Abstract
Over the last decade, metabolomics has continued to grow rapidly and is considered a dynamic technology in envisaging and elucidating complex phenotypes in systems biology area. The advantage of metabolomics compared to other omics technologies such as transcriptomics and proteomics is that these later omics only consider the intermediate steps in the central dogma pathway (mRNA and protein expression). Meanwhile, metabolomics reveals the downstream products of gene and expression of proteins. The most frequently used tools are nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS). Some of the common MS-based analyses are gas chromatography-mass spectrometry (GC-MS) and liquid chromatography-mass spectrometry (LC-MS). These high-throughput instruments play an extremely crucial role in discovery metabolomics to generate data needed for further analysis. In this chapter, the concept of metabolomics in the context of systems biology is discussed and provides examples of its application in human disease studies, plant responses towards stress and abiotic resistance and also microbial metabolomics for biotechnology applications. Lastly, a few case studies of metabolomics analysis are also presented, for example, investigation of an aromatic herbal plant, Persicaria minor metabolome and microbial metabolomics for metabolic engineering applications.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Biomarker discovery
- Mass spectrometry
- Metabolite profiling
- Metabolic pathway
- Metabonomics
- Phytochemical analysis
4.1 Introduction
Metabolomics is an emerging technology that concerns with the understanding of living organisms at the metabolic level . As a definition, the metabolome represents the whole metabolites within a biological system [1]. It represents the crucial phenotype of cells inferred by the perturbation of gene expression and protein functions, due to the environmental effects or mutations. Gene expression and protein function also can be influenced by metabolome changes. Consequently, metabolomics plays an important role in cellular system and gene functions.
4.1.1 Different Aspects of Metabolomics
Specifically, the study of small molecule metabolite level is defined as metabolomics which is aimed at the measurement of metabolites present in a cell or tissue under a particular set of conditions. Metabolomics is considered an “omics” for the small molecules that provides information on the metabolome set at the individual level. Metabolomics is known by several names such as metabolic profiling, targeted/untargeted metabolic profiling and metabonomics [2]. Nevertheless, these approaches involved the analysis of metabolite levels from plant/microbe extracts, biofluids and living tissues [3]. While metabolomics is well associated with metabolite analysis of plant/microbes, the term metabonomics is usually applied to studies of human. Analysis of biofluids such as urine and plasma, nutrition, responses to drugs or diseases are some of the examples that are being tackled by metabonomics [4].
4.1.2 Metabolomics in the Context of Systems Biology
According to Weckwerth et al. [5], the principal goal of “omic” is the nontargeted analysis of proteins, transcripts and metabolites in a biological sample . The more inspiring phase of omic technology is the sophisticated exploration of assessable dynamics in biological systems. Utilisation of gas chromatography (GC) and liquid chromatography (LC) coupled with mass spectrometry (MS) is enabling high-throughput identification and quantitation of metabolites from various samples. This is a requirement for the analysis of dynamic systems, and hence, metabolomics is one of the key technologies in systems biology area. The data-driven approach is used to discover novel components such as genes, enzymes and metabolites as well as interactions in large-scale omics datasets . Thus, metabolomics aim is to discover new networks based on omics data for functional study [6]. In addition, Saito [7] has demonstrated the use of systems biology approach in photochemistry field for functional genomics study. In this new study, the systematic integration of different “omics” was used to investigate regulation and functions of metabolites in plant systems. Hypothesis is generated in this integrated approach and hence will be confirmed by reverse genetics, biochemistry and functional genomics (Fig. 4.1).
Workflow of data-driven in systems biology and functional genomics. Multi-omics approaches are used to generate hypothesis in systems biology studies. The figure was modified from “Phytochemical genomics —a new trend” of Kazuki Saito [7]
4.2 Applications of Metabolomics
Metabolomics can be applied in multifunctional projects utilising diverse types of analytical tools depending on designated research aims [8]. It can be divided into different categories: targeted analysis allowing quantitative analysis of targeted compounds, untargeted metabolic profiling focusing at a qualitative analysis of untargeted metabolites and metabolomics to determine an unbiased overview of metabolic configurations. Collectively, metabolic fingerprinting approach is performed for a rapid screening and determination of metabolites, which could reduce the use of analytical platforms for analysis of metabolites with biochemical relevance [8]. Such technique has become a rapid developing method in postgenomic study [6] or for phenotypic characteristic determination for quick identification and discrimination.
On the other hand, with the recent technological advances in the analytical tools, high-throughput profiling of large numbers of metabolites in biological samples has become common and cheaper. Tools such as mass spectrometry have enabled the simultaneous identification of a broad range and diverse metabolites group. In particular, the application of metabolomics has attracted and currently being used in almost all fields including human disease research and plant and microbial studies. The increment of metabolomics application demonstrates the utility of a metabolomics approach for the understanding of biochemical reactions and its impact.
4.2.1 Metabolomics in Human Studies
The metabolomics application is very important in human disease study especially in oncobiology research areas. Tumour cells are highly proliferated and with higher rates of transcription and translation. Therefore, there is a huge challenge in metabolomics-based medicinal field to predict the presence of tumour cells. Previous studies have focused on cancer detection using metabolic markers in preclinical analyses and quantification of the biomarkers in fluids [9]. Currently, many metabolites have been detected and identified, suggesting that they can be used as markers for many tumours (Table 4.1). Subsequently the integrated omics—metabolomics, proteomics and genomics techniques—have accelerated the process of early detection and diagnosis of cancer.
