Abstract
In the first chapter, entitled Fundamentals of Cardiac T1 Mapping, we present an overview of the scientific principles, technical choices and challenges associated with cardiac T1 mapping. This chapter reminds the reader that T1 mapping relies on a physical model of the MR signal, and shows the ways in which this model breaks down when the underlying assumptions are not met.
We start from first principles and introduce the basics of body T1 mapping before moving onto the most widely used cardiac sequences: MOLLI and ShMOLLI (which are based on a Look-Locker sequence) and SASHA (based on a saturation recovery sequence). We take a particularly close look at the confounding factors that might impact T1 mapping results in practice.
Next, we give key insights into the T1 mapping post-processing techniques, which are often used to communicate T1 mapping results to radiologists and cardiologists. Finally, we emphasize the need for proper validation, provide suggestions for standardizing the field, and end the chapter with recommendations for the clinical application of cardiac T1 mapping.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
Magnetic resonance imaging (MRI) has revolutionized the way we visualize and understand the human body. MR image acquisition enables clinicians and scientists to tweak a number of parameters (e.g., repetition time, echo time, flip angle) to generate unique tissue contrast. Each combination of parameters leads to a wealth of information about the tissue microstructure, yet reverse engineering the tissue makeup from MR images is not an easy task.
The fundamental contrast mechanisms that produce MR images are the longitudinal and the transverse relaxations, characterized by the T1 and T2 parameters, respectively. The MR signal is the product of the interaction between billions of spins (atoms with a magnetic moment) over a macroscopic volume (on the order of millimeters cubed). T1 and T2 share a complex relationship, making it difficult to isolate their individual contributions to the MR signal and understand exactly how many different spin populations are being imaged, as well as what their relative contributions are.
Quantitative MRI attempts to make sense of this wealth of information using biophysical models that relate the MR signal to the tissue makeup. In this chapter we will focus on the fundamentals of T1 mapping.
T1
Hydrogen atoms (H) exhibit nuclear magnetic resonance, and water molecules (H2O) are the source of most of the signal in MRI. We distinguish between free water, where motion is unhindered, structured water, where water is bound to a macromolecule by a single hydrogen atom, and bound water, where water is bound to a macromolecule by both hydrogen atoms [1].
In the absence of an external magnetic field, spins are randomly oriented and the net magnetization is zero. In an applied magnetic field (1.5T or 3T for clinical scanners), spins align with the applied field, contributing to a non-zero magnetization. From a classical physics perspective, spins precess around the applied field at the Larmor frequency, which is proportional to the field strength. If an ensemble of spins is now excited by a radiofrequency (RF) pulse at the Larmor frequency, the magnetization is perturbed. In a classical representation, the RF pulse tips the magnetization away from equilibrium. The longitudinal component of the magnetization exponentially returns to equilibrium it relaxes with a time constant T1. Precession and relaxation are embedded in the Bloch equations, which describe the evolution of the magnetization over time. It is important to keep in mind that the Bloch equations are phenomenological: they agree with experience but cannot be entirely derived from first principles. They are also macroscopic, which is why we always refer to an ensemble of spins.
T1 is known as the longitudinal relaxation time constant, the relaxation in the z-direction, or the spin-lattice relaxation time constant. The term “lattice” comes from the early days of Nuclear Magnetic Resonance (NMR), where relaxation back to equilibrium was explained in solids in terms of interactions between the nuclear spins and the crystal lattice. We can still talk about T1 as a spin-surroundings relaxation time where the surroundings are the local environment of the spin. T1 relaxation occurs because of local magnetic field fluctuations due to molecular motion (tumbling) and is the most efficient (shortest T1) when the fluctuations are near the Larmor frequency, which is the case for soft tissue (structured water) but not for liquids (free water) or solids (bound water). For bound water, T1 decreases when the temperature increases because the higher temperature breaks the bonds and allows faster molecular motion. For in vivo imaging of soft tissue at common field strengths, T1 increases when the temperature increases [1]. T1 decreases in the presence of gadolinium-based contrast agents because gadolinium creates strong local magnetic field fluctuations at the Larmor frequency.
