDs of ABCB19 by phot1 marks inhibition of transport activity, suggesting an analogous mode of ABCB regulation via protein phosphorylation where TWD1 would function in recruiting individual AGC3 kinases. Please note that TWD1–phot1 interaction awaits confirmation but is supported by the finding that TWD1–ABCB19 interaction, shown to as well positively enhance B19 activity, was ameliorated in phot1 plants. Functional domains of TWD1 are in blue (FKBD), red (TPR), yellow (calmodulin-binding domain), and gray (in-plane membrane anchor); question marks label functionalities that need experimental approval
Interestingly, TWD1–ABCB19 interaction, shown as well to positively enhance B19 activity, was ameliorated by light irradiation but enhanced in phot1 plants, supporting the concept that phot1-catalyzed ABCB19 phosphorylation blocks TWD1–ABCB19 interaction. Alternatively, phot1 might simply compete for TWD1 docking surfaces at the ABCB19 NBD2.
Using co-immunoprecipitation and shotgun LC–MS/MS analysis, PID was identified as a valid partner in interaction with TWD1. PID interaction was verified by BRET analysis in planta and in vitro pulldowns. In vitro and yeast expression analyses indicated that PID specifically modulates ABCB1-mediated auxin efflux in an action that is dependent on its kinase activity and that is reverted by quercetin binding and thus inhibition of PID auto-phosphorylation. ABCB1/PID co-transfection in tobacco revealed that PID enhances ABCB1-mediated auxin efflux in the absence of TWD1, while PID had a negative impact on ABCB1 in yeast. As discussed above, the most likely explanation is that ScFKBP12 is able to functionally complement TWD1 in yeast as has been suggested for TWD1 modulation of ABCB1 (Bailly et al. 2008; Bouchard et al. 2006). Interestingly, triple ABCB1/PID/TWD1 co-transfection in tobacco revealed that PID blocks ABCB1-mediated auxin efflux in the presence of TWD1 (see Fig. 2a), suggesting that TWD1 might function as a recruiting factor for ABCB1 phosphorylation. The fact that ABCB1 phosphorylation in the presence of TWD1 has the opposite effect on ABCB1 transport capacity than TWD1–ABCB1 interaction per se argues for the idea that protein phosphorylation is not the primary mode of TWD1 activation. Obviously, both modes of ABCB1 regulation—directly via TWD1 interaction and PID phosphorylation—might also take place in parallel or in competition, resulting in fine-tuning of ABCB activity as reported for mammalian ABCBs. Alternatively, ABCB1 phosphorylation in the presence of TWD1 might disrupt TWD1–ABCB1 interaction, leading to ABCB1 inhibition (see below).
Phospho-proteomics analyses identified S634 as a key residue of the regulatory ABCB1 linker, which was verified by mutation analyses in yeast and tobacco. In the absence of TWD1, PID does phosphorylate S634, resulting in ABCB1 activation. On the other hand, negative ABCB1 regulation in the presence of TWD1 argues for a second, PID-specific ABCB1 phosphorylation site that does not essentially need to be part of the linker.
Currently, PID is seen as a positive regulator of NPA-sensitive PAT (Lee and Cho 2006). This is based on the correlation of the pid mutant shoot phenotype that—in analogy to the more drastic one of pin1 (Palme and Galweiler 1999)—can be widely phenocopied by NPA treatment (Wisniewska et al. 2006) and supported by the fact that pid shoots (Bennett et al. 1995) and roots (Sukumar et al. 2009) show reductions of acropetal and basipetal PAT, respectively. A current study, however, shows that PID phosphorylation of the ABCB1 linker might modulate not only ABCB1 activity but also NPA binding capacities (Henrichs et al. 2012). This implies that enhanced (reduced) NPA (quercetin) binding to PID gain-of-function microsomes might be a direct result of altered ABCB1 phosphorylation at S634 by PID. These findings, however, also suggest that the pinoid phenotype and repression of PID-OX defects by NPA are at least to a certain magnitude taken over by PIN-independent transport mechanisms, such as ABCBs. This is also supported by additive, drastic developmental defects of pin1 pid alleles (Furutani et al. 2007). NPA action might be therefore mediated by closely related AGC3 kinases, like PID2 or WAG1/WAG2, that have been shown to share the regulation of identical NPA-sensitive PAT pathways (Dhonukshe et al. 2010; Santner and Watson 2006).