One of the major cases of tumours is the hepatocellular carcinoma (HCC) . The identification of cytokine biomarkers using microarrays has been previously reported [10]. Moreover, some studies have been conducted to profile metabolites in patient with HCC. Patterson et al. utilised UPLC-TOF mass spectrometry and discovered the increased of glycodeoxycholate, deoxycholate-3-sulphate and bilirubin in the patient’s biofluid samples [11]. Other polar metabolites such as arginine, alanine and lysine were found to be altered in liver cancer [12]. Other studies also reported that taurine, choline, glycerophosphocholine and phosphocholine were altered in breast cancers. In addition, myo-inositol levels were found to be increased in colon adenocarcinoma, prostate cancer and ovarian carcinoma [9, 13,14,15].
However, there are several hindrances in the cancer metabolome research. Data analysis difficulty in justifying the group of tumours due to the profile of metabolites could be varied among different types of tumour. The characterisation of metabolites that response to tumour is also difficult due to variation of sample to sample, the accuracy and sensitivity of the analytical systems and the physiological characteristics of the tumour [15].
Many studies have been carried out to search for new metabolic biomarkers for tumour disease. However, the metabolomics approach is still not yet established. In order to understand the metabolic changes and pathway alteration, it is important to integrate omics approaches—metabolomics, proteomics, transcriptomics and genomics. This will provide an appropriate information and better selection of biomarkers for potential application in prevention and diagnosis. The finding will result in a better selection of the potential biomarkers for prevention and diagnosis of the disease [29].
4.2.2 Metabolomics in Plant Studies
Metabolomics is predominantly significant in the plant study, because plants produce a huge number of metabolites—extremely more than are produced by animals and microorganisms [6]. Secondary metabolites that were produced by plant provide many used in response to abiotic stress. Generally, secondary metabolites will be produced by plants in response to the environment stress. Environmental metabolomics is a promising area of study in plant physiology to answer environmental or ecological factors in relation to plant metabolite changes [30, 31]. Metabolomics approaches have been an emerging technology to understanding plant behaviour towards environmental and ecological factors.
The study on plant metabolomics also covers plant responses towards stress and abiotic resistance. Stress in plants involves any significant changes in plant growth conditions that could disturb certain metabolic pathways . Metabolomics could significantly contribute to the stress study in plant biology by identifying metabolites/compounds, stress metabolism by-products, adaptation response of plants and signal transduction of plant molecules. Some examples of plant metabolites associated with biotic and abiotic stresses are polyols mannitol and sorbitol; glycine betaine; sugars such as sucrose, trehalose and fructan; or amino acids. Proline and ectoine involved as osmolytes and osmoprotectant for plant protection under high salt concentration, drought and desiccation stresses [32].
Metabolomics also leads to possible pathway identification of food metabolites for human health benefit. The modification of certain pathway has led to the improvement of plant metabolites with nutritional value. One of the successful stories is the study of genetically modified golden rice (GR), with the β-carotene accumulating in the endosperm [33]. Many metabolites involve in the defence against some human diseases; however, the production level in natural plants is insufficient to bestow optimal benefits. Metabolic engineering was established to enhance the production of important secondary metabolites such as anthocyanins in tomato [34]. The study has reported this new tomato variety has a strong purple coloration with threefold improved antioxidant capability. Moreover, they also fed the cancer-susceptible mice with this tomato variety and proved that this tomato could prolong the lifespan of cancer-susceptible mice.
4.2.3 Microbial Metabolomics
Microbes have impacted our daily life in many ways, giving constant interaction in a delicate and complex interrelationship. Typically microorganism is known for their ability to modulate their metabolic composition to tackle fluctuations in environmental conditions [35]. Changes in the metabolome set of microbes offer not only understanding of phenotypic characteristics but also bioactive compounds that are useful to human and animals. On the other hand, numerous studies have indicated the usefulness of microbes as renewable sources for fine chemical productions [36]. In order to enhance the productivity and yield of the desired compounds, understanding and improvement of the cellular system of particular microbes are required [37]. Microbial metabolomics is one of the many platforms dedicated for the study of microbial system. Microbial metabolomics emphasises on the collection of low-molecular-weight metabolites through separation, identification and quantification using series of analytical platforms from any microbes [38, 39]. Namely, microbial metabolomics tackles two aspects of microbial systems. Firstly, metabolic footprinting or exo-metabolomics that focuses on the excretion of cells into the extracellular surrounding. The second aspect is known as metabolic fingerprinting or intracellular metabolites analysis which deals with the cellular metabolism or metabolites found inside the cell. Both aspects are equally important to provide key information that contributes to the understanding of any microbial systems [40]. However, in comparison with plant metabolomics, the use of metabolomics to study microorganisms is relatively late. Nevertheless, analysis of microbial system by methods of metabolomics has gained increased attention and has considered essential for understanding the cellular functions [41, 42].
