Abstract
Circadian clocks drive the daily rhythms in our physiology and behaviour that adapt us to the 24-h solar and social worlds. Because they impinge upon every facet of metabolism, their acute or chronic disruption compromises performance (both physical and mental) and systemic health, respectively. Equally, the presence of such rhythms has significant implications for pharmacological dynamics and efficacy, because the fate of a drug and the state of its therapeutic target will vary as a function of time of day. Improved understanding of the cellular and molecular biology of circadian clocks therefore offers novel approaches for therapeutic development, for both clock-related and other conditions. At the cellular level, circadian clocks are pivoted around a transcriptional/post-translational delayed feedback loop (TTFL) in which the activation of Period and Cryptochrome genes is negatively regulated by their cognate protein products. Synchrony between these, literally countless, cellular clocks across the organism is maintained by the principal circadian pacemaker, the suprachiasmatic nucleus (SCN) of the hypothalamus. Notwithstanding the success of the TTFL model, a diverse range of experimental studies has shown that it is insufficient to account for all properties of cellular pacemaking. Most strikingly, circadian cycles of metabolic status can continue in human red blood cells, devoid of nuclei and thus incompetent to sustain a TTFL. Recent interest has therefore focused on the role of oscillatory cytosolic mechanisms as partners to the TTFL. In particular, cAMP- and Ca2+-dependent signalling are important components of the clock, whilst timekeeping activity is also sensitive to a series of highly conserved kinases and phosphatases. This has led to the view that the ‘proto-clock’ may have been a cytosolic, metabolic oscillation onto which evolution has bolted TTFLs to provide robustness and amplify circadian outputs in the form of rhythmic gene expression. This evolutionary ascent of the clock has culminated in the SCN, a true pacemaker to the innumerable clock cells distributed across the body. On the basis of findings from our own and other laboratories, we propose a model of the SCN pacemaker that synthesises the themes of TTFLs, intracellular signalling, metabolic flux and interneuronal coupling that can account for its unique circadian properties and pre-eminence.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Intracellular
- Circadian rhythms
- Signal transduction
- Metabolic regulation
- SCN
- Post-translational
- Cytoscillator
1 Circadian Rhythms in Health and Disease, In Vivo and In Vitro
Circadian rhythms are biological oscillations with periods of approximately 1 day. They are manifest in the temporal organisation of behavioural, physiological, cellular and neuronal processes—influencing phenomena as diverse as sleep/wake cycles, glucose homeostasis, innate immunity and cell division. Because this endogenous timekeeping interacts with myriad biological systems, circadian disruption has significant impacts upon human health and the diseased state; e.g. whilst acute clock disruption can result in the short-term side effects known as jet lag, long-term shift workers (~15 % workforce in developed nations) suffer from chronic circadian dysregulation associated with an increased susceptibility to cardiovascular disease, type II diabetes and various cancers (Reddy and O’Neill 2010). In addition, many identified ‘clock genes’ could equally well be described as tumour suppressors, had they been first studied by oncologists, since their genetic lesion can lead to mis-regulation of both the cell and circadian cycle (Reddy et al. 2005). Put simply, because bodily processes change dramatically and predictably between day and night, there exists clear translational potential in elucidating the mechanistic basis for circadian timekeeping from the perspectives of novel therapeutic targets and therapeutic efficacy.
1.1 Why Be Rhythmic?
The capacity to anticipate temporal environmental cycles, brought about by the Earth’s daily rotation, is thought to have conferred a constant selective pressure over evolutionary timescales such that, essentially, all eukaryotes and many prokaryotes exhibit intrinsic circadian timekeeping (Roenneberg and Merrow 2002). Whether or not circadian rhythms arose divergently, or several times, and converged across the different kingdoms of life is not presently clear. Certainly, however, there appear to be advantages associated with the temporal organisation of physiology and metabolism, as evidenced by the yeast metabolic oscillation which separates in time biochemically incompatible catabolic and anabolic phases (Robertson et al. 2008). A well-known mammalian circadian equivalent is the clock-driven up-regulation of gluconeogenic transcripts in hepatocytes in anticipation of the nightly fast, regardless of feeding history (Akhtar et al. 2002). Whilst modern humans attempt to populate a 24-h society, our late Pleistocene genome still encodes the clock of a diurnal hunter-gatherer, resulting in hyperglycaemia and increased risk of the metabolic syndrome for those that indulge in midnight feasting. Even fruit flies maintained for 1,000 generations in constant darkness continued to be rhythmic demonstrating how deeply circadian rhythms are hardwired into cellular metabolism. Within this vein, the observation that populations of blind cavefish (Phreatichthys andruzzii) which have existed in constant darkness for ~2 million years have extremely long periods [nearly 2 days (Cavallari et al. 2011)] suggests that whilst rhythms are not readily dispensable, they can adapt under appropriate selection.
1.2 Pharmaceutically Relevant Circadian Principles
In mammals, daily timekeeping is now well established as a cell-intrinsic phenomenon, being observed in cultured cells and tissues over many days, in vitro. In spite of this, the language used to describe biological clocks was developed decades ago, when behavioural rhythms in experimental organisms were the most commonly studied output. As such, several criteria need to be defined with reference to drug action. First, a circadian rhythm is one exhibiting a period of approximately 24 h under constant conditions, i.e. in the absence of external time cues. Each point on the cycle can be assigned a unique phase, commonly expressed as hours of circadian time. Recent widespread application of genetically encoded real-time reporters has identified a small number of compounds that significantly and dose-dependently shift the cycle to a new phase and/or shorten or lengthen free-running period in cultured tissues and cells. Of these, few have been tested for their effect upon behavioural rhythms in mice, in vivo, but it is interesting to note that some of the earliest studies upon circadian rhythms were investigations into the period increase elicited by simple inorganic compounds, e.g. lithium salts and heavy water (Engelmann et al. 1976; Pittendrigh et al. 1973) (Fig. 1).
Schematic of how drug treatment may affect circadian timekeeping. Using an appropriate reporter, drug treatment (blue, green) can elicit differential effects upon the properties of circadian rhythms (period, phase, amplitude) that persist in organisms and cultured cells (red, untreated). An appropriate reporter might include behavioural activity, metabolite concentration, gene expression, bioluminescence, fluorescence, etc.
The biological clock is not a disembodied timer, but responds appropriately to its environment, and thereby allows organisms to cope with the varying day length and light quality associated with time of day, seasons and the weather. Relevant external cues entrain the phase of the oscillation, so that in nature the internal cycle is subtly reset each day (e.g. by dawn and dusk illumination, time of feeding, temperature cycles, etc.) in order to resonate with external solar cycles. For humans, light is the strongest entraining stimulus (or Zeitgeber), with intrinsic circadian phase being able to advance or delay by up to 60 min per day when light is experienced during late or early night, respectively (Skene and Arendt 2006). By exposing test subjects or model organisms to light pulses around the circadian cycle, a phase response curve may be described (Johnson 1999). This is relevant because similar phase-dependent shifts may be evoked in cultured tissues (Fig. 1), and animal behavioural rhythms, by pulsed drug application at particular circadian phases. Such experiments provide insight into the mechanisms that facilitate phase shifts, in vivo, as well as offering the potential for coordinated pharmacological manipulation of human rhythms in order to reduce the adverse long-term health effects that result from circadian misalignment with the external world. When organisms are under entrainment such as light/dark cycles, external time is reported with respect to the Zeitgeber time, ZT, such that ZT0 = dawn, ZT12 = dusk. Under constant conditions, circadian time, CT, is used and refers to endogenous timekeeping, i.e. CT0 corresponds to the phase that diurnal animals become active, whereas CT12 represents onset of activity in nocturnal species. These terms remain in use, although recently alternative models have been proposed (Roenneberg 2010).
An additional feature common to all circadian rhythms is that they are temperature compensated (Q10 ~ 1), the adaptability of which is intuitive since a clock that ran faster on warm days would be of little utility but unusual since most biological and chemical reactions double in rate for every 10 °C temperature increase. In other contexts, such compensation can result as an emergent network property or intramolecularly but generally involves mutually antagonistic processes that are each temperature dependent (Ruoff et al. 2007). Pharmacological manipulation of temperature compensation has been reported (Dibner et al. 2009), but it is the canonical clock property that seems least well understood in terms of mechanism.
2 Chronopharmacology
Over the last half century, models for the basis of rhythmicity have cycled from the biochemical to the electrophysiological, to the genetic and back (Edmunds 1983; King and Takahashi 2000; Njus et al. 1976). What is uncontested is that to produce any oscillation, delayed negative feedback is required. For its amplitude not to damp out, some positive feedforward is also needed (Lenz and Sogaard-Andersen 2011). Regardless of timekeeping mechanism, therefore, and whether the application is medicinal or scientific, the action of any drug upon a biological target could be considered as an interaction between its pharmacology and biological timing, since the cell exists in a state of cyclical flux that affects many aspects of gene expression, macromolecular turnover and metabolism. ‘Chronopharmacology’ might be considered to consist of the following three separable, but related factors [which are discussed in greater detail in Ortiz-Tudela et al. (2013), Antoch and Kondratov (2013), and Musiek and Fitzgerald (2013)].
2.1 Chronopharmacokinetics
Many xenobiotic uptake, detoxification and clearance pathways are circadian regulated, meaning that the absorption, distribution, metabolism and elimination of drugs and their secondary metabolites, as well as accompanying side effects, may be affected by the circadian phase of administration. For example, evening dosing resulted in the lowest observed toxicities for NSAIDs in osteoarthritic patients (Levi and Schibler 2007).
2.2 Chronopharmacodynamics
If the concentration or activity of a drug target is circadian regulated (e.g. enzyme activities can be modulated at the level of transcription/translation, post-translational modification or spatial localisation, or secretion if extracellular), then this can be expected to have concomitant effects upon drug efficacy. For example, ‘statins’ have been known for years to be most efficacious when administered during subjective night (Muck et al. 2000), since the pharmacodynamics vary over the circadian cycle.
2.3 Chronoactivity
A relatively small number of drugs have been shown to affect cellular rhythms, in vitro, presumably by directly or indirectly modulating the activity of cellular components that play a role in timekeeping or pathways of entrainment. Whilst being useful as research tools, understanding the circadian action of such drugs in vivo is particularly complex, since tissues may respond differently and effects upon intrinsic timekeeping might be undesirable. For example, many aerosolised asthma medications contain glucocorticoids, but glucocorticoids have also been shown to effect phase resetting in several tissues (Reddy et al. 2007). Thus, whilst their use may be life-saving in the short term, it is conceivable that some adverse side effects associated with their long-term use may be related to internal desynchronisation. In contrast, several psychiatric conditions are associated with disordered sleep/wake cycles, i.e. poorly organised behavioural rhythms. In the case of schizophrenics, a commonly prescribed mood stabiliser is lithium salts. Therapeutic doses (1–2 mM serum concentration) are around the level where a lengthening of free-running period may be observed in experimental organisms, and it has been proposed by several that the action of lithium on the biological clock may contribute to its positive treatment outcomes (Schulz and Steimer 2009).
2.4 Clinical Relevance
Clearly, in light of the massive undertaking represented by a modern clinical trial, it would be prohibitively expensive to investigate systematically all chronopharmacological aspects of a new or existing drug. Indeed, many initial observations were made anecdotally (Muck et al. 2000). The potential medical benefits offered by chronopharmacology, such as increased efficacy or reduced side effects through timing of treatment, are sufficient, however, to ensure that such factors will not be neglected indefinitely (Minami et al. 2009). The most cost-effective solution will surely be to reduce the complexity of the problem by approaching it in the light of existing knowledge and testing predictions in cellular and animal models first, because of the profound conservation of circadian principles between cells, tissues and species. As such, in the rest of this chapter, the state of existing knowledge about timekeeping mechanisms will be discussed. Particular emphasis will be placed upon various non-transcriptional mechanisms, since many of these are thought essential for circadian timekeeping and, more importantly, are likely to be more amenable to pharmacological manipulation than transcription.