T1 Outside of the Heart
T1-weighted imaging provides an image with arbitrary units, where contrast can only be described by comparison between different tissues or with respect to a reference tissue in that same image, in a qualitative manner. T1 mapping produces an image, the map, in which each pixel represents the measured T1 value at that location (measured in milliseconds, see Fig. 1.7), where contrast can be described in an absolute, quantitative manner.
Fast, accurate and precise T1 mapping is never and nowhere trivial. Nevertheless, T1 mapping has been successfully used in the brain to study patients with Parkinson’s disease, multiple sclerosis, stroke, schizophrenia and HIV [2]. It has yet to become part of routine clinical evaluation, partially due to the long scan times. Recently, there have been efforts to standardize the field through the implementation of vendor-specific relaxometry techniques. Cardiac T1 mapping has been leading the way in these efforts, even though cardiac and respiratory motion present major challenges. This apparent paradox might be because the arsenal of pulse sequences is more limited in the heart, making it more difficult to relate the MR signal to physiology without explicit quantification.
T1 in the Heart
Figure 1.1 shows a schematic of the composition of the heart tissue. Myocytes (muscle cells) make up 75% of the volume of the heart. The remaining 25% constitute the extracellular space, which is made of fibroblasts and collagen, other glycoproteins and proteoglycans, as well as blood vessels (smooth muscle cells and endothelial cells) [3, 4]. Each of these components has a specific T1 and contributes in a unique way to the MR signal.
Native T1
Cardiac MRI has seen tremendous growth over the past 20 years, with a recent focus put on the ability to relate macroscopic changes in the MRI signal to tissue pathology.
In the myocardium, many factors (e.g., cell volume, edema, infiltration, scarring, fibrosis) contribute to the MR signal, so it is difficult to determine the specificity of T1 to any particular spin population. Even in a single voxel, multiple tissue compartments (homogeneous “buckets” of spins) contribute to the signal. For example, studies have explored the relationship between T1 and collagen in the myocardium of canine specimens and found a moderate negative correlation (r = −0.45) between T1 and the hydroxyproline concentration (a measure of collagen) [5]. However, this correlation is primarily driven by bulk differences in the collagen concentration between the atria and the ventricles, so it is not clear whether (native T1) can be used to discriminate between more subtle changes in collagen content that occur during scarring and fibrosis. Additionally, T1 is significantly influenced by water content, so any T1 measurements need to be controlled for inflammation and hydration.
Native T1 can still be helpful clinically, even when the underlying contrast mechanism is not fully understood. For example, recent data indicates that native T1 may allow to differentiate normal myocardium not only from acute injury with edema but also from scarring in myocardial infarction and myocarditis [6, 7]. It is also the method of choice when the risks of contrast enhancement (gadolinium side effects) outweigh the benefits. For example, contrast enhancement is contraindicated in patients with chronic kidney disease, but it is precisely these patients that are at a much higher risk of cardiovascular disease than the general population.
Late Gadolinium Enhancement
Unlike native T1 mapping that provides a narrow dynamic range in the myocardium, late gadolinium enhancement (LGE) can produce significant shortening of T1 in regions of infarction [8,9,10]. Gadolinium is considered an extracellular contrast agent due to its ability to diffuse from the vascular space into the extracellular tissue fluid without affecting the intracellular space. As gadolinium accumulates in infarcted tissue, it can be used as a tracer sensitive to collagen and therefore scarring and fibrosis. The decrease of T1 associated with gadolinium administration has been used to measure the extracellular volume (ECV) [11], as described below, as well as to correlate ECV with the collagen volume fraction [12].