E. coli is among the earliest microorganisms that has been used in numerous biotechnology approaches and plays an important role in the development of microbial metabolomics studies [43]. E. coli has served as a valuable model for microbial system, and several metabolomics studies aimed at the nontargeted approach to study metabolites in E. coli have been reported [44]. Meanwhile lactic acid bacteria (LAB) have gained significant interests in systems biology studies because of its generally recognized as safe (GRAS) status and extensive usage in fermentation process. More importantly, various beneficial effects to human health such as anti-inflammatory and allergic have been attributed by LAB via its probiotic effects [45]. Specifically, the global metabolomics studies of LAB have been frequently reported [46,47,48]. For example, the differences in the phenotypic characteristics of Lactococcus lactis strains isolated from different environments showed that each strain requires different amino acids for proper growth [49].
Beside E. coli and LAB, yeasts such as Saccharomyces cerevisiae [50] and filamentous fungi are also known for their impact on human and have been manipulated for the production of bioactive secondary metabolites using methods of metabolomics [51]. Recently, the interest of using filamentous fungi to produce high-valued metabolites has been reported. The various ecological niches and different life styles of filamentous fungi have highlighted the potential of this microorganism to produce a great number of secondary metabolites [52]. For example, the filamentous fungi are source of bulk and fine chemicals of therapeutic compounds that can be used to treat human and plant diseases [53, 54]. However, the global metabolite profiling of yeasts and filamentous fungi has not been frequently described. Furthermore, like other microorganisms, understanding and utilisation of yeasts and fungi require comprehensive strategy such as identification and informatics to increase its usefulness in biotechnology applications.
4.3 Metabolomics Workflow
In the past 10 years, metabolomics has progressively made improvements related to metabolomics software as well as hardware with an increasing complexity of tools and applications. For the discovery study without prior knowledge on the composition of metabolites in the sample, untargeted analytical techniques could be applied to detect a plethora of metabolites. In this case, metabolite identification is crucial following data procurement and processing. Nowadays, the identification of metabolites in untargeted metabolomics studies is a significant bottleneck in metabolomics. To grab the whole concepts of metabolomics, it is important to highlight the workflow of metabolomics.
4.3.1 Experimental Design
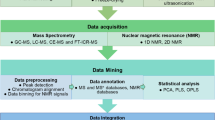
Figure 4.2 shows the metabolomics workflow, which starts with a biological question, followed by experimental design , data collection and analysis and finally biological interpretation. Data collection and analysis are the key components in metabolomics workflow. The two components tackle both qualitative and quantitative issues that strengthen the biological interpretation of the final results.
Metabolomics approach is carried out to estimate the effects of treatment and differences between groups and subsequently to know the cause of such differences. Some typical questions that are tackled in metabolomics studies are the following:
-
Are there any differences between samples?
-
What are the differences between samples?
-
What are the reasons for the differences?
To tackle such problems, metabolomics requires maximum information extracted from the subject, taking into account all factors including biasness and variation. Designing an effective experimental workflow to confer both biological questions and variation is one of the vital aspects in metabolomics. Besides that, experimental workflow should also focus on the sample preparation and extraction, which deal with complex biological matrices and sample-size determination.
4.3.2 Data Acquisition
Metabolomics approach has been applied in various fields including medical, synthetic biology, plant, animal and microbial systems. The metabolome represents the changes in phenotype and function in a biological system. Metabolomics studies involve high-resolution analysis using high-throughput analytical tools for simultaneous determination and quantification analysis of metabolites [38]. In particular, the advent of analytical tools has contributed to the generation of biochemical information or metabolome data [55]. The use of analytical tools in metabolomics contributes towards classification and quality control based on the chemical components reported in the chromatogram and mass spectrum [56]. Since the goal of metabolomics is to profile all metabolites in biological samples, metabolomics greatly depends on the application of analytical instruments for acquisition and identification of metabolome data [57]. Gas chromatography-mass spectrometry (GC-MS) is the most favourable analytical tool used in metabolomics studies. GC-MS gives high peak capacity, excellent repeatability, vast and readily available electron ionised (EI) compound libraries and relatively easy instrument handling. GC-MS requires samples to be derivatised or volatilised for detection. Compound identification using GC-MS is straightforward due to the availability of compound library. Identification is based on the generated mass spectrum comparison against standard mass spectrum from other libraries such as the National Institute of Standards and Technology (NIST) mass spectral library.
On the other hand, compared to GC-MS, liquid chromatography-mass spectrometer (LC-MS) detects a wider range of metabolites, from high- to low-molecular-weight compounds as well as hydrophilic and hydrophobic metabolites. LC-MS largely depends on column specification and mobile phase selection for the separation of compounds. Electrospray ionisation (ESI) is generally used for ionisation source in the LC-MS instrument. Moreover, LC-MS does not require derivatisation or special preparation for sample analysis as needed by GC-MS. However, LC-MS gives insufficient peak capacity and stability, mainly due to matrix effect. Despite the disadvantages, LC-MS has been widely opted for simultaneous analysis of both primary and secondary metabolites. Apart from chromatographic techniques, capillary electrophoresis coupled to mass spectrometer (CE-MS) [5] has been introduced to examine metabolites of amino acids, glycolytic system, pentose phosphate pathway and tricarboxylic acid (TCA) cycle. CE-MS is favourable over normal high-pressure liquid chromatography (HPLC) due to its separation capacity. However, CE-MS is also known to have low repeatability and easily influenced by temperature changes. Spectroscopy-based analytical tools such as nuclear magnetic resonance (NMR) and Fourier-transform infrared spectroscopy (FTIR) have also been applied in metabolomics studies. These tools detect specific resonance absorption profiles of metabolites using magnetic field and infrared (IR), respectively [58].