3 Models and Mechanisms of Circadian Timekeeping
3.1 The SCN: First Amongst Equals
The suprachiasmatic nucleus (SCN) of the hypothalamus was long thought the centre of mammalian timekeeping, since its surgical ablation in rodents abolishes circadian rhythms in behaviour, body temperature and the secretion of endocrine factors such as melatonin and cortisol (Welsh et al. 2010). Moreover, SCN-ablated rodents receiving a surgical graft of foetal SCN tissue take on the circadian behavioural phenotype of the donor strain (King et al. 2003; Ralph et al. 1990). Recently, however, it has become apparent that circadian rhythms are a property inherent to most, if not all, mammalian cells—being observed to persist throughout the body in vivo and in isolated tissues/cells for many days, in vitro (Welsh et al. 2004; Yoo et al. 2004). 10–20 % of mammalian genes are expressed rhythmically in one or more tissues (Reddy 2013), although specific ‘clock-controlled genes’ (CCGs) vary between tissues, appropriately to organ function (Deery et al. 2009; Doherty and Kay 2010; Reddy et al. 2006). The observation that some genes are expressed rhythmically has enabled the widespread application of real-time bioluminescent reporters for cellular rhythms, whereby firefly luciferase fusions with relevant genomic sequences enable non-invasive, long-term recordings of rhythmic bioluminescence to be made (Yamaguchi et al. 2000). Using reporters such as the PERIOD2::LUCIFERASE (PER2::LUC) knock-in mouse has enabled single-cell time-lapse imaging of molecular circadian rhythms in culture. These revealed that the accuracy and robustness of timekeeping in dissociated SCN neurons, and cultured fibroblasts, are poor compared with rhythms from intact SCN slices or whole animals [cycle-to-cycle period variation of ~2 %, 3 % and 9 % in mice, slices and neurons, respectively (Herzog et al. 2004)]. Thus, whilst circadian rhythms are a cellular phenomenon, clearly there are emergent network properties at higher levels of biological scale that make the whole greater than the sum of its parts.
Our view of the SCN has therefore evolved to become that of the primary locus for coordinating rhythms generated by innumerable subordinate cellular clocks distributed throughout the rest of the body. Comprising around 10–20,000 neurons, the SCN employs both humoral factors and axonal projections to other brain regions to maintain stable phase relationships between peripheral tissues (Welsh et al. 2010). Sited above the optic chiasm, a proportion of SCN neurons have excitatory glutamatergic innervations from the retinohypothalamic tract, receiving photic cues from both image- and non-image-forming photoreceptor cells, as well as non-photic signals from the brainstem (5HT, NPY) (Brown and Piggins 2007; Leak et al. 1999). Concomitant with biological function, therefore, the SCN is a heterogeneous assemblage of cells that integrates multiple inputs to maintain its phase of oscillation and in turn populates a diversity of output pathways to convey temporal cues appropriate to each target site (see also Slat et al. 2013). This complex cellular heterogeneity and circuit structure enables additional systems-level functionality, such as encoding of day length, to emerge (VanderLeest et al. 2007).
3.2 Level of Abstraction
To understand any biological system, it is helpful to reduce its complexity to the simplest level where the phenotype of interest may be observed. In circadian rhythms, traits such as free-running period of behavioural activity (under constant conditions), or the relative phases of gene expression, are used as proxies for timekeeping. This can cause problems, however, since animals which are behaviourally arrhythmic cannot be assumed to be deficient in cellular timekeeping, i.e. mice with no legs are also arrhythmic when assayed with running wheels. Similarly, circadian rhythms in mammals should not really be studied at levels of scale below cellular until and unless the oscillation can be reconstituted biochemically in vitro (as has been demonstrated for the cyanobacterial oscillator, where a mixture of three Kai proteins plus ATP exhibit circadian cycles of auto-phosphorylation).
Since the correct level of biological abstraction at which to study circadian timekeeping is probably the cell (Noble 2008), it may be puzzling to scientists outside the circadian field that mechanisms of timekeeping within the SCN remain the subject of such intense scrutiny. Other than the momentum that gathers around any successful experimental model, this can be explained in several ways. First, the SCN is bona fide master clock—without it experimental rodents become behaviourally arrhythmic, and whilst timekeeping in peripheral tissues continues, they become desynchronised one from another and free-run with their own intrinsic period (Tahara et al. 2012). Therefore, we need to understand the SCN in order to place timekeeping within a physiological context. Second, unlike many neuronal cultures, organotypic SCN slices remain viable for many months in vitro and maintain many of their in vivo circadian properties, e.g. robustness, accuracy and interneuronal coupling. In addition, the SCN is amenable to media changes without perturbing the ongoing oscillation, and its circadian period is reflective of its genetic background. Finally, although the SCN is a highly specialised timekeeping organ, it appears to use several timekeeping mechanisms which are general to mammalian cells but with higher amplitude and are therefore more readily detectable.
3.3 The State of the Art: Circadian Timing by Transcriptional Feedback in Mammals
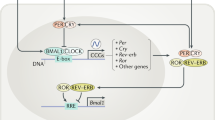
Based on both forward and reverse genetics, recent models of cellular timekeeping have focused on transcriptional/translational feedback mechanisms whereby positive activators (e.g. BMAL1 & CLOCK) bind to commonly occurring regulatory promoter elements (e.g. E-boxes) of many circadian-regulated genes, including those that encode transcriptional ‘clock gene’ repressors, e.g. PERIOD1/2 (PER1/2) and CRYPTOCHROME1/2 (CRY1/2), facilitating transcriptional activation around anticipated dawn (CT0). The repressor proteins are processed post-translationally, eventually accumulating to form complexes later in the day, prior to nuclear entry around anticipated dusk (CT12). At night, these repressive complexes inhibit CLOCK/BMAL1-driven transcription in many CCG promoters, including their own, and are then progressively degraded. This relaxes the transcriptional repression before dawn, licensing the cycle to begin anew (Reppert and Weaver 2002). As more clock gene transcription factors and their co-complexes have been identified, sophisticated models of cycling transcriptional activation/repression with concomitant chromatin remodelling/histone modification have been developed and account for many experimental observations (Ukai and Ueda 2010). Indeed, the circadian system has been a successful means of giving new life to well-established knowledge of transcriptional mechanisms, repackaging everyday factors into a clock context. Significantly, some clock genes, e.g. Period1/2, are immediate-early transcription factors, whose promoters also contain functional cAMP/Ca2+ response elements (CREs). In the SCN, in vivo and in vitro, appropriate activation of cAMP/Ca2+ signalling by extracellular (EC) stimuli induces Period gene expression and thereby facilitates clock resetting at night (entrainment) (Obrietan et al. 1999; Tischkau et al. 2003). These ideas and findings are reviewed elsewhere within this book (Buhr and Takahashi 2013; Sahar and Sassone-Corsi 2013), but in summary, the overarching hypothesis that has emerged to account for cellular timekeeping over the last two decades is one that posits cycling ‘clock gene’ transcription at its mechanistic core, with ancillary roles for post-translational mechanisms.
3.4 The Plot Thickens
Several recent findings have challenged whether cycling transcription is sufficient or even necessary to account for cellular timekeeping:
3.4.1 Transcriptional/Translational Feedback Loops Are Common
Transcriptional/translational feedback is a very common motif in cell biology (Kholodenko 2010) and could be described as the way a cell achieves proteostasis, i.e. a sufficient complement of protein activity to meet its requirements. In a signalling context, for example, a recurrent pattern is the rapid degradation of unstable inhibitors, a requirement for signal transmission, followed by their transcriptional up-regulation; this facilitates signal termination and a return to baseline (Legewie et al. 2008). For such oscillatory gene expression feedback loops, the time course is generally much shorter than 24 h, e.g. ERK signalling (2–3 h) (Kholodenko 2010) and NF-κB pathway (3–4 h) (Nelson et al. 2004), whilst the developmental segmentation clock has a period of 2–6 h (Jiang et al. 2000), reflecting a summation of the individual steps of gene expression (transcriptional activation/chromatin remodelling ⇒ elongation/splicing/5′-capping ⇒ termination/polyadenylation/nuclear export ⇒ translation ⇒ transport/translocation). Therefore, without positing a major contribution from post-translational processes, it is unclear why any transcriptional/translational cycle followed by cellular ‘clock proteins’ should take 24 h—it could all be done much more quickly.
3.4.2 Mismatches Between Transcriptome, Proteome and Protein Activity
With a couple of important exceptions, proteins mediate every cellular process of consequence. An uncontested biological principle is that protein sequences encoded by DNA are transmitted through messenger RNA intermediates, leading to the common inference that cellular mRNA transcript levels correlate with the levels of protein that they encode. Recently, however, understanding of post-transcriptional regulation has advanced to the point that this cannot be assumed to be the case. Indeed, there are now numerous clock-relevant examples whereby protein activity is regulated post-transcriptionally, e.g. via interfering microRNA-mediated mRNA silencing (Cheng et al. 2007), alternative splicing (McGlincy et al. 2012), transcript-specific translational rate (Kim et al. 2007, 2010), global translational rate (Cao et al. 2011) or post-translationally through phosphorylation-directed, ubiquitin-mediated, proteasomal degradation (Eide et al. 2005; Reischl et al. 2007).
From a global cellular perspective, the evidence supporting a pre-eminent role for post-transcriptional regulation is compelling. Using microarray-based techniques numerous groups have reported that ~10 % of total mRNA transcripts across a range of tissues are circadian regulated. More recently, the cytosolic soluble proteome of murine liver and SCN was investigated across circadian time and showed that 10–20 % of proteins vary significantly over the circadian cycle. Strikingly though, no obligatory correlation was observed between a gene cycling at the mRNA vs. protein level (Deery et al. 2009; Reddy et al. 2006), i.e. rhythmic transcripts can encode proteins whose level is constant, rhythmic protein levels can be observed from transcripts of constant level and so on (Robles and Mann 2013).
The liver study also identified a number of proteins that were subject to rhythmic post-translational modification, e.g. peroxiredoxin (PRX) 6 exhibited a modification rhythm in anti-phase to protein and transcript levels. Such observations are critical since many protein activities are ultimately regulated by a cascade of covalent modification, and therefore, for genes implicated in clock mechanism, rhythmic transcript levels cannot be assumed to be of functional relevance since it is ultimately the spatio-temporal dynamics of protein activity that mediate biological responses.
3.4.3 Stochastic Effects/Gene Dosage
At the level of a single cell, transcription from a given locus is inherently noisy due to a combination of there only being 2 (or 1 on the X chromosome) copies/cell, the poor efficiency of successful transcriptional initiation and the frequency of RNA polymerase stalling (Blake et al. 2003; Wu and Snyder 2008). Such burst kinetics were recently shown directly for transcription from the Bmal1 promoter (Suter et al. 2011). These stochastic effects might be expected to lead to highly variable cycle-to-cycle variation in period length rather than the ~10 % observed in isolated cells, in vitro. Moreover, dividing cells in G2 and ~40 % hepatocytes are polyploid (≥4N chromosomes) (Gentric et al. 2012). Clearly, this would be expected to lead to gene dosage effects upon circadian period if the timing mechanism was mostly reliant on the timing of transcription. This has not, however, been observed calling into question a direct dependence of periodicity upon gene expression.