Extracellular Volume Calculation
ECV is computed by measuring T1 before and after gadolinium administration. The difference in T1 pre- and post-contrast is interpreted as a measure of the amount of collagen present in the myocardium, because other tissues in the myocardium are not affected by gadolinium, which stays in the extracellular space. Therefore, an ECV map is physiologically relevant and easier to interpret than a T1 map. The ECV measurement assumes that the transfer rate of gadolinium between blood and tissue is much faster than the removal of the gadolinium from the blood pool. The post-contrast T1 value is highly dependent on (1) gadolinium dose, (2) time post bolus, which is hard to control in practice, (3) clearance rate, which is influenced by the cardiac output, and (4) hematocrit (ratio of the volume of red blood cells to the total volume of blood), because gadolinium is present in plasma but does not enter red blood cells. The latter is particularly important for normalizing the ECV and producing values that are comparable across patients. Recently, a method has been proposed where the hematocrit can be determined based on blood T1 [13], but it is typically obtained through an independent blood test.
In a 2-compartment model, ECV can be computed according to
where Hct stands for hematocrit.
Because of imperfections in T1 mapping, the ECV calculation is sequence-dependent and therefore difficult to standardize. In practice, however, ECV appears more robust than post-contrast T1, and it has shown prognostic value [14]. Therefore, efforts have been made towards its standardization, as evidenced by the consensus statement of the Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement [15]. A scientific consensus has the potential to make ECV the gold standard for the assessment of focal fibrosis. ECV can also help discriminate between non-focal expansion of the extracellular space and a sequence-dependent bias in T1, making it a candidate for evaluating diffuse fibrosis [16].
T1 Mapping Gold Standard
Inversion recovery T1 mapping was first performed in the late 1940s for NMR experiments. It consists in inverting the longitudinal magnetization and sampling it as it recovers toward equilibrium with a time constant T1. There is a consensus among researchers that gold standard T1 mapping uses inversion recovery (IR) pulse sequences with long repetition times. However, many different sequences and fitting techniques are used in practice, which can lead to a wide range of “gold standard” T1 values. This discrepancy in the gold standard makes it difficult to validate new T1 mapping sequences, as there is a large variability of reference T1 values in literature. We have investigated this problem in detail, developing a robust methodology for in vivo T1 mapping, open source code for data fitting, as well as reference data sets [17]. We expect to see additional efforts in this direction, in keeping with the concepts of open science and reproducible research [18].
The key in the development of T1 mapping techniques is to start from the Bloch equations and derive the complete signal equation keeping simplifications to a minimum. All assumptions should be stated so that further simplifications can be justified. Such simplifications are often needed to come up with a model that can be more easily fitted. Anyone using a given T1 mapping technique should ensure that the assumptions are met. For example if the model assumed TR much greater than T1, one should at least check that the value of T1 obtained is indeed much smaller than TR. This check is a necessary, but not a sufficient, condition to ensure that the model holds. The new technique should also be compared to the gold standard in simulations and phantom scans, so that expected precision and accuracy are known.
Let us illustrate this approach for gold standard T1 mapping. Consider a spin echo IR sequence\( \left\{{\theta}_1- TI-{\theta}_2-\frac{TE}{2}-{\theta}_3-\left( TR- TI-\frac{TE}{2}\right)\right\} \) whereθ 1, θ 2 and θ 3 are RF pulses, typically prescribed as 180°, 90° and 180°, respectively, TI is the inversion time, TR the repetition time, and TE the echo time. If we assume instantaneous pulses, perfect spoiling of M xy after θ 1 and no off-resonance effects, then sampling the magnetization at different inversion times TI n leads to the following data equation for the received signal:\( S\left(T{I}_n\right)={e}^{i\phi}\left({r}_a+{r}_b{e}^{-\kern0.5em \frac{T{I}_n}{T1}}\right) \), which has fourreal-valued unknown parameters (ϕ, r a, r b, T1). This signal equation can be simplified to a different four-parameter model if TR is much greater than T1 or to a three-parameter model if TR is much greater than T1 and θ 1 = 180°.