4.3.3 Data Analysis
Data analysis is a crucial process in metabolomics studies. The process aims at finding significant changes and validates the obtained data. Large data or often referred to as metadata are obtained using various analytical platforms and tools. Data analysis strategies in metabolomics studies can be divided into two, namely, the nontargeted and targeted approaches [2]. Additionally, nontargeted may be referred to as chemometric metabolomics where patterns from a sample are processed and significant differences are identified. Targeted approach is mainly referred to as quantitative analysis. Compounds are firstly identified and quantified [59].
4.3.4 Nontargeted Approach
In this step, all possible metabolites are processed prior to identification. Nontargeted or untargeted approach deals with detection of as many groups of metabolites as possible to get patterns or fingerprints. An organised two-dimensional data matrix consisting of metabolites id (retention time (Rt), m/z value) and quantitative variables (peak height, peak area, etc.) needs to be generated. To obtain this information, data alignment is required. Alignment is usually carried out using Rt or binning of m/z values. In addition, alignment using internal standard Rt may reduce variation among samples and subsequently missing values. Generally, nontargeted approach deals with extremely large data matrices. Therefore, visualisation using multivariate statistical analysis (MVA) is performed to extract interesting and meaningful metabolites from the data matrices. Principal component analysis (PCA) is a non-supervised MVA approach that has been used in data mining of metabolomics . PCA uses score and loading matrices to explain the basis of the variance within the data. By comparing these matrices, the relationship and identification of metabolites that contribute to the differences among samples can be obtained. Apart from that, partial least squares discrimination analysis (PLS-DA) and orthogonal partial least squares discrimination analysis (OPLS-DA) could be used to discriminate important metabolites with biomarker potential [60].
4.3.5 Targeted Approach
Metabolites obtained from nontargeted approach can potentially be false positive or false negative due to analysis procedure and bias. Therefore, extraction of significant metabolites using appropriate methods is required for biological interpretation. On the other hand, targeted approach requires confirmation of significance and reliability of peaks that have been annotated according to its metabolite id. High reproducibility of data matrices using quantification method is important for targeted approach. Quantification can be carried out using calibration curve of pure standard or calculation against internal standard. Similar to nontargeted approach, tools of MVA are also employed for analysis of targeted approach. PCA, PLS-DA and OPLS-DA are used as major MVA tools in metabolomics studies. Finally, in order to obtain the biological interpretation from MVA, additional bioinformatics tools are being integrated into metabolomics studies. Metabolite set enrichment analysis (MSEA), for example, is being utilised to identify and interpret patterns of metabolite changes in biologically meaningful context [61].
4.4 Metabolomics Case Studies
4.4.1 Metabolomics of Kesum Herbs
Saito and Matsuda [6] have described in details how metabolomics technology is emerging and how functional genomics and plant biotechnology can benefit metabolomics technology. Metabolome data driven by systems biology approach have unravelled the secrets of plant cell systems and how it could contribute to the plant biotechnology application. Study on plant species has been carried out in nearly 300,000 species, and yet about 100,000 species still remain unexplored [62]. The overall number of metabolites that exist in plant is about one million [63] showing that it has a plethora of compounds that could be beneficial for pharmaceutical industries. Yet, only a few plants were further explored particularly on their biological activities and chemical constituents of their metabolites [64]. Hence, more research is required in cataloguing the metabolome of unexplored plant species.
Persicaria minor syn. Polygonum minus Huds. or locally known as kesum is often used in various traditional medicines, formulations as well as local cuisines. Throughout the years, compounds from the essential oil of this local herb have been successfully profiled. Metabolites produced by this plant were analysed and identified by analytical pipelines, such as gas chromatography-mass spectrometry (GC-MS) , GC-MS with flame ionisation detector (GC-FID), two-dimensional gas chromatography time-of-flight mass spectrometry (GC × GCTOF MS) and liquid chromatography time-of-flight mass spectrometry (LC-TOF) . A total of 48 compounds using GC × GCTOF MS were successfully identified. By using GC-MS technique, a total of 42 compounds were identified. Hence, the techniques used were also efficient and reliable to identify and quantify the metabolites from Polygonum minus Huds [65].