3.4.4 Lessons from Gene Over-Expression and Knockout
Over-expression or knockout of most so-called clock genes has negligible effects on the behavioural period in mice and cultured SCN (longer/shorter by <10 %) (Hastings et al. 2008), leading to the common interpretation that there is substantial redundancy between them (Welsh et al. 2010). In some instances (Bmal1 −/−, Per2 over-expression, Cry1 −/− /2 −/−), mice are behaviourally arrhythmic (Bunger et al. 2000; Chen et al. 2009; Vitaterna et al. 1999), but this does not demonstrate that cellular rhythms have also been abolished. Indeed, where studied (Bmal1 −/−, Cryptochrome1 −/− /2 −/−), rhythms of circadian bioluminescence have been observed to persist in organotypic PER2::LUC SCN slices from these animals (Ko et al. 2010; Maywood et al. 2011). This robustness to genetic lesion implies that, at least in the SCN, the circadian circuitry is competent to maintain rhythmic PER2::LUC expression via other promoter elements and/or post-transcriptional regulation, even when rhythmic E-box activation is absent. Clearly, however, some basal activity of certain general transcription factors appears to be necessary for ‘normal’ cellular rhythms, but whether or not their rhythmic abundance is a prerequisite for timekeeping remains to be seen. These mammalian data have parallels with experiments performed in the fungus, Neurospora crassa, where the absolute necessity of identified ‘clock genes’ for cellular timekeeping was previously called into question (Lakin-Thomas 2006; Merrow et al. 1999; Granshaw et al. 2003).
3.4.5 Other Circadian Mutants: A Focus on Enzymes
It is notable that the circadian mutant mice with strongest period phenotypes carry dominant, apparently anti-morphic, mutations in genes that encode enzymes with roles in post-translational modification. For example, the Clock −/− mouse has a subtly shorter circadian period than wild type, whereas a mutation resulting in truncation of exon 19 that abolishes CLOCK’s acetyltransferase activity results in a much longer period (~28 h in homozygotes) both in behavioural activity and cultured SCN (Debruyne et al. 2006; Vitaterna et al. 1994). Similarly, mice with homozygous deletions of casein kinase 1ε (CK1ε) have slightly longer circadian period (~0.5 h), whereas mice homozygous for the Tau (R182C) mutation exhibit a very short circadian period (20–21 h) (Meng et al. 2008). We infer from this that timekeeping competence is more susceptible to genetic perturbation of enzyme activity than direct disruption of identified transcriptional components.
3.4.6 Circadian Reporters: What Do They Report and Is It Enough?
Bioluminescent reporters for ‘core’ clock gene activity have been indispensable tools for delineating the complex, semi-redundant circuitry that facilitates temporal coordination of physiology. In cases where genetic/pharmacological manipulation has resulted in the apparent loss of bioluminescence rhythms (arrhythmicity), however, absence of evidence is not evidence of absence. Indeed, one must consider whether the reporter, or the transcriptional circuit, and not the cellular clock per se has been affected. For example, the discrepancy in findings (arrhythmic vs. no effect) between different groups that have over-expressed the transcriptional repressor CRY1 in mammalian cell lines seems to be largely attributable to the length and nature of the promoter sequence that was used (Chen et al. 2009; Fan et al. 2007; Ueda et al. 2005). The emergent consensus is that cycling levels of most ‘clock proteins’ (e.g. CRY, BMAL1) are not required for timekeeping, but cycling PER is essential (Lee et al. 2011). This interpretation would be more compelling, however, were it supported by data using a clock reporter outside the genetic circuitry within which PER normally operates. In this context, the development of real-time post-translational reporters, not dependent on nascent gene expression, is highly desirable.
3.4.7 Clocks Continue Despite the Inhibition of Transcription
In all recent cellular clock models, the rate and timing of transcription constitute a key state variable. It was surprising, therefore, to learn that cellular rhythms in NIH3T3 fibroblasts are extremely robust to global inhibition of cellular mRNA production (using RNA polymerase II inhibitors, α-amanitin and actinomycin D). In this elegant study, >70 % of mRNA production was abolished during 3 days following drug treatment, and yet circadian period was shortened by <10 %; furthermore, the shortening was attenuated at lower temperatures, suggesting a hitherto unsuspected aspect of temperature compensation (Dibner et al. 2009). Similarly, using the PER2::LUC translational reporter, we observed that organotypic SCN slices exhibited at least one additional peak of bioluminescence during chronic treatment with α-amanitin oleate. At these concentrations (10–100 nM), we observed >70 % reduction in total 3H-uridine incorporation in culture (Fig. 2a–c).
Circadian rhythms persist in culture when transcription is inhibited or absent. (a, b) Representative and expanded plot showing rhythms in PER2::LUC bioluminescence persist in organotypic SCN slices during chronic treatment with α-amanitin oleate (AM, n = 4); arrow denotes start of drug treatment; (c) dose response for effect of AM upon nascent RNA synthesis using 3H-uridine incorporation; (d) representative time course showing rhythms of PRX over-oxidation persist in isolated human erythrocytes (from O’Neill and Reddy 2011)
The above data are similar to recent findings in the marine alga Ostreococcus tauri (O’Neill et al. 2011). When cultures expressing either a transcriptional or translational clock gene::luciferase fusion were incubated with saturating concentrations of cordycepin (an inhibitor of total RNA synthesis), an additional cycle of correctly phased gene expression was observed only in the translational reporter cell line: not the transcriptional reporter (O’Neill et al. 2011). Taken together, these data might suggest that, provided the cell has sufficient mRNA in existence, non-transcriptional mechanisms are competent to sustain an additional cycle of clock-regulated protein synthesis. Indeed, in Ostreococcus, all transcriptional contributions to timekeeping seem to be restricted to subjective morning (CT0–8), since the reversible application of cordycepin to cultures outside this window has no effect on circadian phase following drug wash-off.
3.4.8 Clocks Continue Despite the Inhibition of Translation
In at least three unrelated experimental organisms, chemical ‘wedge’ experiments have been performed using the ubiquitous ribosomal inhibitor cycloheximide. The null hypothesis in such time courses posits that translation has no contribution to timekeeping at any point during the circadian cycle; any systematic deviations from the null hypothesis imply that it does. To perform these laborious experiments, translation is inhibited for increasing durations, beginning at different phases throughout the circadian cycle. In all three experimental models (mouse bioluminescent SCN slices, Bulla gouldiana ocular electrophysiology, Ostreococcus culture bioluminescence), approximately two-thirds of the circadian cycle (≥16 h) was insensitive to translational inhibition, again implying that the majority of timekeeping function is not reliant on nascent gene expression (O’Neill et al. 2011; Khalsa et al. 1996; Yamaguchi et al. 2003).
3.4.9 Post-Translational Oscillations in Non-mammalian Systems
Extending these observations, there now exist several paradigms for cellular timekeeping in the complete absence of nascent gene expression. In O. tauri, it was recently shown that PRX post-translational rhythms persist for several cycles both in constant darkness (when transcription completely shuts down) and also in the presence of inhibitors of gene expression (O’Neill et al. 2011). This follows on from previous work in another alga Acetabularia mediterranea, where circadian rhythms of chloroplast movement were observed to persist when the nucleus of the cell was removed (Woolum 1991). The landmark observations, however, were performed in the cyanobacteria Synechococcus elongatus, a prokaryote. Here it was shown that the ~24-h rhythm of KaiA/B/C protein phosphorylation and complex formation that occurs in living cells and normally interacts reciprocally with genome-wide transcriptional regulation could be reconstituted in vitro using just the three recombinant proteins (KaiA, B & C) with ATP (Nakajima et al. 2005). Bacterial expression systems tend to work on a 1 protein ⟹ 1 function principle, whilst mammalian proteins tend to encode multiple domains with multiple, context-dependent cellular functions. We therefore think it unlikely that a directly equivalent experiment can be performed for mammalian timekeeping. It does raise the possibility, however, that the smallest functional circadian timekeeping unit may not include the nucleus.
3.4.10 Circadian Rhythms in Human Erythrocytes
Recently, the absolute requirement for nascent gene expression in mammalian cells was investigated in vitro. The ultimately cytotoxic effects of chronic inhibition of gene expression often confound pharmacological approaches to this problem. To circumvent this, preparations of human red blood cells (which are naturally anucleate) were employed. The rhythmic post-translational PRX modification, first observed in mouse liver, was used as a rhythmic marker. Briefly, the peroxiredoxin family constitutes a major part of the cellular defence against reactive oxygen species (ROS), specifically H2O2, which are an unavoidable by-product of aerobic metabolism. Erythrocytes express PRX at high levels (~1 % total protein), presumably due to the high ROS generation resulting from haemoglobin auto-oxidation. 2-Cys PRXs exist primarily as dimers that catalyse their own oxidation by H2O2 at conserved peroxidatic cysteine residues. The resultant sulphenic acid (CysP-SOH) may be reduced by a resolving cysteine on the opposing monomer (CysP-S-S-CysR) and ultimately reduced to the free thiol (SH) by the thioredoxin system. The kinetics of the resolving cysteine attack is quite slow, however, and in the presence of additional H2O2, over-oxidation to the sulphenic (CysP-SO2H) or even sulphonic (CysP-SO3H) form occurs (reversible through sulphiredoxin-catalysed, ATP-dependent mechanisms). By performing anti-2-Cys PRX-SO2/3 immunoblots upon time courses of erythrocytes, isolated in a minimal glucose/salt buffer under constant conditions, circadian rhythms of PRX oxidation were observed (Fig. 2d). These rhythms were temperature compensated, entrainable by temperature cycles and robust to inhibitors of gene expression. In addition, the concentrations of several cellular metabolites ([ATP], [NADH], [NADPH]) appeared to be rhythmically modulated, as did an indirect fluorescence assay for haemoglobin multimeric state (O’Neill and Reddy 2011).
These data might suggest an underlying rhythmic capacity exists in the cytoplasm, not directly reliant on nascent gene expression, similar to the proposed ‘cytoscillator’ that we hypothesised previously (Hastings et al. 2008). At present, it is unclear whether the metabolic rhythms observable in isolated erythrocytes are of direct physiological relevance, since other previously reported metabolic rhythms in cultured fibroblasts and mouse tissues, e.g. NAD+ concentration, were attributed a transcriptional basis (Ramsey et al. 2009). It is interesting to note, however, that whilst cycles of PRX oxidation could be observed in transcriptionally arrhythmic mouse embryonic fibroblasts from CRY1/2 null mice, they were clearly perturbed compared to the more robust oscillation observable in the wild-type control. Although more work is needed, the implication is that in nucleated cells some post-translational metabolic rhythm interacts with (and probably reciprocally regulates) the defined transcriptional elements relevant to timekeeping.
3.4.11 High-Throughput Screening for Clock Regulators
A recent unbiased genome-wide, RNAi screen identified a number of genes whose downregulation significantly affected the period or amplitude of cellular rhythms (using two different bioluminescent reporters). Whilst RNAi-based approaches are frequently problematic due to off-target effects, it is telling that a significant proportion of ‘hits’ were identified as components of well-characterised metabolic and signalling pathways. Indeed, of the 12 strongest period phenotypes that were investigated in detail, knockdown of POLR3F and ACSF3 had no apparent effect on ‘clock gene’ expression, even though circadian period was increased or shortened, respectively (Zhang et al. 2009).
Several groups have also employed drug discovery approaches to identify compounds that affect timekeeping in cell culture. Although larger (often proprietary) library screens are still in progress, several data sets have been published (Chen et al. 2012; Hirota et al. 2008, 2011; Isojima et al. 2009) with roughly 1 % of compounds being observed to significantly affect circadian period. Several of these compounds confirmed the contribution of post-translational mechanisms already implicated in cellular rhythms (e.g. CK1δ/ε, GSK3β, adenylyl cyclase). In addition, novel regulatory mechanisms have been identified (e.g. CK1α; Hirota et al. 2011), as well as a number of inhibitors, agonists and antagonists that are reported to target proteins with no established role in cellular timekeeping (Isojima et al. 2009). Many of this latter group comprise membrane and intracellular signalling proteins. The implication of these ‘discovery science’ approaches is that significant numbers of cellular systems that contribute to the fidelity of timekeeping have not yet been integrated into any coherent model of cellular rhythms.