Protein phosphatases for ABCB1 and B19 dephosphorylation have not yet been identified, but indirect evidence suggests that PP2A, RCN1, might be involved: rcn1 abcb1 abcb19 triple mutants exhibited strong embryonic and postembryonic auxin-related phenotypes (Mravec et al. 2008). Moreover, PP2C (At2g30020) was identified as putative TWD1 interactor (Henrichs et al. 2012).
In summary, these two analogous sets of data on ABCB19 and ABCB1 regulation by AGC kinases, phot1 (Christie et al. 2011) and PID, imply that AGC kinases, besides their function as a molecular switch of PIN polarity, have a direct impact on auxin efflux (ABCB) activity. Although phosphorylated residues in ABCB19 by phot1 have not yet been identified and a phot1–TWD1 interaction has not been proven (see Fig. 2), both modes of actions show common features: In the presence of TWD1, both phosphorylation events catalyzed by phot1 and PID lead to an inhibition of ABCB activity. In both cases, ABCB phosphorylation might result in a block of ABCB–TWD1 interaction, which would be a plausible ratio for a loss of functionality. However, this mode of action, although intriguing and worth testing, is in contrast to the current picture that ABCB–TWD1 interaction is of transient nature, which is obviously supported by the low TWD1/ABCB expression stoichiometry (Bailly et al. 2008, Wang et al. 2013). Moreover, this regulatory circuit would suggest a paradox situation in which TWD1 recruits its individual AGC kinase for ABCB phosphorylation, resulting in TWD1–ABCB separation (Fig. 2).
5 Interaction and Polarity Loops Are Interconnected
Auxin transport is thought to employ two main regulatory feedback loops, a transcriptional loop and a posttranscriptional PIN polarity loop (Benjamins and Scheres 2008). In the polarity loop, PIN polarity is regulated by its phosphorylation status provided by protein kinases and phosphatases, represented by PID and PP2A (see Sect. 4; Fig. 3b).
Interaction and polarity loops of ABCB and PIN-mediated auxin transport are interconnected. (a) In a novel interaction loop, TWD1 and PIN proteins positively contribute to ABCB-mediated auxin transport by protein–protein interaction. Polar PINs provide vectorial auxin streams, while TWD1 acts a positive modulator of nonpolar ABCB fluxes. ATIs, such as flavonols, disrupt ABCB1–TWD1 interaction probably by binding to ABCB1, resulting in transport inhibition. Arrows denote positive and bars negative regulation at the transport level. (b) In a previously suggested polarity loop (dashed line; modified from Benjamins and Scheres (2008)), PIN polarity is coordinated by the phosphorylation status of PINs controlled by PID/PP2A action. PID interferes with all major components of interaction and polarity loops in an action that is inhibited by ATI/quercetin binding. Note that the negative impact of PID on TWD1–ABCB1 interaction by ABCB1 phosphorylation has not yet been demonstrated
However, a whole series of recent data imply a third protein–protein interaction feedback loop of auxin transport (see Fig. 3a). Therein, distinct ABCB–PIN and ABCB–TWD1 pairings of a putative multi-protein auxin efflux complex are building the basis for a plastic control of auxin streams during PAT. This complex probably does not involve direct TWD1–PIN interaction, which is also supported by the finding that expression and locations of PIN1 and PIN2 are unchanged in twd1 (Bouchard et al. 2006; Wu et al. 2010).
Endogenous auxin transport inhibitors, that for flavonoids have been suggested and partially also verified, are also part of this regulatory network. The integrative, sometimes even confusing, modulatory effect of flavonoids on auxin transport might result from combinatory effect on ABCB activity and its interaction with TWD1 and additionally on PIN gene expression and cellular trafficking (Peer et al. 2001; Peer and Murphy 2006, 2007; Santelia et al. 2008). Very recent findings suggest that polarity and interaction loops are interconnected via the action of AGC kinases, such as PID (Fig. 3b): Besides decoding PIN polarity, PID also interacts with TWD1, thus negatively regulating ABCB1 activity (Henrichs et al. 2012). At present it is unclear if ABCB1 inhibition is of direct nature or causes indirectly by loss of interaction with TWD1.