The effects of temperature in regulating major metabolic profiles in the herbal plants have also been reported. A set of simulated reciprocal transplant experiments was performed to understand the temperature effects on plant metabolites. P. minus plants were harvested after growing in growth chambers in a controlled environment with the temperature set to mimic lowland and highland conditions. GC-MS and LC-TOF were utilised to identify and classify the metabolites that response to the separation between the treatments. A total of 37 volatile compounds by GC-MS analysis were obtained. Meanwhile, LC-TOF successfully identified a total of 85 flavonoids. Aldehydes and terpene groups were accumulated in highlander’s population at treatment of higher temperature. Larger amounts of flavonols were also detected at higher temperature treatment. However, anthocyanin compounds decreased in this treatment. It is possible that the chemical composition was influenced by the effects of temperature on the plant origin [66]. Fourier-transform infrared spectroscopy (FTIR) was also used to characterise different populations of the above-mentioned plant grown in different controlled environments. The plant population was discriminated by using a thermal perturbation technique of 2D-IR correlation spectra (Fig. 4.3). The study implied that IR fingerprinting could directly differentiate the populations of plant origin and also the effect of temperature on the plant’s growth [67].
Contour plot of 2D correlation IR synchronous spectroscopy. Two-dimensional spectra from P. minor treated in different temperature treatments were analysed in the region of 2,000–450 cm−1. (a) Lower growth temperature treatment, (b) control, (c) higher growth temperature treatment. Square boxes indicate autopeaks. Circles indicate crosspeaks. Red colour indicates strong peak, while light blue indicates weak peak. The figure is reprinted from “Direct discrimination of different plant populations and study on temperature effects by Fourier transform infrared spectroscopy”, by K. Khairudin, N. Sukiran, H-H. S.N. Baharum and N. Noor, 2014, Metabolomics, p. 203–211. Copyright 2016 by Springer. Adapted with permission
The metabolite profiles were further investigated based on the different tissues of this herbal plant. Techniques such as solid-phase microextraction (SPME) and hydrodistillation were used before the extracts were subjected to GC-MS analysis. The compounds that contribute to the aroma and flavours of this species have been successfully profiled, which were about 77 metabolites. High levels of terpenoids were also detected in leaves, yet much less were detected in stem and root [68].
Even though the volatile compounds from the essential oil of P. minor have been intensively studied, information contributed to the aroma-active compounds of this plant species was still lacking. Therefore, the aroma-active compounds of the compelling aroma of P. minor were investigated and characterised by using GC-MS/olfactometry (GC-MS/O) and aroma extraction dilution analysis (AEDA) [69]. Based on this finding, several decanal, dodecanal, 1-nonanal, farnesol and α-bergamotene were identified as the key compounds that contributed to the characteristic fragrance of this plant. The finding is important to unravel the information on biosynthesis of aromatic compounds in this herb. Furthermore, this finding also can be further applied in the flavour and fragrance industries.
Further investigation on biological properties of P. minor was also carried out [70, 71]. The study was performed by using essential oil and solvent extracts. Antimicrobial, antioxidant and anticholinesterase activities were investigated, and the plant showed high activity towards antioxidants, particularly in DPPH scavenging activity. Meanwhile, aqueous and methanol extracts from leaf samples were shown to have the best acetylcholinesterase inhibitory activity. The highest antimicrobial activity was detected against methicillin-resistant Staphylococcus aureus (MRSA). These findings could provide us the first phase of phytochemical profiling for the exploration of the value-added pharma-cognitive properties of this plant species.
4.4.2 Lactococcus lactis as a Model for Metabolic Engineering Application
L. lactis has been subjected to several stressful conditions including oxidation, heating and cooling, acid, high osmolality and starvation . Understanding the stress response behaviour is important, not only for strain optimisation but also to manipulate L. lactis as host for heterogeneous compound production. Moreover the stress response mechanism in L. lactis is of the fundamental interest for metabolic engineering application. L. lactis strains are by far the most extensively studied lactic acid bacteria (LAB). Over the last decades, several genetic tools have been developed for this strain. L. lactis strains pose an attractive metabolic regulation that suit the development of effective cell factories [72]. L. lactis strains are suitable for metabolic engineering manipulation because they do not produce endotoxins and are classified as GRAS. L. lactis strains are recognised in two major subspecies: L. lactis subsp. lactis and L. lactis subsp. cremoris. Like other LAB, L. lactis subsp. cremoris is known for its limited biosynthetic capacity to produce essential metabolites and thus explained its complex nutritional requirement [49]. In order to investigate the capability of L. lactis subsp. cremoris to produce specific metabolites to fulfil its growth requirement, the strain was exposed to different temperatures and agitation. Initially, the extracellular profiling of L. lactis cultivated at 30 °C was carried out using Fourier-transform infrared spectroscopy (FTIR). Further investigation was performed on the bacterium that contributes to the production of organoleptic properties by using headspace and gas chromatography-mass spectrometry (HSGC-MS). We found that L. lactis subsp. cremoris produced high levels of 3-methylbutanal, 2-methylbutanal and 2-methylpropanal. These volatile compounds have been reported to influence the aroma, taste and quality of cheese production. Metabolite profiling was carried out on intra- and extracellular of L. lactis when exposed to different conditions. Both temperatures, 37 °C without agitation and 30 °C with agitation (150 rpm), were chosen as the preferred conditions to grow L. lactis because these conditions have not been tested for its metabolite profiling but have been explored for transcriptome analysis of L. lactis [47].