3.4.12 Conservation of Post-Translational Mechanisms Across Taxa
Across the eukaryotes, transcription factors implicated in timekeeping mechanisms are poorly conserved between phyla (Hastings et al. 2008; O’Neill et al. 2011). In contrast, a number of ubiquitous post-translational mechanisms are apparently utterly conserved in their timekeeping roles, e.g. CK1, CK2, GSK3β, PP1/2A and proteasomal degradation (Hastings et al. 2008). Inhibitors of these enzymes have the same effects on cellular rhythms in the alga, O. tauri, as they do in mammalian cells (O’Neill et al. 2011), despite their divergence ~1.5 billion years ago. Whether this remarkable degree of conservation reflects a general requirement for certain housekeeping mechanisms, e.g. for targeted protein degradation, or conversely that these enzymes constitute part of a conserved post-translational timing mechanism that targets more recently occurring transcription factors is unclear. The paradigm of the conserved cell cycle role of eukaryotic cyclin-dependent kinases, and not their transcriptional targets, argues for the latter (Hastings et al. 2008). However, the striking similarity between the post-translational processing of clock proteins, e.g. PER2, with components of the wnt signalling pathway, e.g. β-catenin (Del Valle-Perez et al. 2011), argues for the former. Although there is some functional redundancy within each enzyme family, there being multiple isoforms, the activity of each is ultimately essential for cellular viability due to their participation in myriad cellular processes (see below). It is no surprise, therefore, that they are constitutively expressed. Interestingly though, some are reported to be rhythmically active, e.g. GSK3β due to their own circadian pattern post-translational modification (Iitaka et al. 2005).
As a marker for circadian timekeeping, intriguingly, the PRX oxidation rhythm appears to be particularly highly conserved, being observable in representative organisms from across the domains of life (Bacteria, Archaea, Eukaryota), unlike any TTFL component. Whilst PRX itself does not appear to play a critical timekeeping role, the redox rhythm it reports persists (albeit perturbed) in organisms that are deficient in ‘core’ TTFL components. We think it plausible that this remarkable conservation reflects either some underlying and ancient metabolic oscillation, which remains deeply embedded in the cellular machinery, or else an evolutionary convergence upon rhythmic redox regulation to facilitate temporal segregation of mutually antagonistic metabolic processes (Edgar et al. 2012).
4 Signalling Pathways and Metabolism
Whilst it is evident that contributions from transcriptional cycles to timekeeping are necessary for coordinated temporal organisation of normal physiology and behaviour and that transcription per se is ultimately required for life so that proteins/RNA can be synthesised, based on the points above, it is reasonable to posit that transcriptional cycles are not the mechanistic basis whereby circadian cycles take 24 h to complete. We will thus consider what other cellular processes might be relevant. The majority of implicated mechanisms, which are not classical transcription factors, are largely involved with signalling and/or metabolism; a few are discussed below, although many more components of well-known signalling pathways, e.g. mTOR and insulin/PI3K, are also increasingly being shown to play a role.
4.1 Second Messenger Pathways
Intracellular second messenger pathways are generally perceived to mediate rapid transmission of extracellular message to effector targets and thereby elicit the appropriate biological responses, e.g. change in ion channel activity, endo-/exocytosis, metabolic flux, transcriptional regulation, etc. Strikingly, however, circadian modulation of ubiquitous signalling systems, e.g. Ca2+, cAMP, cGMP and nitric oxide, has been observed in a range of contexts (Hastings et al. 2008; Golombek et al. 2004), and their pharmacological manipulation has been demonstrated to affect timekeeping function. For technical reasons, it has not yet been possible to ascertain whether these reflect global changes in basal concentration as opposed to some clock-relevant subcellular spatio-temporal pattern of transients. Circadian crosstalk between these pathways, common in other signalling contexts, has not yet been investigated to any degree. Whilst previously considered an output from the ‘core’ oscillator and/or a means of entrainment, recent findings suggest that second messenger signalling makes direct mechanistic contributions to timekeeping itself (O’Neill et al. 2008). Since the majority of these experiments were performed in SCN organotypic slices, they will be discussed in the final section.
4.2 Phosphorylation and Other Post-Translational Modifications
From acetylation to sumoylation, from glycosylation to cysteine oxidation, all the major classes of post-translational protein modification have been implicated as regulating and/or being regulated by circadian timekeeping (Doi et al. 2006; Durgan et al. 2012; Gupta and Ragsdale 2011; Lee et al. 2008). This should be no surprise since if the cell is the clock, why should it restrict itself to any particular subset, of the biochemical tools at its disposal, with which to sculpt the spatio-temporal dynamics of whichever protein activities are relevant to timekeeping. Since protein phosphorylation is the best characterised of these, what follows is a brief description of the key components that have been identified to date.
4.2.1 CK1
CK1 family members are conserved, ubiquitously expressed Ser/Thr kinases that exist in an auto-phosphorylated inactive state, until dephosphorylated through activation of specific protein phosphatases. They have a wide range of cellular targets, both cytosolic and nuclear and regulate processes as diverse as membrane trafficking, DNA replication, wnt signalling and RNA metabolism. CK1 has a noted preference for phosphate-primed phosphorylation sites (Cheong and Virshup 2011).
Early mutagenesis screens in Drosophila revealed casein kinase1 (CK1) as a regulator of circadian period length, with different doubletime mutations leading either to shortened or lengthened periods of rest-activity rhythms in flies (Kloss et al. 1998). In a remarkable parallel set of studies, the spontaneous Tau mutation of the Syrian hamster revealed the first circadian mutation in mammals, and this was later shown to involve an arginine to cysteine substitution within CK1ε, which caused a shortening of circadian period by 2 h for each copy of the mutated allele (Lowrey et al. 2000). In a further landmark discovery in humans, a group of familial sleep disorders characterised by early awakening were shown to segregate with mutations in human CK1δ or putative phosphorylation sites in human PER2. Subsequent genetic engineering of hamster and human mutations into mice has demonstrated how gain of function mutations of CK1δ and ε can enhance the rate of PER protein degradation, thereby accelerating the circadian cycle (Kloss et al. 1998). More recent genetic manipulations have shown that CK1δ and ε share overlapping roles in the pacemaker, and in the absence of both enzymes, the canonical transcriptional oscillator stops completely (Lee et al. 2011; Etchegaray et al. 2011; Meng et al. 2010). The development of selective inhibitors against CK1δ and ε has now made it possible, at least in animal studies and in tissue culture, to pharmacologically regulate circadian period, extending it to 30 h in a wild-type background and, by using a suitable dose, correcting to wild-type a shortened circadian period in CK1 mutants (Meng et al. 2010). Phosphorylation sites on PER2 regulated by wild-type and mutant CK1 are poorly characterised, but it is clear that, as with β-catenin, they license PER proteins for ubiquitinylation by the F-box protein β-TRCP and consequent proteasomal degradation (Reischl et al. 2007; Xu et al. 2009). Recently, pharmacological screening also implicated a hitherto unsuspected role for CK1α in the clock (Hirota et al. 2011).
4.2.2 CK2
CK2 is another ubiquitous and highly conserved protein Ser/Thr kinase that plays a central role in the control of a variety of pathways in cell proliferation, transformation, apoptosis and senescence (Montenarh 2010). It is composed of a catalytic dimeric α-subunit and a regulatory dimeric β-subunit. This complex is strongly implicated in regulating circadian rhythms in Arabidopsis thaliana (plant), Neurospora crassa (fungus) and Drosophila melanogaster (insect) and was recently identified in a large-scale functional RNAi screen to bind, phosphorylate and destabilise PER proteins in mammalian cells, probably acting synergistically with CK1 (Maier et al. 2009). Pharmacological inhibition of CK2 increases circadian period (Tsuchiya et al. 2009). Although many modes for activation have been reported, the upstream pathways for CK2 activation are unclear at present (Montenarh 2010).
4.2.3 GSK3
Glycogen synthase kinase-3 (GSK3) is a conserved and ubiquitously expressed multifunctional Ser/Thr kinase that was originally identified as a regulator of glycogen metabolism. It plays a key role in numerous signalling pathways including regulation of the cell cycle, inflammation and cell proliferation (Xu et al. 2009). GSK3 is inactivated through phosphorylation by AKT/PKB in the insulin/PI3K signalling pathway, and spontaneous circadian cycling of GSK3 phosphorylation has been observed in cultured fibroblasts. This enzyme was originally implicated in timekeeping by over- and under-expression mutants in Drosophila that, respectively, decrease and increase circadian period (Martinek et al. 2001). There are two mammalian isoforms: α and β, with the GSK3β−/− mouse being embryonically lethal, due to this enzyme’s essential role in development. In mammalian cells, pharmacological inhibition of GSK3 dose-dependently shortens circadian period (Hirota et al. 2008). GSK3β is reported to interact with clock proteins BMAL1, CLOCK, CRY2, PER2 and REV-ERBα, phosphorylate them and thereby regulate stability (Iitaka et al. 2005; Yin et al. 2006; Kurabayashi et al. 2010; Sahar et al. 2010; Spengler et al. 2009).
4.2.4 AMPK
5′-AMP-activated protein kinase is a ubiquitous and conserved, energy level sensor that acts as a metabolic switch that regulates several intracellular systems including the cellular uptake of glucose, the β-oxidation of fatty acids and mitochondrial biogenesis (Hardie 2011). It is a heterotrimer protein (α/β/γ, each having multiple isoforms). The α-subunit is catalytic (phosphorylating Ser/Thr), with the γ-subunit directly sensing AMP + ADP:ATP ratios (Xiao et al. 2011) but requiring additional phosphorylation by an upstream AMPK kinase for activity. Recently, AMPK was shown to phosphorylate and destabilise CRY1 and induce CK1-mediated degradation of PER2 in mammalian cells, with its activity and localisation being rhythmic in mouse liver (Lamia et al. 2009; Um et al. 2007).
4.2.5 Protein Phosphatases
Protein phosphorylation exists in dynamic equilibrium with phosphatase-mediated dephosphorylation. To date, the conserved and ubiquitously expressed protein phosphatase PP1 has been reported to regulate PER proteins (Lee et al. 2011; Schmutz et al. 2011), with PP5 being reported to modulate CK1ε activity in a CRY-dependent fashion (Partch et al. 2006). Based on observations in Drosophila and Neurospora, it seems likely that PP2A also plays some role in mammalian timekeeping (Sathyanarayanan et al. 2004; Yang et al. 2004).
4.3 Proteasomal Degradation
The functional contribution to cellular timekeeping made by the ubiquitous enzymes mentioned above has been interpreted in the context of net increases in site-specific clock protein phosphorylation occurring over the circadian cycle, with some phosphorylation events promoting nuclear entry, but protein hyperphosphorylation licensing proteins for ubiquitin-mediated proteasomal degradation (Virshup et al. 2007). Because these means of regulating protein turnover are a well-established principle in cell biology and by no means unique to clocks (Xu et al. 2009; Westermarck 2010), it seems entirely plausible and is well supported by genetic and biochemical evidence.