Interestingly, flavonoids, such as quercetin, but not NPA are able to interfere with both interaction and polarity loops on different levels (1) direct inhibition of ABCB activity, (2) disrupting TWD1–ABCB interaction, and (3) inhibition of PID that itself has an indirect impact on ABCB1 phosphorylation/activity or PIN polarity. As such, these partially opposite effects reflect pretty well the complexity of flavonol action.
In summary, it appears that auxin controls besides its own biosynthesis and homeostasis also its own transport by interconnected regulatory feedback loops (Benjamins and Scheres 2008). Auxin, in interplay with ATIs, is able to compose the set of its own regulatory machinery according to the developmental stage of the cell and the environmental conditions of the plant. As a consequence, individual transporter and regulatory proteins interact, interfere, and regulate each other in order to allow a fine-tuning of an auxin distribution pattern.
6 Outlook
Upstream events regulating PID—and most probably also TWD1—activity reveal interesting links to known but widely unclear regulatory mechanisms of the auxin transport machinery: Since the 1970s, it is well known that gravity perception and PAT are tightly controlled by intracellular calcium levels (Dela Fuente and Leopold 1973; Toyota et al. 2008a). However, the order and relationship between both signaling pathways are unclear. However, recent work provided evidence that gravistimulation-induced calcium increases constitute an upstream event of PAT (Toyota et al. 2008b).
This is supported by the finding that PID is negatively and positively regulated by protein–protein interaction with calcium-binding proteins, TCH3 and PBP1, respectively. TCH3 is a calmodulin-related protein, and as a consequence, calmodulin inhibitors enhance PID activity (Sistrunk et al. 1994). In this respect, it might be worth recalling that TWD1 itself is calmodulin binding, although the in vivo relevance of this interaction is still unclear (Fig. 4) (Geisler et al. 2003; Kamphausen et al. 2002). Studies on the human homologue of TWD1, FKBP38, show that anti-apoptotic function of HsFKBP38 requires a priori activation by calmodulin activating the cis–trans peptidyl-prolyl isomerase (PPIase) activity of HsFKBP38 (Edlich et al. 2005, 2007). Taking the human FKBP38 as an example, a scenario becomes likely that conformational changes of TWD1 are induced by calcium-dependent binding of a calmodulin-like proteins, such as TCH3, that might affect directly ABCB activity or PID activity and thus indirectly ABCB auxin transport.
Overview of protein–protein interactions regulating PIN- and ABCB-mediated auxin efflux. As explained in detail in the text, reversible protein phosphorylation by AGC3 kinases, represented here by PID, have a dual effect on PIN polarity (4) and ABCB transport capacity (3). PID activity is calcium-dependently regulated inversely by the calcium-binding proteins pinoid-binding protein1 (PBP1) and the calmodulin-like TOUCH3 (TCH3) (2). Dephosphorylation of PIN and ABCB proteins by protein phosphatases, such as phosphatase PP2A (5), leads to a top-to-bottom switch of PIN proteins and reversal of PID-mediated ABCB regulation. Note that many of these regulatory processes are directly or indirectly controlled by changes of intracellular calcium concentration provided by plasma membrane calcium channels that itself are under control of auxin-related responses, such as gravitropism (1). Further, besides PID also TWD1 was shown to bind calmodulin with its C-terminal calmodulin-binding domain (6) although the functional relevance of this interaction is still unknown
Although several lines of evidence also indicate a function for WAG kinases during auxin transport, the regulatory impact of WAGs on the auxin transport machinery remains less clear. Enhanced NPA sensitivity, the fact that WAG kinases, like PID, are polar and plasma membrane-associated, and their enhanced expression in the root tips suggest that PID and WAG kinases act in the same or in a parallel pathway (Santner and Watson 2006), probably by regulation of NPA-binding proteins, like ABCB1, or interactors, like TWD1. Taking into account that members of the AGC3 clade show redundancy during development and toward their calcium-dependent regulation by TCH3 and PBP1 (Dhonukshe et al. 2010; Huang et al. 2010; Robert and Offringa 2008), the specific impact of individual AGC3 kinases in regulation of auxin transport needs to be explored. Therefore, the effect of WAG kinases on PIN protein polarity and ABCB activity needs to be investigated. WAG kinases are rapidly downregulated by light and show a more pronounced effect on root growth; they probably play a role during gravitropism or root development, responses where PID plays a limited role.