The growth of L. lactis subsp. cremoris MG1363 at 30 °C with and without agitation and at 37 °C was monitored using plate counts (cfu/mL) and optical density (OD600). Specifically, the growth curve for all growth conditions can be characterised into mid- exponential, around 3–5 h, early stationary phase after 6–7 h and finally entered stationary phase around 8 h. During exponential phase, the carbon source, glucose, was converted into biomass and fermentation products of lactate, acetate, ethanol and carbon dioxide (CO2). Decreasing exponential phase took place after 6 h after cultivation. The exponential growth phase is generally considered as the linear part of the growth curve. During this stage, cells are assumed to be in steady state. The exact period of the exponential growth phase determined the right sampling point/time for metabolite and fluxome analysis because, during this stage, all of the intermediate concentration and fluxes are assumed constant. Therefore, in batch cultures, a metabolic steady state exists during exponential growth where the growth rate is constant. In addition, a few studies have shown that exponentially growing cells are much more sensitive to environmental stresses such as during starvation, temperature response, accumulation of acid and ethanol and osmotic and oxidative stresses that their stationary phase counterparts.
Generally, the main factors that affect the characteristics of the metabolic contents in fermented products are the ingredients, fermentation techniques and manufacturing practice. In this study these factors are characterised by the used medium , M17, the tested conditions (30 °C with and without agitation and 37 °C without agitation) and finally detection tools. Nonetheless, the metabolic contents should be strictly associated with the physiochemical composition of the medium. Meanwhile, the environmental adaptability of microbial is resulted from the genetic information, controlled by the complex regulatory networks that enable adaptation to a variety of environments. In this study, 61 metabolites were detected using trimethylsilyl (TMS) derivatisation and 44 metabolites using methyl chloroformate (MCF) derivatisations. However only 47 metabolites were statistically validated (P < 0.05), comprising of 13 amino acids, fermentation by-products of acetate and lactate, propanoate metabolism products of acetone and propanoate, butanoate metabolism product of 2,3 butanediol, lipids of butanoic acid, palmitic acid and hexanoic acid.
In order to understand the metabolic changes of L. lactis when exposed to different conditions, correlation analysis was performed to investigate the pairwise relationship of profiled metabolites. Generally, metabolites that are biosynthetically linked were clustered together. Figure 4.4 describes the correlation levels between L. lactis extracellular with significance (P < 0.05). Red colour indicates positive correlation, while green colour exhibits low and negative correlation. Three clusters of metabolites (A, B, C) were obtained from the correlation network. Specifically clusters A and cluster B were linked by butanoic acid. Cluster A was composed of metabolites that are associated with phenylalanine metabolism, such as aniline, benzoate and phenylacetic acid; amino acids and biogenic amines of aspartate, serine, GABA and putrescine; fatty acid group of 1-hexadecanol; and fermentation by-products of lactate and ethanol . Cluster B contained metabolites such as decanoic acid, heptadecane, 2,4-dihydroxyacetophenone and thioglycolic acid. Meanwhile, cluster C comprised of pyruvic acid, threonine and D-arabinono-1,4-lactone together while by-product fermentation of lactate, ethanol and acetate. In particular, metabolites in cluster C were also of interest because the correlated metabolites were identified as by-products of fermentation and may play major contribution towards the phenotypic characteristic of L. lactis.
Correlation analysis of the 47 extracellular metabolites (P < 0.05) found in all conditions. Green represents correlation coefficient value of >0.7, black represents correlation coefficient value of >0.6 and <0.7 and red represents correlation coefficient value of <0.6. A represents cluster A, B represents cluster B and C represent cluster C
References
Oldiges M et al (2007) Metabolomics: current state and evolving methodologies and tools. Appl Microbiol Biotechnol 76:495–511
Putri SP et al (2013) Current metabolomics: practical applications. J Biosci Bioeng 115:579–589
Theodoridis G, Gika HG, Wilson ID (2008) LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. TrAC Trends Anal Chem 27:251–260
Lindon JC, Nicholson JK (2008) Analytical technologies for metabonomics and metabolomics, and multi-omic information recovery. TrAC Trends Anal Chem 27:194–204
Weckwerth W, Morgenthal K (2005) Metabolomics: from pattern recognition to biological interpretation. Drug Discov Today 10:1551–1558
Saito K, Matsuda F (2010) Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol 61:463–489 (Merchant S, Briggs WR, Ort D, eds). Annual Reviews. issn 1543–5008, Palo Alto, California, United State of America.