4.3.1 F-Box and Leucine-Rich Repeat Protein 3: The After-Hours Mutation
By analogy with the observation that mutations affecting the phosphorylation of Per and Cry alter circadian period and then also changes in ubiquitinylation, the intermediary between (some) phosphorylations and proteasomal degradation should have a similar effect. This was demonstrated by two independent mutagenesis screens, which revealed long circadian periods in mice carrying point mutations in the C-terminal leucine-rich region of the F-box and leucine-rich repeat protein 3 (FBXL3), a component of SCF ubiquitinylation complexes (E3 ligases). In both mutations, Fbxl3 Afterhours and Fbxl3 Overtime, circadian period of behavioural cycles and SCN bioluminescence rhythms is extended by ca. 1 h and 3 h in heterozygotes and homozygotes, respectively. This prolongation is ascribed to a reduced rate of CRY degradation, itself a consequence of a reduced affinity between mutant FBXL3 and its CRY substrates, which in turn slows down proteasomal targeting of CRY proteins (Godinho et al. 2007; Siepka et al. 2007). More recently, a second F-box protein, FBXL21, has also been implicated in the circadian clock as it also binds to, and directs for degradation, CRY proteins. It also compromises the negative-feedback actions of CRY on transactivation by CLOCK-BMAL1 complexes and is both highly enriched and rhythmically expressed in the SCN (Dardente et al. 2008). Interestingly high-throughput drug screening has recently revealed FBXL-mediated degradation of CRY as a novel target for pharmacological modulation of cellular timekeeping (Hirota et al. 2012).
4.4 Rhythmic Regulation of Protein Stability/Activity
In the context of the TTFL that has been proposed to account for cellular rhythms, current data suggest that a dynamic interplay between clock protein phosphorylation and dephosphorylation by these enzymes acts as an interval timer to regulate the kinetics of complex formation, protein degradation and nuclear entry, with certain specific serine/threonine residues on each clock protein substrate being implicated in tipping the balance between degradation and nuclear import (Virshup et al. 2007). An essentially identical model is proposed for wnt signalling, however, with the critical difference being that an upstream activating signal is required for stabilisation of β-catenin and its nuclear entry (Del Valle-Perez et al. 2011). No such signal has been identified for clock protein regulation, although presumably something must act upstream to elicit the observed rhythms in kinase activity/clock protein stability.
Intuitively, any non-housekeeping protein must possess specific pathways for its synthesis and degradation to avoid erroneous expression and accumulation of oxidised/misfolded proteins. Being intrinsic to so many aspects of cellular signalling and metabolism, however, it is inconceivable to us that the kinases and phosphatases mentioned above could have specific roles purely in the regulation of clock gene transcription factors, since their hundreds of other cellular targets are not observed to be rhythmically regulated post-translationally. Given the known synergistic action of these and other clock-implicated kinases in the context of other protein substrates with multisite phosphorylation domains (Salazar and Hofer 2009), as also found in PER, it seems reasonable to us that most of the known transcription factor clock proteins act as cooperative, coincidence-detecting substrate effectors to amplify a low-amplitude modulation of enzyme activity within cellular signalling and metabolic systems, resulting in rhythmic clock protein activity, localisation and stability.
Again, by analogy with the cell cycle, a teleological justification for phase-specific activation and irreversible protein degradation is appealing since it would impart directionality to transcriptional elements of the circadian cycle. In the absence of external factors, which might stimulate additional clock protein synthesis, the slower kinetics of gene expression would impart robustness against perturbation to any purely post-translational oscillation we presume persists in isolated erythrocytes. In this context, transcriptional feedback repression of clock proteins would not be required for rhythmicity, but clearly would offer the advantage of positive signal amplification—since no signal can be transduced when the protein substrate is absent. This still leaves the question of what might act upstream to license post-translational rhythms in enzyme activity, which we posit are essential for circadian-regulated transcription factor activity/stability.
4.5 Metabolic Interactions
In a large number of experimental organisms, including mammalian cells and tissues, circadian rhythms in redox balance (e.g. NAD+:NADH ratio), metabolite concentrations and coordinated metabolic processes (e.g. autophagy) have been reported (Minami et al. 2009; Merrow and Roenneberg 2001; Brody and Harris 1973; Powanda and Wannemacher 1970; Dallmann et al. 2012; Ma et al. 2011). For example, more than 20 years ago, reduced glutathione levels were reported to be rhythmic in isolated platelets, in vitro (Radha et al. 1985), and whilst platelets do still contain organelles, e.g. mitochondria and ribosomes, again the implication is that circadian rhythms can persist in the absence of (cycling) nuclear transcription, but not in the absence of metabolism, which is essential for cellular life.
It is interesting to note that several of the identified ‘clock gene’ transcription factors are haem-binding proteins and exhibit reciprocal regulation between rhythmic haem metabolism and the haem protein’s redox/ligand status (Yin et al. 2007; Kaasik and Lee 2004; Dioum et al. 2002), e.g. haem binding and thus activity of the nuclear receptor REV-ERBβ are governed by a redox-sensitive cysteine (Gupta and Ragsdale 2011). Furthermore, the transcriptional activity of complexes containing the acetyltransferase CLOCK with BMAL1 and the antagonistic deacetylase SIRT1 is differentially regulated by the redox state of their NAD cofactors (Rutter et al. 2001; Nakahata et al. 2008; Asher et al. 2008). Thus, the activities of clock-relevant transcriptional factors would appear to be reliant upon metabolic state, whereas their localisation/stability would appear to be governed by intracellular signalling systems. Moreover, there are many established reciprocal pathways connecting redox balance and cellular metabolism with the activity of the various signalling mechanisms discussed above, e.g. (Cheong and Virshup 2011; Montenarh 2010; Hardie 2011; Vander Heiden et al. 2009; Sethi and Vidal-Puig 2011; Dickinson and Chang 2011; Metallo and Vander Heiden 2011). It is therefore entirely plausible to us that rhythms in the cytosol persist through cyclical, distributed crosstalk between multiple metabolic and signalling networks, with transcriptional clock components acting as coincidence-detecting substrate effectors that integrate the state of the network as a whole. In this context, irrelevant network perturbations would be ignored and appropriate extracellular cues responded to in a phase-dependent fashion. Rhythmic licensing of transcription, with its slower kinetics, would impart robustness to the ‘cytoscillator’ by rhythmic modulation of protein/transcript levels. Critically a rhythmic transcriptional contribution would not be required for oscillator competence, but the additional repression of clock protein activity upon its cognate gene and CCGs would facilitate signal amplification (Fig. 3).
A general model for cellular timekeeping. Circadian timekeeping is functionally distributed within the cell’s metabolic and signalling networks and does not require nascent gene expression. In most (nucleated) cells, however, the integrated output from these networks is apparent in the circadian cycles of protein activity/stability/localisation observed, for example, in canonical clock protein transcription factors which act as ‘coincidence detectors’ for network state. These rhythmically modulate chromatin structure and facilitate coordinated temporal regulation of downstream transcriptional networks, including their own cognate clock gene circuitry, resulting in signal amplification. Rhythmic modulation of ‘clock-controlled genes’ facilitates coordinated temporal regulation of physiology and feeds forward into metabolic/signalling networks, modulating expression of some component mechanisms, e.g. rhythmic NAMPT expression facilitates rhythmic activity of the NAD+ salvage pathway (Ramsey et al. 2009), and PDE1B degrades cAMP and affects rhythmic amplitude (Zhang et al. 2009). The circadian state of the signalling network modulates communication with local and distant targets, whilst selectively and temporally gating the capacity of relevant extracellular signals to affect circadian phase
5 SCN-Specific Timekeeping Mechanisms
5.1 SCN Physiology
SCN neurons exhibit several unusual features. Whilst circadian rhythms are ubiquitous in mammalian cells, the SCN exhibits much greater amplitude, robustness and accuracy resulting in, and from, increased interneuronal synchrony, i.e. amplitude and synchrony are mutually interdependent (Hastings et al. 2008; Abraham et al. 2010). For example, unlike other cultured tissues, the SCN appears to be resistant to entrainment by temperature cycles, unless its interneuronal communication is compromised (Buhr et al. 2010). What follows is a discussion of the genetic and pharmacological approaches that have been employed to delineate what makes the SCN so special, with particular emphasis on Ca2+ and cAMP signalling.
SCN electrophysiology is overtly rhythmic, most neurons being more depolarised (~−50 mV) and spontaneously firing action potentials (APs) (ca. 10 Hz) during circadian day but hyperpolarised (~−60 mV) and silent (<1 Hz) at night (Pennartz et al. 2002; Colwell 2011). Blockade of neurotransmission (e.g. with tetrodotoxin, TTX) abolishes electrical rhythms and induces rapid damping of amplitude of circadian gene expression with progressive interneuronal desynchronisation (Yamaguchi et al. 2003). By implication, electrical excitability is required for coupling between individual cellular oscillators, making the whole greater than the sum of its parts. The axons of most SCN neurons project outwards to communicate with surrounding brain regions. Intra-SCN communication originates predominantly from exocytosis of dense-cored vesicles, principally from dendritic sites (Castel et al. 1996). Vesicle release is mostly non-synaptic or parasynaptic, is Ca2+ dependent and may involve retrograde transmission facilitated by neural back-propagation (Gompf et al. 2006) and follow slower kinetics than for most excitable cells.
Metabotropic neuropeptide signalling appears to be essential to SCN timekeeping. Although functional electrical synapses exist, they are not required for timekeeping (Long et al. 2005). Similarly, no ionotropic neurotransmitter receptor has been demonstrated to be indispensable for SCN-intrinsic timekeeping, in vitro. For example, although intra-SCN synapses are mainly GABAergic, with most neurons synthesising/releasing GABA and expressing GABAA receptors, chronic inhibition of GABAergic signalling with bicuculline does not significantly affect timing (Gompf et al. 2006; Aton et al. 2006). GABA signalling does contribute to entrainment (Ehlen and Paul 2009) and modulate amplitude, however, possibly by restricting the extent of resting membrane depolarisation during the day whilst hyperpolarising it at night (Aton et al. 2006), perhaps acting in concert with a nightly K+ channel efflux (Colwell 2011).
Several neuropeptides mediate SCN interneuronal communication, the foremost being vasoactive intestinal peptide (VIP) which binds the VIP/PACAP receptor (VPAC2), primarily signalling through adenylyl cyclase (AC) via Gsα (An et al. 2011). Auxiliary roles exist for gastrin-releasing peptide (GRP) and arginine vasopressin (AVP), both also signalling through their respective G-protein-coupled receptors to activate phospholipase C (PLC) (Gamble et al. 2007). VIP, GRP and AVP are expressed differentially in subpopulations throughout the SCN, although their receptors (particularly VPAC2) are more widely distributed (Welsh et al. 2010). Most likely, neuropeptide release occurs during the day in response to increased electrical activity facilitating vesicular exocytosis, thus allowing localised paracrine communication within the SCN network (Maywood et al. 2011).
Mice with homozygous deletion of genes encoding VIP or VPAC2 exhibit severely disrupted behavioural rhythms. The resting membrane potential in SCN neurons from these knockout mice (in vitro) is hyperpolarised, exhibiting reduced electrical activity, compared with wild type (Aton et al. 2005; Maywood et al. 2006). In SCN slices from homozygous VIP or VPAC2-null mice, molecular rhythms are profoundly affected. Most notably, the number of detectable bioluminescent neurons is substantially reduced relative to wild type, and rhythms in those neurons are stochastic, low amplitude and desynchronised from each other—similar to dissociated neurons or fibroblasts. Critically, several cycles of higher amplitude, synchronised rhythms can be rescued in VIP-null SCN by exogenous VIP (Aton et al. 2005). Similar observations have been made in VPAC2-null slices, treated with forskolin to directly activate AC. Rhythmic amplitude can similarly be rescued by GRP application, or by elevated intracellular Ca2+ (high [K+]EC) (Maywood et al. 2006). This implies that the timekeeping deficit in these animals can be ascribed to deficits in cAMP/Ca2+ signalling.