Finally, and connected to the above, there is a need to understand how nature was able to dually utilize and separate functionality of AGC kinases, such as PID, for regulation of ABCB activity and PIN polarity. PID coevolved relatively early during plant evolution together with PINs during the origin of land plants, while phot1 seemed to have appeared later during development of seed plants.
References
Bailly A, Sovero V, Geisler M (2006) The twisted dwarf's ABC: how immunophilins regulate auxin transport. Plant Signal Behav 1:277–280
Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M (2008) Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem 283:21817–21826
Bandyopadhyay A, Blakeslee JJ, Lee OR, Mravec J, Sauer M, Titapiwatanakun B, Makam SN, Bouchard R, Geisler M, Martinoia E, Friml J, Peer WA, Murphy AS (2007) Interactions of PIN and PGP auxin transport mechanisms. Biochem Soc Trans 35:137–141
Barbez E, Kubes M, Rolcik J, Beziat C, Pencik A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, Zazimalova E, Petrasek J, Geisler M, Friml J, Kleine-Vehn J (2012) A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 485:119–122
Barik S (2006) Immunophilins: for the love of proteins. Cell Mol Life Sci 63:2889–2900
Benjamins R, Scheres B (2008) Auxin: the looping star in plant development. Annu Rev Plant Biol 59:443–465
Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R (2001) The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128:4057–4067
Bennett S, Alvarez J, Bossinger G, Smyth D (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Development 128:4057–4067
Benschop JJ, Mohammed S, O'Flaherty M, Heck AJ, Slijper M, Menke FL (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6:1198–1214
Bernasconi P, Patel BC, Reagan JD, Subramanian MV (1996) The N-1-naphthylphthalamic acid-binding protein is an integral membrane protein. Plant Physiol 111:427–432
Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, Geisler M, Nagashima A, Sakai T, Martinoia E, Friml J, Peer WA, Murphy AS (2007) Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19:131–147
Blecher O, Erel N, Callebaut I, Aviezer K, Breiman A (1996) A novel plant peptidyl-prolyl-cis-trans-isomerase (PPIase): cDNA cloning, structural analysis, enzymatic activity and expression. Plant Mol Biol 32:493–504
Bosco CD, Dovzhenko A, Liu X, Woerner N, Rensch T, Eismann M, Eimer S, Hegermann J, Paponov IA, Ruperti B, Heberle-Bors E, Touraev A, Cohen JD, Palme K (2012) The endoplasmic reticulum localized PIN8 is a pollen specific auxin carrier involved in intracellular auxin homeostasis. Plant J 71:860–870
Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, Schulz B, Geisler M (2006) Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem 281:30603–30612
Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK (2001) Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol 126:524–535
Buer CS, Muday GK, Djordjevic MA (2007) Flavonoids are differentially taken up and transported long distances in Arabidopsis. Plant Physiol 145:478–490
Burgardt NI, Linnert M, Weiwad M, Geisler M, Lucke C (2012) NMR assignments of the FKBP-type PPIase domain of FKBP42 from Arabidopsis thaliana. Biomol NMR Assign 6:185–188
Chambers TC, McAvoy EM, Jacobs JW, Eilon G (1990) Protein kinase C phosphorylates P-glycoprotein in multidrug resistant human KB carcinoma cells. J Biol Chem 265:7679–7686
Chen J, Lalonde S, Obrdlik P, Noorani Vatani A, Parsa SA, Vilarino C, Revuelta JL, Frommer WB, Rhee SY (2012) Uncovering Arabidopsis membrane protein interactome enriched in transporters using mating-based split ubiquitin assays and classification models. Front Plant Sci 3:124
Cheng Y, Qin G, Dai X, Zhao Y (2008) NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci USA 105:21017–21022
Christie JM, Yang H, Richter GL, Sullivan S, Thomson CE, Lin J, Titapiwatanakun B, Ennis M, Kaiserli E, Lee OR, Adamec J, Peer WA, Murphy AS (2011) phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol 9:e1001076