Saito K (2013) Phytochemical genomics—a new trend. Curr Opin Plant Biol 16:373–380
Fiehn O (2002) Functional genomics (Town C, ed, Ch. 11). Springer, Dordrecht, pp 155–171
Serkova NJ, Glunde K (2009) Methods Mol Biol 520:273–295
Liu T et al (2011) Rapid determination of serological cytokine biomarkers for hepatitis B virus-related hepatocellular carcinoma using antibody microarrays. Acta Biochim Biophys Sin 43:45–51
Patterson AD et al (2011) Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res 71:6590–6600
Chen J et al (2009) Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal Chim Acta 650:3–9
Lee JH et al (2011) P117. H+-myo-inositol transporter SLC2A13 as a potential marker for cancer stem cells in an oral squamous cell carcinoma. Oral Oncol 47(Supplement 1):S111
Serkova NJ et al (2008) The metabolites citrate, myo-inositol, and spermine are potential age-independent markers of prostate cancer in human expressed prostatic secretions. Prostate 68:620–628
Spratlin JL, Serkova NJ, Eckhardt SG (2009) Clinical applications of metabolomics in oncology: a review. Clin Cancer Res 15:431–440
Kovac JR, Pastuszak AW, Lamb DJ (2013) The use of genomics, proteomics, and metabolomics in identifying biomarkers of male infertility. Fertil Steril 99:998–1007
Hamamah S et al 1H nuclear magnetic resonance studies of seminal plasma from fertile and infertile men. Int J Gynecol Obstet 43:96–97
Rocha CM et al (2011) Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. J Proteome Res 10:4314–4324
Xu X-H, Huang Y, Wang G, Chen S-D (2012) Metabolomics: a novel approach to identify potential diagnostic biomarkers and pathogenesis in Alzheimer’s disease. Neurosci Bull 28:641–648
Trushina E, Dutta T, Persson X-MT, Mielke MM, Petersen RC (2013) Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and Alzheimer’s disease using metabolomics. PLOS ONE 8:e63644
Carraro S et al (2007) Metabolomics applied to exhaled breath condensate in childhood asthma. Am J Respir Crit Care Med 175:986–990
de Laurentiis G et al (2008) Metabonomic analysis of exhaled breath condensate in adults by nuclear magnetic resonance spectroscopy. Eur Respir J 32:1175–1183
Henry PG, Mochel F (2012) The search for sensitive biomarkers in presymptomatic Huntington disease. J Cereb Blood Flow Metab 32:769–770
Smolinska A, Blanchet L, Buydens LMC, Wijmenga SS (2012) NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: A review. Anal Chim Acta 750:82–97
Wang-Sattler R et al (2012) Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol 8:1–11
Liu G, Snapp HM, Ji QC, Arnold ME (2009) Strategy of accelerated method development for high-throughput bioanalytical assays using ultra high-performance liquid chromatography coupled with mass spectrometry. Anal Chem 81:9225–9232
Kim K et al (2011) Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS 15:293–303
Gomez-Casati DF, Zanor MI, Busi MV (2013) Metabolomics in plants and humans: applications in the prevention and diagnosis of diseases. Biomed Res Int 2013:792527
Herder C, Karakas M, Koenig W (2011) Biomarkers for the prediction of type 2 diabetes and cardiovascular disease. Clin Pharmacol Ther 90:52–66
Brunetti C, George RM, Tattini M, Field K, Davey MP (2013) Metabolomics in plant environmental physiology. J Exp Bot 64:4011–4020
Viant MR, Sommer U (2012) Mass spectrometry based environmental metabolomics: a primer and review. Metabolomics 9:144–158
Mittler R (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7:405–410
Paine JA et al (2005) Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat Biotech 23:482–487
Butelli E et al (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotech 26:1301–1308
Merlo ME, Jankevics A, Takano E, Breitling R (2011) Exploring the metabolic state of microorganisms using metabolomics. Bioanalysis 3:2443–2458
Gaspar P, Carvalho AL, Vinga S, Santos H, Neves AR (2013) From physiology to systems metabolic engineering for the production of biochemicals by lactic acid bacteria. Biotechnol Adv 31:764–788
Toya Y, Shimizu H (2013) Flux analysis and metabolomics for systematic metabolic engineering of microorganisms. Biotechnol Adv 31:818–826
Mozzi F, Ortiz ME, Bleckwedel J, De Vuyst L, Pescuma M (2013) Metabolomics as a tool for the comprehensive understanding of fermented and functional foods with lactic acid bacteria. Food Res Int 54:1152–1161
Zhang W, Li F, Nie L (2010) Integrating multiple ‘omics’ analysis for microbial biology: application and methodologies. Microbiology 156:287–301
Mapelli V, Olsson L, Nielsen J (2008) Metabolic footprinting in microbiology: methods and applications in functional genomics and biotechnology. Trends Biotechnol 26:490–497
Liebeke M, Dörries K, Meyer H, Lalk M (2012) Functional genomics: methods and protocols (Kaufmann M, Klinger C, eds). Springer, New York, pp 377–398
Mashego MR et al (2006) Microbial metabolomics: past, present and future methodologies. Biotechnol Lett 29:1–16
Rabinowitz JD (2007) Cellular metabolomics of Escherichia coli. Expert Rev Proteomics 4:187–198
Winder CL et al (2008) Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites. Anal Chem 80:2939–2948
Ménard S et al (2004) Lactic acid bacteria secrete metabolites retaining anti-inflammatory properties after intestinal transport. Gut 53:821–828