Model | Assumptions |
|---|---|
\( S\left(T{I}_n\right)={e}^{i\phi}\left({r}_a+{r}_b{e}^{-\kern0.5em \frac{T{I}_n}{T1}}\right) \) | • Instantaneous pulses • Perfect spoiling of M xy after θ 1 • No off-resonance effects |
\( S\left(T{I}_n\right)=c\left(1-\left(1-\cos \left({\theta}_1\right)\right){e}^{-\kern0.5em \frac{T{I}_n}{T1}}\right) \) | • All of the above • TR much greater than T1 • Knowledge of the absolute timing of TI n |
\( S\left(T{I}_n\right)=c\left(1-2{e}^{-\kern0.5em \frac{T{I}_n}{T1}}\right) \) | • All of the above • θ 1 = 180° |
Let us, for example, examine the assumption θ 1 = 180° (from the three-parameter model) a bit closer. Even if an adiabatic RF pulse is used [19], the effective flip angle depends on T1 and T2, making it impossible to obtain a perfect 180° inversion. Figure 1.2 illustrates the effects of T1 and T2 for muscle at 1.5T: the flip angle is about 155° instead of the prescribed 180°, which translates into the correct signal equation being \( S\left(T{I}_n\right)=c\left(1-1.9{e}^{-\kern0.5em \frac{T{I}_n}{T1}}\right) \) vs\( S\left(T{I}_n\right)=c\left(1-2{e}^{-\kern0.5em \frac{T{I}_n}{T1}}\right) \); the effect is therefore farfrom negligible! In addition the transition bands of the inversion pulse are often partially included in the imaging slice, and the effective flip angle should be taken as the integral of the inversion profile over the slice thickness.
A Silver-Hoult adiabatic inversion pulse of length 8.64 ms was simulated, with a prescribed slice thickness of 2 mm. T1 and T2 values of muscle at 1.5T were used [20]. The pulse profile is shown (left), with a zoom on the passband (right), illustrating a ~15% discrepancy introduced by ignoring T1 and T2 effects
Once an appropriate signal equation is used, the fitting procedure should be carefully considered. Often a Levenberg-Marquardt algorithm is used and initialization of the parameters is required, which may bias the results [21]. We have proposed an alternative algorithm, which optimizes the precision and accuracy of the T1 estimation and is much faster than Levenberg-Marquardt [17]. The quality of the fit should be checked visually in different regions of interest (Fig. 1.3) to make sure that the fitting line indeed goes through (or is close to) the sampling points. Alternatively, metrics like the goodness of fit or the error (uncertainty) map can be inspected.
It is important to understand the influence of the signal-to-noise ratio (SNR) on the fitting performance. For in-vivo experiments, time is always critical and SNR is the obvious trade-off. Many sampling strategies overlook the fact that having more points on the curve may not help if each point has a lower SNR. For gold standard T1 mapping, we recommend using four points corresponding to inversion times TIs of 50, 400, 1100 and 2500 ms. One should also keep in mind that in a given T1 map the precision of the T1 estimation is worse for tissues with large T1 values.
T1 Mapping in the Heart
Figure 1.4 summarizes four T1 mapping techniques, one that is considered the gold standard (inversion recovery spin echo), and three that are commonly used for cardiac T1 mapping (MOLLI, ShMOLLI and SASHA). While there are a number of other cardiac T1 mapping sequences being developed, we focus on those that are readily available as product sequences on a clinical MRI scanner. With an inversion recovery gold standard pulse sequence, a single line of k-space is acquired every TR and TR is long (on the order of T1, typically a few seconds) to enable sufficient recovery of the magnetization before each inversion pulse [17]. For example, with a TR of 2550 ms, and a 192 × 144 matrix size (typical for a cardiac T1 mapping acquisition), a single slice gold standard acquisition takes approximately 6 min per inversion time, i.e., 6 min per point on the curve that will be fitted to derive T1.
To perform T1 mapping in the heart, both cardiac and respiratory motion have to be taken into account. If the acquisition is gated, the same phase of the cardiac cycle can be obtained every TR, and cardiac motion is less of an issue as long as the patient does not suffer from arrhythmias. Respiratory motion is mitigated by breath holding, which constrains the full acquisition (i.e., the acquisition of all the points) to be shorter than a breath hold, i.e., less than about 20 s. Another mitigation strategy is to resolve the respiratory motion either prospectively or retrospectively. If a prospective approach is taken, the sequence can be gated. If a retrospective approach is taken, one has to either (1) ensure that k-space was fully sampled or (2) use compressed sensing or a similar technique for reconstruction [22]. Approaches combining prospective and retrospective navigation schemes have also been proposed [23].