Conseil G, Baubichon-Cortay H, Dayan G, Jault JM, Barron D, Di Pietro A (1998) Flavonoids: a class of modulators with bifunctional interactions at vicinal ATP- and steroid-binding sites on mouse P-glycoprotein. Proc Natl Acad Sci USA 95:9831–9836
Conseil G, Perez-Victoria JM, Jault JM, Gamarro F, Goffeau A, Hofmann J, Di Pietro A (2001) Protein kinase C effectors bind to multidrug ABC transporters and inhibit their activity. Biochemistry 40:2564–2571
Consortium AIM (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333:601–607
Cox DN, Muday GK (1994) NPA binding activity is peripheral to the plasma membrane and is associated with the cytoskeleton. Plant Cell 6:1941–1953
De Bodt S, Hollunder J, Nelissen H, Meulemeester N, Inze D (2012) CORNET 2.0: integrating plant coexpression, protein-protein interactions, regulatory interactions, gene associations and functional annotations. New Phytol 195:707–720
de la Fuente van Bentem S, Anrather D, Roitinger E, Djamei A, Hufnagl T, Barta A, Csaszar E, Dohnal I, Lecourieux D, Hirt H (2006) Phosphoproteomics reveals extensive in vivo phosphorylation of Arabidopsis proteins involved in RNA metabolism. Nucleic Acids Res 34:267–3278
Dela Fuente RK, Leopold AC (1973) A role for calcium in auxin transport. Plant Physiol 51:845–847
Dhonukshe P, Huang F, Galvan-Ampudia CS, Mahonen AP, Kleine-Vehn J, Xu J, Quint A, Prasad K, Friml J, Scheres B, Offringa R (2010) Plasma membrane-bound AGC3 kinases phosphorylate PIN auxin carriers at TPRXS(N/S) motifs to direct apical PIN recycling. Development 137:3245–3255
Ding Z, Galvan-Ampudia CS, Demarsy E, Langowski L, Kleine-Vehn J, Fan Y, Morita MT, Tasaka M, Fankhauser C, Offringa R, Friml J (2011) Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis. Nat Cell Biol 13:447–452
Ding Z, Wang B, Moreno I, Dupláková N, Simon S, Carraro N, Reemmer J, Pěnčík A, Chen X, Tejos R, Skůpa P, Pollmann S, Mravec J, Petrášek J, Zažímalová E, Honys D, Rolčík J, Murphy A, López AO, Geisler M, Friml J (2012) ER-localized auxin transporter PIN8 regulates auxin homoeostasis and male gametophyte development in Arabidopsis. Nat Commun 3:941. doi: 10.1038/ncomms.1941
Edlich F, Lucke C (2011) From cell death to viral replication: the diverse functions of the membrane-associated FKBP38. Curr Opin Pharmacol 11:1–6
Edlich F, Weiwad M, Erdmann F, Fanghanel J, Jarczowski F, Rahfeld JU, Fischer G (2005) Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J 24:2688–2699
Edlich F, Maestre-Martinez M, Jarczowski F, Weiwad M, Moutty MC, Malesevic M, Jahreis G, Fischer G, Lucke C (2007) A novel calmodulin-Ca2+ target recognition activates the Bcl-2 regulator FKBP38. J Biol Chem 282:36496–36504
Feraru E, Friml J (2008) PIN polar targeting. Plant Physiol 147:1553–1559
Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, Hooykaas PJ, Palme K, Offringa R (2004) A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306:862–865
Furutani M, Kajiwara T, Kato T, Treml BS, Stockum C, Torres-Ruiz RA, Tasaka M (2007) The gene MACCHI-BOU 4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and phototropism at the molecular level. Development 134:3849–3859
Galvan-Ampudia CS, Offringa R (2007) Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci 12:541–547
Geisler M, Bailly A (2007) Tête-à-tête: FKBPs function as key players in plant development. Trends Plant Sci 12:465–473
Geisler M, Murphy AS (2006) The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett 580:1094–1102
Geisler M, Kolukisaoglu HU, Bouchard R, Billion K, Berger J, Saal B, Frangne N, Koncz-Kalman Z, Koncz C, Dudler R, Blakeslee JJ, Murphy AS, Martinoia E, Schulz B (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol Biol Cell 14:4238–4249
Geisler M, Girin M, Brandt S, Vincenzetti V, Plaza S, Paris N, Kobae Y, Maeshima M, Billion K, Kolukisaoglu UH, Schulz B, Martinoia E (2004) Arabidopsis immunophilin-like TWD1 functionally interacts with vacuolar ABC transporters. Mol Biol Cell 15:3393–3405
Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, Ejendal KF, Smith AP, Baroux C, Grossniklaus U, Muller A, Hrycyna CA, Dudler R, Murphy AS, Martinoia E (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J 44:179–194
Geisler-Lee J, O'Toole N, Ammar R, Provart NJ, Millar AH, Geisler M (2007) A predicted interactome for Arabidopsis. Plant Physiol 145:317–329
Gil P, Dewey E, Friml J, Zhao Y, Snowden KC, Putterill J, Palme K, Estelle M, Chory J (2001) BIG: a calossin-like protein required for polar auxin transport in Arabidopsis. Genes Dev 15:1985–1997
Granzin J, Eckhoff A, Weiergraber OH (2006) Crystal structure of a multi-domain immunophilin from Arabidopsis thaliana: a paradigm for regulation of plant ABC transporters. J Mol Biol 364:799–809
Harrar Y, Bellec Y, Bellini C, Faure JD (2003) Hormonal control of cell proliferation requires PASTICCINO genes. Plant Physiol 132:1217–1227
Hemenway CS, Heitman J (1996) Immunosuppressant target protein FKBP12 is required for P-glycoprotein function in yeast. J Biol Chem 271:18527–18534
Henrichs S, Wang B, Fukao Y, Zhu J, Charrier L, Bailly A, Oehring SC, Linnert M, Weiwad M, Endler A, Nanni P, Pollmann S, Mancuso S, Schulz A, Geisler M (2012) Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J 31:2965–2980
Huang F, Zago MK, Abas L, van Marion A, Galvan-Ampudia CS, Offringa R (2010) Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22:1129–1142
Jacobs M, Rubery PH (1988) Naturally-occurring auxin transport regulators. Science 241:346–349
Jurgens G, Geldner N (2007) The high road and the low road: trafficking choices in plants. Cell 130:977–979
Kamphausen T, Fanghanel J, Neumann D, Schulz B, Rahfeld JU (2002) Characterization of Arabidopsis thaliana AtFKBP42 that is membrane-bound and interacts with Hsp90. Plant J 32:263–276
Kim JY, Henrichs S, Bailly A, Vincenzetti V, Sovero V, Mancuso S, Pollmann S, Kim D, Geisler M, Nam HG (2010) Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. J Biol Chem 285:23309–23317
Kleine-Vehn J, Friml J (2008) Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol 24:447–473
Kleine-Vehn J, Huang F, Naramoto S, Zhang J, Michniewicz M, Offringa R, Friml J (2009) PIN auxin efflux carrier polarity is regulated by PINOID kinase-mediated recruitment into GNOM-independent trafficking in Arabidopsis. Plant Cell 21:3839–3849
Lalonde S, Sero A, Pratelli R, Pilot G, Chen J, Sardi MI, Parsa SA, Kim DY, Acharya BR, Stein EV, Hu HC, Villiers F, Takeda K, Yang Y, Han YS, Schwacke R, Chiang W, Kato N, Loque D, Assmann SM, Kwak JM, Schroeder JI, Rhee SY, Frommer WB (2010) A membrane protein/signaling protein interaction network for Arabidopsis version AMPv2. Front Physiol 1:24
Lee SH, Cho HT (2006) PINOID positively regulates auxin efflux in Arabidopsis root hair cells and tobacco cells. Plant Cell 18:1604–1616
Lee K, Thorneycroft D, Achuthan P, Hermjakob H, Ideker T (2010) Mapping plant interactomes using literature curated and predicted protein-protein interaction data sets. Plant Cell 22:997–1005
Li JF, Bush J, Xiong Y, Li L, McCormack M (2011) Large-scale protein-protein interaction analysis in Arabidopsis mesophyll protoplasts by split firefly luciferase complementation. PLoS One 6:e27364
Lomax TL, Muday GK, Rubery PH (1995) Auxin transport. In: Davies PJ (ed) Plant hormones: physiology, biochemistry and molecular biology. Kluwer, Dordrecht, pp 509–530
Luschnig C (2001) Auxin transport: why plants like to think BIG. Curr Biol 11:R831–R833
Merks RM, Van de Peer Y, Inze D, Beemster GT (2007) Canalization without flux sensors: a traveling-wave hypothesis. Trends Plant Sci 12:384–390
Michalke W, Gerard FK, Art EG (1992) Phytotropin-binding sites and auxin transport in Cucurbita pepo: evidence for two recognition sites. Planta 187:254–260
Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130:1044–1056
Morel J, Claverol S, Mongrand S, Furt F, Fromentin J, Bessoule JJ, Blein JP, Simon-Plas F (2006) Proteomics of plant detergent-resistant membranes. Mol Cell Proteomics 5:1396–1411
Morris DA (2000) Transmembrane auxin carrier systems–dynamic regulators of polar auxin transport. Plant Growth Regul 32:161–172