5.2 SCN Second Messenger Signalling
Second messenger signalling has long been viewed as an important means of cellular entrainment, e.g. in vitro, Glu elicits Ca2+-mediated phase shifts in SCN, as does VIP acting via VPAC2/Gsα/AC/cAMP (Welsh et al. 2004; Brown and Piggins 2007; An et al. 2011). Moreover, circadian modulation of second messenger signalling has been reported as a rhythmic cellular output, e.g. in the SCN, both in vivo and in vitro, [cAMP]cyto varies ~fourfold, peaking shortly after projected dawn [~CT2 (O’Neill et al. 2008; Doi et al. 2011)]. Similarly, fluorescent probes reveal SCN [Ca2+]cyto to be robustly rhythmic, again peaking shortly after projected dawn [~CT2 (Ikeda et al. 2003)]. It is significant that rhythms in both [Ca2+] and resting membrane potential are unaffected by TTX treatment (Pennartz et al. 2002). Intriguingly, the morning peaks and nightly nadirs in cytosolic cAMP and Ca2+ are coincident: with maximal activity occurring in advance of the peak in electrical activity (~CT6, midday) and so cannot be driven by it (Ikeda et al. 2003). Conversely, cAMP and Ca2+ are required for SCN electrophysiological excitability (Atkinson et al. 2011; Shibata et al. 1984). Signal transduction pathways have been established between elevated cAMP/Ca2+ and transcriptional activation via CREs, e.g. in the Period1/2 promoter, so logically if second messenger signalling is both rhythmic output from, as well as input to, some hypothetical core clock mechanism, then dynamic cAMP/Ca2+ signalling becomes indistinguishable from that core mechanism (Hastings et al. 2008). Therefore, appropriate manipulation of cAMP and/or Ca2+ signalling should determine the key properties of cellular rhythms, i.e. amplitude, phase and period.
5.2.1 Effects upon Amplitude
Treatments that chronically elevate (forskolin + IBMX, pertussis toxin) or reduce (MDL12,330A) [cAMP]cyto induce dose-dependent damping of SCN rhythms and progressive interneuronal desynchronisation, in many respects phenocopying the VIP or VPAC2-null SCN. Normal rhythms return gradually following wash-off (O’Neill et al. 2008; Aton et al. 2006), revealing the self-organising properties of the SCN cells and circuit. Treatments that chronically elevate (ryanodine, high [K+]EC) or inhibit [Ca2+]IC (Ca2+ chelators, low [Ca2+]EC, Ca2+ channel inhibitor cocktails) also dose-dependently reduce amplitude, with presumed desynchronisation (Maywood et al. 2006; Ikeda et al. 2003; Shibata et al. 1984; Lundkvist et al. 2005).
Chronically elevated/reduced cytosolic cAMP/Ca2+ levels also increase/decrease PER2::LUC baseline bioluminescence, respectively, stressing the critical contribution that CRE activation makes to clock gene regulation. Such manipulations reveal the dependence upon dynamic second messenger-mediated interneuronal coupling for the reciprocal interaction between amplitude and synchrony inherent to SCN timekeeping (Abraham et al. 2010).
5.2.2 Effects upon Phase
Following wash-off of forskolin + IBMX and subsequent decline of cAMP levels, SCN slices, regardless of prior phase adopt a common new phase, resetting to dusk (~CT12), when [cAMP]cyto normally approaches its nadir (coincident with the peak of PER2::LUC activity) (O’Neill et al. 2008). Glu-induced Ca2+-mediated phase resetting of SCN has been reported extensively, being mimicked by Glu receptor agonists and blocked by antagonists (Kim et al. 2005). Ca2+ influx also resynchronises neurons in VPAC2-null SCN, and release of SCN slices from media with 0 mM KCl (low [Ca2+]cyto) resets internal phase to just after dawn (~CT3) when the cytosolic Ca2+ peak is normally observed (Maywood et al. 2006; Lundkvist et al. 2005). Thus, pharmacologically enforced cAMP/Ca2+ transitions override prior phase, forcing SCN phase to whenever such transitions would normally occur within the self-sustained circadian cycle.
5.2.3 Effects upon Period
Non-competitive (p-site) inhibitors of AC dose-dependently suppress SCN cAMP signalling (An et al. 2011), and reversibly, increase circadian period (to >31 h) in every tissue tested, in vitro, and are additive to manipulations that increase SCN period by other mechanisms (O’Neill et al. 2008). Increased mouse behavioural period, in vivo, was also observed when a p-site inhibitor (THFA) was delivered continuously and directly to the SCN via osmotic minipump (O’Neill et al. 2008). Whilst equivalent SCN experiments have yet to be performed for Ca2+, the period of rat liver explants ex vivo was reversibly lengthened by pharmacological inhibition of endoplasmic reticulum (ER) Ca2+ store release and import, as well as membrane-permeable Ca2+ chelators (Baez-Ruiz and Diaz-Munoz 2011). Similar results may be expected from SCN slices.
5.3 SCN Timing and Second Messenger Crosstalk
Manipulation of SCN cAMP/Ca2+ signalling generally results in more marked SCN phenotypes than mutation/knockout of identified clock genes. Therefore, in addition to their other myriad biological roles (Hastings et al. 2008), dynamic cAMP/Ca2+ signalling critically contributes to SCN timekeeping, begging the question: which signalling proteins are involved, and how might crosstalk with circadian transcriptional elements be achieved?
Wild-type SCN slices exhibit daily cycles of elevated cAMP/Ca2+ culminating in rhythmic CRE activation at Per gene promoters, acting synergistically with rhythmic E-box activation, and it is presumed, thereby amplifying the oscillation. Within the CREB transcription factors, ATF4 has recently been implicated as one such terminal effector (Koyanagi et al. 2011). The SCN-specific pathways that facilitate CRE activation are ill defined, but cAMP transduction certainly involves EPAC with an auxiliary role for PKA (O’Neill et al. 2008). For Ca2+, the effectors CaMKII, MAPK and PKC have been similarly implicated (Welsh et al. 2010; Lee et al. 2010). Based on the extensive literature concerning synergistic effector regulation between the cAMP and Ca2 signalling systems (Welsh et al. 2010), it is likely that the two normally operate in tandem to facilitate maximal CRE activation in the SCN.
It is unknown whether any AC isoforms preferentially participate in cellular rhythms, although the daily increase in SCN Ca2+ cyto signalling likely initiates primarily from intracellular stores. Whilst Ins(1,4,5)P3 (IP3) and ryanodine receptors (IP3R, RyR) are certainly involved, the relative contributions made by different Ca2+-mobilising messengers (IP3, cADPR, NAADP) and Ca2+-induced Ca2+ release remain poorly characterised. Although plasma membrane Ca2+ flux is required for SCN timekeeping, this is very likely an indirect consequence of the requirement for EC Ca2+ in vesicular exo-/endocytotosis (Schweizer and Ryan 2006), and for replenishing depleted intracellular stores during store-operated Ca2+ entry (SOCE) (Cohen and Fields 2006). Direct crosstalk between intracellular messenger systems, e.g. cAMP modulation of IP3R activity (Schweizer and Ryan 2006), has not been investigated in the SCN.
The transcriptional clock circuitry within the SCN modulates cAMP/Ca2+ signalling through several means. For example, gene expression rhythms are observed for several SCN neuropeptides, neuropeptide receptors and AC/RyR isoforms, and functional contributions to timekeeping have been established, i.e. factors which enable rhythmic transcription (input) are themselves rhythmically expressed (output) (Welsh et al. 2010). Furthermore, daytime repression of Gi/o has been reported, through rhythmic expression of RGS16 (Doi et al. 2011). Most significantly, CRY1 was recently reported to directly inhibit Gsα activity in vitro and in mouse liver in vivo (Zhang et al. 2010); if this mechanism operates in the SCN, then it may well contribute to the decline of cAMP/Ca2+ late in the day, when CRY levels are increasing.
5.4 Daily Paracrine Positive-Feedback Coupling Within the SCN
It is known that elevated cytosolic cAMP and Ca2+ are competent to activate plasma membrane cation channels, e.g. cAMP regulates CNG channels to conduct mixed cation influx (Kaupp and Seifert 2002); SOCE induces Ca2+ and mixed cation influx via ORAI and TRPC channels, respectively (Cheng et al. 2011). Critically, persistent subthreshold cation channels exhibiting some similar properties to CNG/TRPC are observed in SCN slices (Kononenko et al. 2004), and to reiterate, compromised cAMP or Ca2+ signalling disrupts spontaneous electrical activity in SCN organotypic slices (Atkinson et al. 2011; Shibata et al. 1984), We propose it is therefore most plausible that shortly after (projected) dawn, prior timekeeping mechanisms facilitate elevated cytosolic cAMP/Ca2+ signalling. This increases the ‘open’ probability of cAMP/Ca2+-sensitive cation channels, thereby depolarising the resting membrane potential (~10 mV) and increasing AP firing probability. AP firing increases neuropeptide release, and neuropeptides act locally to stimulate further cAMP/Ca2+ signalling in neighbouring neurons (feedforward), which respond similarly. Clearly, this leads to positive feedback, sustained auto-amplification of cAMP/Ca2+ signalling within the SCN network, amplifying PER1/2 expression in the process, in the same phase as E-box activation. It is presumed that this sustained second messenger activity is relaxed by some combination of vesicular neuropeptide depletion, receptor desensitisation/internalisation and clock-driven modulation of the Gs/Gq/Gi/o transduction pathways, e.g. CRY1 and RGS16. Some redundancy must exist between Ca2+ and cAMP signalling for timekeeping within individual neurons, but we speculate that SCN-intrinsic encoding of projected dawn must rely upon the coincident detection of both second messenger systems being more active within the SCN network as a whole (see Fig. 4).
Schematic of SCN paracrine positive-feedback coupling from a single neuron perspective. Cellular timekeeping normally results from reciprocal crosstalk between transcriptional/translational feedback loops (1) with extranuclear oscillations (2) in signalling and metabolism to facilitate rhythmic regulation of clock-controlled genes (3), e.g. AVP, and also increased cAMP/Ca2+ signalling around anticipated dawn (4). cAMP/Ca2+ depolarises resting membrane potential (V m), thereby increasing electrical activity and neuropeptide release (5), further elevating cAMP/Ca2+ signalling within the network, eliciting further neuropeptide release (6). Neuropeptide receptor activation amplifies cAMP/Ca2+ signalling (7) and activates downstream effectors (8). Later in the day, cAMP/Ca2+ signalling decreases through some combination of neuropeptide depletion and/or receptor desensitisation/internalisation and/or change in gene expression of inhibitors of G-protein signalling (9)
In a circadian context, second messenger signalling has not been studied extensively outside the SCN, although current data suggest that in fibroblast cultures also, a critical timekeeping role for dynamic changes in cAMP, Ca2+ and membrane potential exists (O’Neill et al. 2008; Noguchi et al. 2012). The above findings certainly provoke further questions, however. For example, is the timekeeping role of intracellular cAMP/Ca2+ rhythms ubiquitous in mammalian cells and simply with higher amplitude in the SCN, or specific to SCN timekeeping? What licenses the increased cAMP/Ca2+ signalling after dawn, and can inhibitors of gene expression block it? What specific spatio-temporal dynamic of SCN cAMP/Ca2+ signalling encodes timekeeping information: is it amplitude or frequency modulated (Berridge 1997); global, local or microdomain dependent? What is the contribution of other intracellular messengers, e.g. cGMP, and the crosstalk between them?
5.5 SCN Signalling in Context
Initiated by cell-intrinsic mechanisms, neuropeptide-mediated paracrine positive feedback between the extensively coupled neurons within the SCN facilitates a sustained increase in cAMP/Ca2+ signalling during the day. This accounts for the enhanced amplitude, robustness and precision of the SCN compared with other tissues that lack such coupling. Accordingly we presume SCN amplitude will be further reinforced, in vivo, when retinorecipient neurons receive photic inputs that evoke appropriate Ca2+ transients during the day. On the other hand, when those same transients occur at night, recipient neurons will phase-shift relative to non-recipient neurons, resulting in an increased phase distribution across the SCN. Due to SCN positive-feedback coupling, however, this transitory phase dispersal is integrated to become more coherent during the following day, resulting in a modest phase shift by the network as a whole. Thus, whilst our understanding of intracellular timekeeping is still lacking in mechanistic detail, the reason why the SCN does it better is becoming increasingly clear.