Another challenge specific to T1 mapping in the heart is the need to be accurate over the wide range of T1 and T2 values found in blood and tissue pre- and post-contrast.
Look-Locker Techniques: MOLLI and ShMOLLI
The Look-Locker (LL) method is a rapid technique that measures T1 from a single recovery of longitudinal magnetization. It alleviates the limitation of the conventional IR method of requiring a long delay (on the order of T1) for longitudinal magnetization to recover until the next inversion pulse is played for subsequent readout. This approach was first theorized by Look and Locker and later implemented in the form of TOMROP (T One by Multiple Read Out Pulses) [24, 25]. The basic sequence diagram is shown in Fig. 1.4. It consists of a single inversion pulse followed by a series of very small angle excitation RF pulses α with gradient echo readouts to sample the recovery curve. Since small angle RF pulses are used, the longitudinal magnetization is only minimally disrupted during T1 recovery and sampling is performed in a continuous manner, i.e., no wait time is necessary until equilibrium is reached. However, if the separation between α pulses is less than T2, the T1 signal is corrupted by residual transverse magnetization gathered from previous α pulses. To avoid this corruption, either the spacing between the α pulses needs to be long (>5T2), or gradient spoiling needs to be employed to crush any residual transverse magnetization. It is also important to note that due to continuous perturbation of the magnetization by successive α pulses, the recovery is driven into equilibrium more quickly, resulting in an “effective” T1 or T1* given by \( S\left(T{I}_n\right)=A-B{e}^{-\frac{T{I}_n}{T{1}^{\ast }}} \),where the T1* calculated from the recovery curveneeds to be converted to the “actual” T1 using \( T{1}^{\ast }=\frac{1}{\frac{1}{T1}-\frac{1}{TR}\kern0.5em \mathit{\ln}\ \mathit{\cos}\ \alpha } \) [26].
Model | Assumptions |
|---|---|
\( S\left(T{I}_n\right)=A-B{e}^{-\kern0.5em \frac{T{I}_n}{T{1}^{\ast }}} \) \( T{1}^{\ast }=\frac{1}{\frac{1}{T1}-\frac{1}{TR}\kern0.5em \mathit{\ln}\ \mathit{\cos}\ \alpha } \) | • T1 much greater than TR • Flip angle α less than 10° • TR much greater than T2 |
Recently, several variants of the LL method have been developed for cardiac T1 mapping. Basic LL cannot be applied due to cardiac and respiratory motion, so a Modified LL Inversion recovery sequence (MOLLI) has been proposed for high-resolution T1 mapping of the heart [27]. MOLLI consists of a series of single-shot images acquired in diastole, separated by inversion pulses. Several variants of the MOLLI sequence try to make the most out of a total number of heartbeats available, limited by the breath-hold duration. The first of these variants was named 3(3)3(3)5 [27]. The name indicates that three images are acquired after the first inversion pulse, three after the second, and five after the third pulse; the number in parenthesis is the number of heartbeats the sequence waits before applying the subsequent inversion pulse. Another common variant of MOLLI is the ShMOLLI sequence (where Sh stands for “Shortened”), which uses nine heartbeats (5(1)1(1)1) [28]. The last two images from ShMOLLI are used only if the T1 of the tissue of interest is short enough to allow near-complete relaxation recovery after the first inversion pulse. For longer T1s, the fitting is done only with the first five images. Note that MOLLI and ShMOLLI use a bSSFP readout and the relationship between T1 and T1* used in LL no longer holds. Empirically, the “effective” T1∗ is converted to the “actual” T1 using\( T1=T{1}^{\ast}\left(\frac{B}{A}-1\right) \) or \( T1=T{1}^{\ast}\left(\frac{B}{A}-1\right)\frac{1}{\theta_1} \) tocompensate for the imperfect adiabatic inversion θ 1 [29, 30].