Morris ME, Zhang S (2006) Flavonoid-drug interactions: effects of flavonoids on ABC transporters. Life Sci 78:2116–2130
Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J (2008) Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development 135:3345–3354
Mravec J, Skupa P, Bailly A, Hoyerova K, Krecek P, Bielach A, Petrasek J, Zhang J, Gaykova V, Stierhof YD, Dobrev PI, Schwarzerova K, Rolcik J, Seifertova D, Luschnig C, Benkova E, Zazimalova E, Geisler M, Friml J (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459:1136–1140
Murphy AS, Hoogner KR, Peer WA, Taiz L (2002) Identification, purification, and molecular cloning of N-1-naphthylphthalmic acid-binding plasma membrane-associated aminopeptidases from Arabidopsis. Plant Physiol 128:935–950
Noh B, Murphy AS, Spalding EP (2001) Multidrug resistance-like genes of Arabidopsis required for auxin transport and auxin-mediated development. Plant Cell 13:2441–2454
Nuhse TS, Stensballe A, Jensen ON, Peck SC (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16:2394–2405
Orr GA, Han EK, Browne PC, Nieves E, O'Connor BM, Yang CP, Horwitz SB (1993) Identification of the major phosphorylation domain of murine mdr1b P-glycoprotein. Analysis of the protein kinase A and protein kinase C phosphorylation sites. J Biol Chem 268:25054–25062
Palme K, Galweiler L (1999) PIN-pointing the molecular basis of auxin transport. Curr Opin Plant Biol 2:375–381
Peck SC (2006) Phosphoproteomics in Arabidopsis: moving from empirical to predictive science. J Exp Bot 57:1523–1527
Peer WA, Murphy AS (2006) Flavonoids as signal molecules targets of flavonoid action. In: Grotewold E (ed) The science of flavonoids. Springer, Berlin, pp 239–268
Peer WA, Murphy AS (2007) Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 12:556–563
Peer WA, Brown DE, Tague BW, Muday GK, Taiz L, Murphy AS (2001) Flavonoid accumulation patterns of transparent testa mutants of arabidopsis. Plant Physiol 126:536–548
Perez-Perez JM, Ponce MR, Micol JL (2004) The ULTRACURVATA2 gene of Arabidopsis encodes an FK506-binding protein involved in auxin and brassinosteroid signaling. Plant Physiol 134:101–117
Petrasek J, Cerna A, Schwarzerova K, Elckner M, Morris DA, Zazimalova E (2003) Do phytotropins inhibit auxin efflux by impairing vesicle traffic? Plant Physiol 131:254–263
Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, Dhonukshe P, Skupa P, Benkova E, Perry L, Krecek P, Lee OR, Fink GR, Geisler M, Murphy AS, Luschnig C, Zazimalova E, Friml J (2006) PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312:914–918
Rakusova H, Gallego-Bartolome J, Vanstraelen M, Robert HS, Alabadi D, Blazquez MA, Benkova E, Friml J (2011) Polarization of PIN3-dependent auxin transport for hypocotyl gravitropic response in Arabidopsis thaliana. Plant J 67:817–826
Robert HS, Friml J (2009) Auxin and other signals on the move in plants. Nat Chem Biol 5:325–332
Robert HS, Offringa R (2008) Regulation of auxin transport polarity by AGC kinases. Curr Opin Plant Biol 11:495–502
Rojas-Pierce M, Titapiwatanakun B, Sohn EJ, Fang F, Larive CK, Blakeslee J, Cheng Y, Cutler SR, Peer WA, Murphy AS, Raikhel NV (2007) Arabidopsis P-glycoprotein19 participates in the inhibition of gravitropism by gravacin. Chem Biol 14:1366–1376
Ruegger M, Dewey E, Hobbie L, Brown D, Bernasconi P, Turner J, Muday G, Estelle M (1997) Reduced naphthylphthalamic acid binding in the tir3 mutant of Arabidopsis is associated with a reduction in polar auxin transport and diverse morphological defects. Plant Cell 9:745–757
Sachs T (1969) Polarity and the induction of organized vascular tissues. Ann Bot 33:263–272
Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, Martinoia E (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 283:31218–31226
Santner AA, Watson JC (2006) The WAG1 and WAG2 protein kinases negatively regulate root waving in Arabidopsis. Plant J 45:752–764