6 General Conclusion
A large number of cellular components relevant to timekeeping (‘clock genes/proteins’) have been identified to date. Of late they have tended to be enzymatic or metabolic in nature, rather than transcriptional. As with most aspects of eukaryotic cell biology, their role is not specific to circadian timekeeping and is usually redundant within it. The era when research papers proclaimed ‘gene X’ or ‘process Y’ plays a role in circadian timekeeping is coming to an end, since the emergent theme seems to be one whereby the activity of some cellular/biochemical process which is rhythmically regulated can in turn feedback into regulating that rhythm and thereby becomes indistinguishable from the core mechanism. In a cycle, it is impossible to separate cause from effect, and yet if the cell itself is the oscillator and can utilise any of the numerous tools at its disposal to sustain rhythms, how do we progress towards a detailed mechanistic understanding? We envisage three approaches:
-
(1)
Pragmatic—It does not matter how the cellular oscillator works. There is a wealth of knowledge concerning how circadian rhythms impact upon biology, and that understanding can be applied productively now.
-
(2)
Systems biology—Circadian rhythms are an emergent property of mammalian cells and cannot be understood at a level simpler than the cell. In order to understand this timekeeping phenomenon, we must understand or have data about all relevant aspects of cell biology. Since the behaviour of a sufficiently complex system cannot be grasped intuitively, the model must be built in silico, being able to make nonintuitive predictions that can be tested and the model refined, iteratively.
-
(3)
Reductionist—There is a core oscillatory mechanism from which all the other identified timekeeping systems are driven, but ultimately feedback into. If this core oscillator can be sufficiently reduced, it can be understood biochemically and a model built from the bottom up.
For the pharmacologist, the first option should be the most attractive. The tools required to test whether a new or existing drug affects cellular rhythms, or whether rhythms affect its pharmacokinetics/dynamics, already exist, and it is hoped that this and the other chapters in this volume will help catalyse such endeavours.
References
Abraham U et al (2010) Coupling governs entrainment range of circadian clocks. Mol Syst Biol 6:438
Akhtar RA et al (2002) Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol 12(7):540–550
An S et al (2011) Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol 105(5):2289–2296
Antoch MP, Kondratov RV (2013) Pharmacological modulators of the circadian clock as potential therapeutic drugs: focus on genotoxic/anticancer therapy. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Asher G et al (2008) SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134(2):317–328
Atkinson SE et al (2011) Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J Biol Rhythms 26(3): 210–220
Aton SJ et al (2005) Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci 8(4):476–483
Aton SJ et al (2006) GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci USA 103(50):19188–19193
Baez-Ruiz A, Diaz-Munoz M (2011) Chronic inhibition of endoplasmic reticulum calcium-release channels and calcium-ATPase lengthens the period of hepatic clock gene Per1. J Circadian Rhythms 9:6
Berridge MJ (1997) The AM and FM of calcium signalling. Nature 386(6627):759–760
Blake WJ et al (2003) Noise in eukaryotic gene expression. Nature 422(6932):633–637
Brody S, Harris S (1973) Circadian rhythms in neurospora: spatial differences in pyridine nucleotide levels. Science 180(85):498–500
Brown TM, Piggins HD (2007) Electrophysiology of the suprachiasmatic circadian clock. Prog Neurobiol 82(5):229–255
Buhr ED, Takahashi JS (2013) Molecular components of the mammalian circadian clock. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Buhr ED, Yoo SH, Takahashi JS (2010) Temperature as a universal resetting cue for mammalian circadian oscillators. Science 330(6002):379–385
Bunger MK et al (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103(7):1009–1017
Cao R et al (2011) Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience 181:79–88
Castel M, Morris J, Belenky M (1996) Non-synaptic and dendritic exocytosis from dense-cored vesicles in the suprachiasmatic nucleus. Neuroreport 7(2):543–547
Cavallari N et al (2011) A blind circadian clock in cavefish reveals that opsins mediate peripheral clock photoreception. PLoS Biol 9(9):e1001142
Chen R et al (2009) Rhythmic PER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Mol Cell 36(3):417–430
Chen Z et al (2012) Identification of diverse modulators of central and peripheral circadian clocks by high-throughput chemical screening. Proc Natl Acad Sci USA 109(1):101–106
Cheng HY et al (2007) microRNA modulation of circadian-clock period and entrainment. Neuron 54(5):813–829
Cheng KT et al (2011) Local Ca(2)+ entry via Orai1 regulates plasma membrane recruitment of TRPC1 and controls cytosolic Ca(2)+ signals required for specific cell functions. PLoS Biol 9(3):e1001025
Cheong JK, Virshup DM (2011) Casein kinase 1: complexity in the family. Int J Biochem Cell Biol 43(4):465–469
Cohen JE, Fields RD (2006) CaMKII inactivation by extracellular Ca(2+) depletion in dorsal root ganglion neurons. Cell Calcium 39(5):445–454
Colwell CS (2011) Linking neural activity and molecular oscillations in the SCN. Nat Rev Neurosci 12(10):553–569
Dallmann R et al (2012) The human circadian metabolome. Proc Natl Acad Sci USA 109(7): 2625–2629
Dardente H et al (2008) Implication of the F-Box Protein FBXL21 in circadian pacemaker function in mammals. PLoS One 3(10):e3530
Debruyne JP et al (2006) A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron 50(3):465–477
Deery MJ et al (2009) Proteomic analysis reveals the role of synaptic vesicle cycling in sustaining the suprachiasmatic circadian clock. Curr Biol 19(23):2031–2036
Del Valle-Perez B et al (2011) Coordinated action of CK1 isoforms in canonical Wnt signaling. Mol Cell Biol 31(14):2877–2888
Dibner C et al (2009) Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28(2):123–134
Dickinson BC, Chang CJ (2011) Chemistry and biology of reactive oxygen species in signaling or stress responses. Nat Chem Biol 7(8):504–511
Dioum EM et al (2002) NPAS2: a gas-responsive transcription factor. Science 298(5602): 2385–2387
Doherty CJ, Kay SA (2010) Circadian control of global gene expression patterns. Annu Rev Genet 44:419–444
Doi M, Hirayama J, Sassone-Corsi P (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125(3):497–508
Doi M et al (2011) Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun 2:327
Durgan DJ et al (2012) O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 286(52):44606–44619
Edgar RS et al (2012) Peroxiredoxins are conserved markers of circadian rhythms. Nature 485(7399):459–464
Edmunds LN Jr (1983) Chronobiology at the cellular and molecular levels: models and mechanisms for circadian timekeeping. Am J Anat 168(4):389–431
Ehlen JC, Paul KN (2009) Regulation of light’s action in the mammalian circadian clock: role of the extrasynaptic GABAA receptor. Am J Physiol Regul Integr Comp Physiol 296(5):R1606–R1612
Eide EJ et al (2005) Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol 25(7):2795–2807
Engelmann W, Bollig I, Hartmann R (1976) The effects of lithium ions on circadian rhythms. Arzneimittelforschung 26(6):1085–1086
Etchegaray JP et al (2011) Casein kinase 1 delta (CK1delta) regulates period length of the mouse suprachiasmatic circadian clock in vitro. PLoS One 5(4):e10303
Fan Y et al (2007) Cycling of CRYPTOCHROME proteins is not necessary for circadian-clock function in mammalian fibroblasts. Curr Biol 17(13):1091–1100
Gamble KL et al (2007) Gastrin-releasing peptide mediates light-like resetting of the suprachiasmatic nucleus circadian pacemaker through cAMP response element-binding protein and Per1 activation. J Neurosci 27(44):12078–12087
Gentric G, Celton-Morizur S, Desdouets C (2012) Polyploidy and liver proliferation. Clin Res Hepatol Gastroenterol 36(1):29–34
Godinho SI et al (2007) The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science 316(5826):897–900
Golombek DA et al (2004) Signaling in the mammalian circadian clock: the NO/cGMP pathway. Neurochem Int 45(6):929–936
Gompf HS, Irwin RP, Allen CN (2006) Retrograde suppression of GABAergic currents in a subset of SCN neurons. Eur J Neurosci 23(12):3209–3216
Granshaw T, Tsukamoto M, Brody S (2003) Circadian rhythms in Neurospora crassa: farnesol or geraniol allow expression of rhythmicity in the otherwise arrhythmic strains frq10, wc-1, and wc-2. J Biol Rhythms 18(4):287–296
Gupta N, Ragsdale SW (2011) Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erb{beta}. J Biol Chem 286(6):4392–4403
Hardie DG (2011) AMP-activated protein kinase–an energy sensor that regulates all aspects of cell function. Genes Dev 25(18):1895–1908
Hastings MH, Maywood ES, O’Neill JS (2008) Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18(17):R805–R815
Herzog ED et al (2004) Temporal precision in the mammalian circadian system: a reliable clock from less reliable neurons. J Biol Rhythms 19(1):35–46
Hirota T et al (2008) A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci USA 105(52): 20746–20751
Hirota T et al (2011) High-throughput chemical screen identifies a novel potent modulator of cellular circadian rhythms and reveals CKIalpha as a clock regulatory kinase. PLoS Biol 8(12):e1000559
Hirota T et al (2012) Identification of small molecule activators of cryptochrome. Science 337(6098):1094–1097
Iitaka C et al (2005) A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem 280(33):29397–29402
Ikeda M et al (2003) Circadian dynamics of cytosolic and nuclear Ca2+ in single suprachiasmatic nucleus neurons. Neuron 38(2):253–263
Isojima Y et al (2009) CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA 106(37):15744–15749
Jiang YJ et al (2000) Notch signalling and the synchronization of the somite segmentation clock. Nature 408(6811):475–479
Johnson CH (1999) Forty years of PRCs–what have we learned? Chronobiol Int 16(6):711–743
Kaasik K, Lee CC (2004) Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430(6998):467–471
Kaupp UB, Seifert R (2002) Cyclic nucleotide-gated ion channels. Physiol Rev 82(3):769–824
Khalsa SB et al (1996) Evidence for a central role of transcription in the timing mechanism of a circadian clock. Am J Physiol 271(5 Pt 1):C1646–C1651
Kholodenko BN, Hancock JF, Kolch W (2010) Signalling ballet in space and time. Nat Rev Mol Cell Biol 11(6):414–426
Kim DY et al (2005) Voltage-gated calcium channels play crucial roles in the glutamate-induced phase shifts of the rat suprachiasmatic circadian clock. Eur J Neurosci 21(5):1215–1222
Kim TD et al (2007) Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev 21(7):797–810
Kim DY et al (2010) hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res 38(20):7068–7078
King DP, Takahashi JS (2000) Molecular genetics of circadian rhythms in mammals. Annu Rev Neurosci 23:713–742
King VM et al (2003) A hVIPR transgene as a novel tool for the analysis of circadian function in the mouse suprachiasmatic nucleus. Eur J Neurosci 17(11):822–832
Kloss B et al (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 94(1):97–107
Ko CH et al (2010) Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol 8(10):e1000513
Kononenko NI, Medina I, Dudek FE (2004) Persistent subthreshold voltage-dependent cation single channels in suprachiasmatic nucleus neurons. Neuroscience 129(1):85–92
Koyanagi S et al (2011) cAMP response element-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J Biol Chem 286:32416–32423
Kurabayashi N et al (2010) DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol 30(7):1757–1768
Lakin-Thomas PL (2006) Transcriptional feedback oscillators: maybe, maybe not. J Biol Rhythms 21(2):83–92
Lamia KA et al (2009) AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science 326(5951):437–440
Leak RK, Card JP, Moore RY (1999) Suprachiasmatic pacemaker organization analyzed by viral transynaptic transport. Brain Res 819(1–2):23–32