Saturation Recovery Techniques: SASHA
Despite several acceleration strategies implemented in MOLLI/ShMOLLI, IR-based techniques require long wait times for magnetization recovery. To avoid these wait times SASHA is based on saturation recovery instead of inversion recovery (Fig. 1.4). The SASHA pulse sequence images a single slice over ten heartbeats, at a specific time of the cardiac cycle. A full image, corresponding to one point on the recovery curve, is acquired every heartbeat. A saturation pulse is added and its position in the cardiac cycle varies from heartbeat to heartbeat so that each image is acquired at a different point of the recovery curve. Using a saturation pulse erases the prior history: there is no need to wait for full recovery as in inversion recovery techniques and TR can therefore be much shorter. In the original SASHA article [31], the saturation recovery times uniformly span the cardiac cycle. No saturation pulse is added before the first image acquisition so that a fully recovered image is obtained. Each image is a single-shot bSSFP image, with an acquisition window of about 175 ms. The signalequation can be written as \( S=A\ \left(1-{\eta}_{apparent}{e}^{-\kern0.5em \frac{TS}{T1}}\right) \) where
-
A is a scaling factor
-
TS is defined as the time between the end of the saturation RF pulse and the center line of k-space
-
\( {\eta}_{apparent}=\frac{a}{a+b}{e}^{\frac{\varDelta }{T1}}{\eta}_{actual} \) is the apparentsaturation efficiency, where
-
η actual is the actual saturation efficiency
-
a and b are functions of the acquisition parameters as well as T1
-
Δ is the time from beginning of imaging to center of k-space
-
η apparent depends on T1, therefore solving for T1 in the equation above ignoring η apparent is not exact. The assumption is that η apparent does not vary much with T1 and the dependency can be neglected.
The signal equation can also be simplified to a 2-parameter model.
Model | Assumptions |
|---|---|
\( S=A\ \left(1-{\eta}_{apparent}{e}^{-\kern0.5em \frac{TS}{T1}}\right) \) | • η apparent does not vary much with T1 |
\( S=A\left(1-{e}^{-\kern0.5em \frac{TS}{T1}}\right) \) | • Ideal saturation |
Depending on the model used, a 3-parameter or a 2-parameter fit is then performed on the magnitude of the signal intensity, with the 3-parameter fit providing greater accuracy [29]. Note that breath holding is required during the entire pulse sequence.
T1 Mapping Limitations
The table below summarizes the sensitivity of MOLLI/ShMOLLI and SASHA, respectively, to various factors [29, 31, 34, 35]. For SASHA, we only consider the three-parameter model, as the two-parameter model introduces lots of inaccuracies.
Factor | MOLLI/ShMOLLI sensitivity | SASHA sensitivity |
|---|---|---|
T2 | Yes, due to SSFP readout and inversion inefficiency (imperfect 180 and non-zero pulse duration) | Negligible |
Heart rate and cardiac motion | Significant effect of irregular heart rate for long T1 values for which sampling occurs over more heartbeats. It can be mitigated by using a single inversion or increasing the time between inversions. ShMOLLI deals with long T1s by ignoring all but the first inversion | Long acquisition window per image (~175 ms) results in the loss of resolution |
SNR | Higher SNR | Lower SNR because the sequence is saturation-based Magnitude fit instead of complex fit introduces a bias |
Off-resonance | Significant, bigger at higher field strengths and larger flip angles. ±100 Hz can result in a 6% error | Negligible. ±100 Hz results in less than 1% error |
Flip angle | T1 underestimated for larger flip angles | Until SAR limits are reached, a larger flip angle increases the SNR |
Inversion/saturation efficiency | Assumes ideal inversion, source of significant bias. Adiabatic pulses can help | Imperfect slice profile for the saturation, which depends on T1 and T2 |
Fitting model | Three-parameter fit uses magnitude data, problematic when zero crossing is close to the inversion time because SNR is low. Estimating zero crossing requires an additional parameter | Two-parameter model introduces biases, resolved when three-parameter model is used |
Inflow effects (for blood T1 estimation) | More sensitive to blood flow, as it samples the recovery curve at several points after inversion. Mainly affects the longer T1s | Less sensitive as it samples the recovery curve before the non-saturated blood has flowed in |
Partial voluming | Significant for small matrix size and/or large slice thickness (~8–10 mm). Blood contamination can affect the myocardium T1 estimate | |