Azizan KA, Baharum SN, Mohd Noor N (2012) Metabolic profiling of Lactococcus lactis under different culture conditions. Molecules 17:8022
Taibi A, Dabour N, Lamoureux M, Roy D, LaPointe G (2011) Comparative transcriptome analysis of Lactococcus lactis subsp. cremoris strains under conditions simulating Cheddar cheese manufacture. Int J Food Microbiol 146:263–275
Tan-a-ram P et al (2011) Assessment of the diversity of dairy Lactococcus lactis subsp. lactis isolates by an integrated approach combining phenotypic, genomic, and transcriptomic analyses. Appl Environ Microbiol 77:739–748
Ayad EHE, Verheul A, de Jong C, Wouters JTM, Smit G (1999) Flavour forming abilities and amino acid requirements of Lactococcus lactis strains isolated from artisanal and non-dairy origin. Int Dairy J 9:725–735
Schneider K et al (2009) Metabolite profiling studies in Saccharomyces cerevisiae: an assisting tool to prioritize host targets for antiviral drug screening. Microb Cell Factories 8:1–14
Smedsgaard J, Nielsen J (2005) Metabolite profiling of fungi and yeast: from phenotype to metabolome by MS and informatics. J Exp Bot 56:273–286
Kluger B, Lehner S, Schuhmacher R (2015) Metabolomics and secondary metabolite profiling of filamentous fungi. In: Zeilinger S, Martín J-F, García-Estrada C (eds) Biosynthesis and molecular genetics of fungal secondary metabolites, vol 2. Springer, New York, pp 81–101
Barkal LJ et al (2016) Microbial metabolomics in open microscale platforms. Nat Commun 7:1–11
Thrane U, Anderson B, Frisvad JC, Smedsgaard J (2007) Metabolomics: a powerful tool in systems biology (Nielsen J, Jewett MC, eds). Springer, Berlin/Heidelberg, pp 235–252
Zurbriggen MD, Moor A, Weber W (2012) Plant and bacterial systems biology as platform for plant synthetic bio(techno)logy. J Biotechnol 160:80–90
Zhao Y et al (2011) Tentative identification, quantitation, and principal component analysis of green pu-erh, green, and white teas using UPLC/DAD/MS. Food Chem 126:1269–1277
Putri SP, Yamamoto S, Tsugawa H, Fukusaki E (2013) Current metabolomics: technological advances. J Biosci Bioeng 116:9–16
Mouwen DJM, Hörman A, Korkeala H, Alvarez-Ordóñez A, Prieto M (2011) Applying Fourier-transform infrared spectroscopy and chemometrics to the characterization and identification of lactic acid bacteria. Vib Spectrosc 56:193–201
Xia J, Wishart DS (2010) MSEA: a web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 38:W71–W77
Zeng M et al (2010) Plasma metabolic fingerprinting of childhood obesity by GC/MS in conjunction with multivariate statistical analysis. J Pharm Biomed Anal 52:265–272
Xia J, Wishart DS (2010) MSEA: web-based tool to identify biologically meaningful patterns in quantitative metabolomic data. Nucleic Acids Res 38:71–77
Sticher O (2008) Natural product isolation. Nat Prod Rep 25:517–554
Dixon RA, Strack D (2003) Phytochemistry meets genome analysis, and beyond. Phytochemistry 62:815–816
Yamada T et al (2008) Mutation of a rice gene encoding a phenylalanine biosynthetic enzyme results in accumulation of phenylalanine and tryptophan. Plant Cell 20:1316–1329
Baharum SN, Bunawan H, Ghani MA, Wan Aida Wan M, Noor NM (2010) Analysis of the chemical composition of the essential oil of Polygonum minus Huds. Using two-dimensional gas chromatography-time-of-flight mass spectrometry (GC-TOF MS). Molecules 15:7006–7015
Goh HH, Khairudin K, Sukiran NA, Normah MN, Baharum SN (2016) Metabolite profiling reveals temperature effects on the VOCs and flavonoids of different plant populations. Plant Biol 18:130–139
Khairudin K, Sukiran N, Goh H-H, Baharum S, Noor N (2013) Direct discrimination of different plant populations and study on temperature effects by Fourier transform infrared spectroscopy. Metabolomics 10:203–211
Ahmad R et al (2014) Volatile profiling of aromatic traditional medicinal plant, Polygonum minus in different tissues and its biological activities. Molecules 19:19220
Azizun Rusdi N, Goh HH, Baharum S (2016) GC-MS/Olfactometric characterisation and aroma extraction dilution analysis of aroma active compounds in Polygonum minus essential oil. Plant Omics 9:289
Hassim N et al (2015) Antioxidant and antibacterial assays on Polygonum minus extracts: different extraction methods. Int J Chem Eng 2015:10
Hassim N, Markom M, Anuar N, Baharum SN (2014) Solvent selection in extraction of essential oil and bioactive compounds from Polygonum minus. J Appl Sci 14:1440–1444
Neves AR, Pool WA, Kok J, Kuipers OP, Santos H (2005) Overview on sugar metabolism and its control in Lactococcus lactis—the input from in vivo NMR. FEMS Microbiol Rev 29:531–554
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Baharum, S.N., Azizan, K.A. (2018). Metabolomics in Systems Biology. In: Aizat, W., Goh, HH., Baharum, S. (eds) Omics Applications for Systems Biology. Advances in Experimental Medicine and Biology, vol 1102. Springer, Cham. https://doi.org/10.1007/978-3-319-98758-3_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-98758-3_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-98757-6
Online ISBN: 978-3-319-98758-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)