Scheidt HA, Vogel A, Eckhoff A, Koenig BW, Huster D (2007) Solid-state NMR characterization of the putative membrane anchor of TWD1 from Arabidopsis thaliana. Eur Biophys J 36:393–404
Schlicht M, Strnad M, Scanlon MJ, Mancuso S, Hochholdinger F, Palme K, Volkmann D, Menzel D, Baluška F (2006) Auxin immunolocalization implicates vesicular neurotransmitter-like mode of polar auxin transport in root apices. Plant Signal Behav 1:122–133
Shirane M, Nakayama KI (2003) Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat Cell Biol 5:28–37
Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J (1994) Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. Plant Cell 6:1553–1565
Stoma S, Lucas M, Chopard J, Schaedel M, Traas J, Godin C (2008) Flux-based transport enhancement as a plausible unifying mechanism for auxin transport in meristem development. PLoS Comput Biol 4:e1000207
Sukumar P, Edwards KS, Rahman A, Delong A, Muday GK (2009) PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiol 150:722–735
Sussman MR, Gardner G (1980) Solubilization of the receptor for N-1-naphthylphthalamic acid. Plant Physiol 66:1074–1078
Szabo K, Bakos E, Welker E, Muller M, Goodfellow HR, Higgins CF, Varadi A, Sarkadi B (1997) Phosphorylation site mutations in the human multidrug transporter modulate its drug-stimulated ATPase activity. J Biol Chem 272:23165–23171
Taylor LP, Grotewold E (2005) Flavonoids as developmental regulators. Curr Opin Plant Biol 8:317–323
Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, Yazaki K (2005) PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell 17:2922–2939
Titapiwatanakun B, Murphy AS (2009) Post-transcriptional regulation of auxin transport proteins: cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J Exp Bot 60:1093–1107
Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, Sakai T, Friml J, Peer WA, Murphy AS (2009) ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J 57:27–44
Toyota M, Furuichi T, Tatsumi H, Sokabe M (2008a) Critical consideration on the relationship between auxin transport and calcium transients in gravity perception of Arabidopsis seedlings. Plant Signal Behav 3:521–524
Toyota M, Furuichi T, Tatsumi H, Sokabe M (2008b) Cytoplasmic calcium increases in response to changes in the gravity vector in hypocotyls and petioles of Arabidopsis seedlings. Plant Physiol 146:505–514
Vanneste S, Friml J (2009) Auxin: a trigger for change in plant development. Cell 136:1005–1016
Vieten A, Sauer M, Brewer PB, Friml J (2007) Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 12:160–168
Wang B, Bailly A, Zwiewka M, Henrichs S, Azzarello E, Mancuso S, Maeshima M, Friml J, Schulz A, Geisler M (2013) Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell (in press)
Weiergraber OH, Eckhoff A, Granzin J (2006) Crystal structure of a plant immunophilin domain involved in regulation of MDR-type ABC transporters. FEBS Lett 580:251–255
Wisniewska J, Xu J, Seifertova D, Brewer PB, Ruzicka K, Blilou I, Rouquie D, Benkova E, Scheres B, Friml J (2006) Polar PIN localization directs auxin flow in plants. Science 312:883
Wu G, Otegui MS, Spalding EP (2010) The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell 22:3295–3304
Zhang J, Nodzynski T, Pencik A, Rolcik J, Friml J (2010) PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc Natl Acad Sci USA 107:918–922
Zourelidou M, Muller I, Willige BC, Nill C, Jikumaru Y, Li H, Schwechheimer C (2009) The polarly localized D6 PROTEIN KINASE is required for efficient auxin transport in Arabidopsis thaliana. Development 136:627–636
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Geisler, M., Henrichs, S. (2013). Regulation of Polar Auxin Transport by Protein–Protein Interactions. In: Chen, R., Baluška, F. (eds) Polar Auxin Transport. Signaling and Communication in Plants, vol 17. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-35299-7_8
Download citation
DOI: https://doi.org/10.1007/978-3-642-35299-7_8
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-35298-0
Online ISBN: 978-3-642-35299-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)