Lee J et al (2008) Dual modification of BMAL1 by SUMO2/3 and ubiquitin promotes circadian activation of the CLOCK/BMAL1 complex. Mol Cell Biol 28(19):6056–6065
Lee Y et al (2010) Coactivation of the CLOCK-BMAL1 complex by CBP mediates resetting of the circadian clock. J Cell Sci 123(Pt 20):3547–3557
Lee HM et al (2011) The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA 108(39): 16451–16456
Legewie S et al (2008) Recurrent design patterns in the feedback regulation of the mammalian signalling network. Mol Syst Biol 4:190
Lenz P, Sogaard-Andersen L (2011) Temporal and spatial oscillations in bacteria. Nat Rev Microbiol 9(8):565–577
Levi F, Schibler U (2007) Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 47:593–628
Long MA et al (2005) Electrical synapses coordinate activity in the suprachiasmatic nucleus. Nat Neurosci 8(1):61–66
Lowrey PL et al (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288(5465):483–492
Lundkvist GB et al (2005) A calcium flux is required for circadian rhythm generation in mammalian pacemaker neurons. J Neurosci 25(33):7682–7686
Ma D, Panda S, Lin JD (2011) Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J 30(22):4642–4651
Maier B et al (2009) A large-scale functional RNAi screen reveals a role for CK2 in the mammalian circadian clock. Genes Dev 23(6):708–718
Martinek S et al (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105(6):769–779
Maywood ES et al (2006) Synchronization and maintenance of timekeeping in suprachiasmatic circadian clock cells by neuropeptidergic signaling. Curr Biol 16(6):599–605
Maywood ES et al (2011) A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci USA 108(34):14306–14311
McGlincy NJ et al (2012) Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol 13(6):R54
Meng QJ et al (2008) Setting clock speed in mammals: the CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 58(1): 78–88
Meng QJ et al (2010) Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc Natl Acad Sci USA 107(34):15240–15245
Merrow M, Roenneberg T (2001) Circadian clocks: running on redox. Cell 106(2):141–143
Merrow M, Brunner M, Roenneberg T (1999) Assignment of circadian function for the Neurospora clock gene frequency. Nature 399(6736):584–586
Metallo CM, Vander Heiden MG (2011) Metabolism strikes back: metabolic flux regulates cell signaling. Genes Dev 24(24):2717–2722
Minami Y et al (2009) Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA 106(24):9890–9895
Montenarh M (2010) Cellular regulators of protein kinase CK2. Cell Tissue Res 342(2):139–146
Muck W et al (2000) Pharmacokinetics of cerivastatin when administered under fasted and fed conditions in the morning or evening. Int J Clin Pharmacol Ther 38(6):298–303
Musiek ES, FitzGerald GA (2013) Molecular clocks in pharmacology. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Nakahata Y et al (2008) The NAD+−dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134(2):329–340
Nakajima et al (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308(5720):414–415
Nelson DE et al (2004) Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science 306(5696):704–708
Njus D et al (1976) Membranes and molecules in circadian systems. Fed Proc 35(12):2353–2357
Noble D (2008) Claude Bernard, the first systems biologist, and the future of physiology. Exp Physiol 93(1):16–26
Noguchi T et al (2012) Fibroblast circadian rhythms of PER2 expression depend on membrane potential and intracellular calcium. Chronobiol Int 29(6):653–664
O’Neill JS, Reddy AB (2011) Circadian clocks in human red blood cells. Nature 469(7331): 498–503
O’Neill JS et al (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320(5878):949–953
O’Neill JS et al (2011) Circadian rhythms persist without transcription in a eukaryote. Nature 469(7331):554–558
Obrietan K et al (1999) Circadian regulation of cAMP response element-mediated gene expression in the suprachiasmatic nuclei. J Biol Chem 274(25):17748–17756
Ortiz-Tudela E, Mteyrek A, Ballesta A, Innominato PF, Lévi F (2013) Cancer chronotherapeutics: experimental, theoretical and clinical aspects. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Partch CL et al (2006) Posttranslational regulation of the mammalian circadian clock by cryptochrome and protein phosphatase 5. Proc Natl Acad Sci USA 103(27):10467–10472
Pennartz CM et al (2002) Diurnal modulation of pacemaker potentials and calcium current in the mammalian circadian clock. Nature 416(6878):286–290
Pittendrigh CS, Caldarola PC, Cosbey ES (1973) A differential effect of heavy water on temperature-dependent and temperature-compensated aspects of circadian system of Drosophila pseudoobscura. Proc Natl Acad Sci USA 70(7):2037–2041
Powanda MC, Wannemacher RW Jr (1970) Evidence for a linear correlation between the level of dietary tryptophan and hepatic NAD concentration and for a systematic variation in tissue NAD concentration in the mouse and the rat. J Nutr 100(12):1471–1478
Radha E et al (1985) Glutathione levels in human platelets display a circadian rhythm in vitro. Thromb Res 40(6):823–831
Ralph MR et al (1990) Transplanted suprachiasmatic nucleus determines circadian period. Science 247(4945):975–978
Ramsey KM et al (2009) Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324(5927):651–654
Reddy AB (2013) Genome-wide analyses of circadian systems. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Reddy AB, O’Neill JS (2010) Healthy clocks, healthy body, healthy mind. Trends Cell Biol 20(1): 36–44
Reddy AB et al (2005) Circadian clocks: neural and peripheral pacemakers that impact upon the cell division cycle. Mutat Res 574(1–2):76–91
Reddy AB et al (2006) Circadian orchestration of the hepatic proteome. Curr Biol 16(11): 1107–1115
Reddy AB et al (2007) Glucocorticoid signaling synchronizes the liver circadian transcriptome. Hepatology 45(6):1478–1488
Reischl S et al (2007) Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms 22(5):375–386
Reppert SM, Weaver DR (2002) Coordination of circadian timing in mammals. Nature 418(6901): 935–941
Robertson JB et al (2008) Real-time luminescence monitoring of cell-cycle and respiratory oscillations in yeast. Proc Natl Acad Sci USA 105(46):17988–17993
Robles MS, Mann M (2013) Proteomic approaches in circadian biology. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Roenneberg T, Merrow M (2002) “What watch?…such much!” Complexity and evolution of circadian clocks. Cell Tissue Res 309(1):3–9
Roenneberg T, Remi J, Merrow M (2010) Modeling a circadian surface. J Biol Rhythms 25(5): 340–9
Ruoff P, Zakhartsev M, Westerhoff HV (2007) Temperature compensation through systems biology. FEBS J 274(4):940–950
Rutter J et al (2001) Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293(5529):510–514
Sahar S, Sassone-Corsi P (2013) The epigenetic language of circadian clocks. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Sahar S et al (2010) Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS One 5(1):e8561
Salazar C, Hofer T (2009) Multisite protein phosphorylation–from molecular mechanisms to kinetic models. FEBS J 276(12):3177–3198
Sathyanarayanan S et al (2004) Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 116(4):603–615
Schmutz I et al (2011) Protein phosphatase 1 (PP1) is a post-translational regulator of the mammalian circadian clock. PLoS One 6(6):e21325
Schulz P, Steimer T (2009) Neurobiology of circadian systems. CNS Drugs 23(Suppl 2):3–13
Schweizer FE, Ryan TA (2006) The synaptic vesicle: cycle of exocytosis and endocytosis. Curr Opin Neurobiol 16(3):298–304
Sethi JK, Vidal-Puig A (2011) Wnt signalling and the control of cellular metabolism. Biochem J 427(1):1–17
Shibata S et al (1984) The role of calcium ions in circadian rhythm of suprachiasmatic nucleus neuron activity in rat hypothalamic slices. Neurosci Lett 52(1–2):181–184
Siepka SM et al (2007) Circadian mutant overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129(5):1011–1023
Skene DJ, Arendt J (2006) Human circadian rhythms: physiological and therapeutic relevance of light and melatonin. Ann Clin Biochem 43(Pt 5):344–353
Slat E, Freeman GM Jr, Herzog ED (2013) The clock in the brain: neurons, glia and networks in daily rhythms. In: Kramer A, Merrow M (eds) Circadian clocks, vol 217, Handbook of experimental pharmacology. Springer, Heidelberg
Spengler ML et al (2009) A serine cluster mediates BMAL1-dependent CLOCK phosphorylation and degradation. Cell Cycle 8(24):4138–4146
Suter DM et al (2011) Mammalian genes are transcribed with widely different bursting kinetics. Science 332(6028):472–474
Tahara Y et al (2012) In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol 22(11):1029–1034
Tischkau SA et al (2003) Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J Biol Chem 278(2):718–723
Tsuchiya Y et al (2009) Involvement of the protein kinase CK2 in the regulation of mammalian circadian rhythms. Sci Signal 2(73):ra26
Ueda HR et al (2005) System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet 37(2):187–192
Ukai H, Ueda HR (2010) Systems biology of mammalian circadian clocks. Annu Rev Physiol 72: 579–603
Um JH et al (2007) Activation of 5'-AMP-activated kinase with diabetes drug metformin induces casein kinase Iepsilon (CKIepsilon)-dependent degradation of clock protein mPer2. J Biol Chem 282(29):20794–20798
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324(5930):1029–1033
VanderLeest HT et al (2007) Seasonal encoding by the circadian pacemaker of the SCN. Curr Biol 17(5):468–473
Virshup DM et al (2007) Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harb Symp Quant Biol 72:413–420
Vitaterna MH et al (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264(5159):719–725
Vitaterna MH et al (1999) Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA 96(21):12114–12119
Welsh DK et al (2004) Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol 14(24): 2289–2295
Welsh DK, Takahashi JS, Kay SA (2010) Suprachiasmatic nucleus: cell autonomy and network properties. Annu Rev Physiol 72:551–577
Westermarck J (2010) Regulation of transcription factor function by targeted protein degradation: an overview focusing on p53, c-Myc, and c-Jun. Methods Mol Biol 647:31–36
Woolum JC (1991) A re-examination of the role of the nucleus in generating the circadian rhythm in Acetabularia. J Biol Rhythms 6(2):129–136
Wu JQ, Snyder M (2008) RNA polymerase II stalling: loading at the start prepares genes for a sprint. Genome Biol 9(5):220
Xiao B et al (2011) Structure of mammalian AMPK and its regulation by ADP. Nature 472(7342): 230–233
Xu C, Kim NG, Gumbiner BM (2009) Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle 8(24):4032–4039
Yamaguchi S et al (2000) The 5' upstream region of mPer1 gene contains two promoters and is responsible for circadian oscillation. Curr Biol 10(14):873–876
Yamaguchi S et al (2003) Synchronization of cellular clocks in the suprachiasmatic nucleus. Science 302(5649):1408–1412
Yang Y et al (2004) Distinct roles for PP1 and PP2A in the Neurospora circadian clock. Genes Dev 18(3):255–260
Yin L et al (2006) Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science 311(5763):1002–1005
Yin L et al (2007) Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318(5857):1786–1789
Yoo SH et al (2004) PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA 101(15): 5339–5346
Zhang EE et al (2009) A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139(1):199–210
Zhang EE et al (2010) Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med 16(10):1152–1156
Acknowledgements
The authors wish to thank Paul Margiotta for graphical support, as well as G. Churchill, E. Herzog, D. Welsh and C. Allen for their helpful discussion and suggestions. JON is supported by The Wellcome Trust [093734/Z/10/Z]. ESM and MHH are funded by the Medical Research Council. No competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
O’Neill, J.S., Maywood, E.S., Hastings, M.H. (2013). Cellular Mechanisms of Circadian Pacemaking: Beyond Transcriptional Loops. In: Kramer, A., Merrow, M. (eds) Circadian Clocks. Handbook of Experimental Pharmacology, vol 217. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-25950-0_4
Download citation
DOI: https://doi.org/10.1007/978-3-642-25950-0_4
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-25949-4
Online ISBN: 978-3-642-25950-0
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)