Magnetization transfer (MT) | Inversion pulses introduce significant MT effect, and T1 is underestimated | Negligible |
Post-Processing
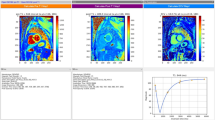
Once a T1 map is obtained, the average T1 value for a region of interest (ROI) is typically computed. Care must be taken in automating the choice of the ROI and the computation of the average T1, so that results are reproducible and no bias is introduced (e.g., due to partial volume effects from blood or epicardial fat). The choice of the segmentation method that provides the ROI should refer to published recommendations. The 17-segment model proposed by the American Heart Association is commonly used [36]. Newer methods based on machine learning look promising but most have yet to be clinically validated [37, 38]. Orientation, number of segments and nomenclature present some variability depending on the application. Figure 1.5 shows a segmental representation of the short-axis view of the left ventricle, as well as the T1 maps obtained using MOLLI, ShMOLLI and SASHA. If an observer notices artifacts (such as susceptibility artifacts), the corresponding ROIs should be excluded from the analysis. Global (averaged over the entire heart) and segmental T1 values can then be compared.
(Top row) Representative cardiac T1 maps in a healthy subject obtained with ShMOLLI, MOLLI and SASHA. (Bottom row) Segmental analysis of the corresponding T1 maps. The details of the acquisition parameters can be found in Teixeira et al. [33]
Even with cardiac gating and breath holding, motion can still be problematic, e.g., because the heart rate is variable, or patients have a hard time holding their breath. Images are often co-registered before the fit, although standard registration techniques may not work well because the different TIs lead to contrast differences. Co-registration is also used before generating an ECV map as the heart position typically moves between pre- and post-contrast acquisitions. Rather than co-registering the T1 maps, co-registration is performed on the individual images before the fit [39].
Validation
Once a new T1 mapping technique has been developed and simulated, its validation is commonly performed with phantoms and histology. Phantom validation should be the first step, and accuracy and precision can be explored using one of the commercially available T1 phantoms, such as the T1MES phantom [32], or Phannie, the phantom developed by the National Institutes of Standards and Technology (NIST) [40]. Note that temperature needs to be controlled when phantoms are scanned as it influences T1.
While most T1 mapping sequences show excellent agreement in phantoms (see Fig. 1.6), this agreement does not necessarily translate to good accuracy and precision in tissue (see Fig. 1.7). A recent study in brain MRI has demonstrated that T1 mapping sequences that agree well in phantoms can show dramatic differences in vivo [41]. These findings have recently been corroborated in the heart [33], demonstrating that in vivo T1 measurements are still confounded by magnetization transfer, RF inhomogeneity and imperfect spoiling, issues that are less prominent in phantoms. Because accuracy and precision are challenging, the focus is often instead put on reproducibility as the minimum requirement for a useful sequence. Reproducibility in cardiac T1 mapping varies but can be controlled for specific tissue/sequence parameters [42]. Precision then allows differentiating between normal and diseased tissue. From a pragmatic standpoint, systematic errors (inaccuracies) can be tolerated as long as the T1 estimate is reproducible and sensitive to pathology.
A comparison of T1 mapping sequences in explanted pig hearts. The IR map was obtained using a turbo spin-echo acquisition and five inversion times, using a slice selective inversion pulse (TI = 33, 100, 300, 900, 2700, 5000 ms; TE/TR = 12 ms/10 s; slice thickness 8 mm; flip angle = 90°; matrix 192 × 144; FOV 360 × 270 mm and turbo factor = 7). The rest of the maps (MOLLI, ShMOLLI and SASHA) were obtained with the protocols published in Teixeira et al. [33]
In vivo validation of T1 mapping sequences is still in its infancy. Late gadolinium enhancement remains the gold standard for non-invasive evaluation of focal fibrosis; native (i.e., non-contrast) T1 mapping and ECV make it possible to quantify the extent of expansion of the extracellular matrix and characterize diffuse fibrosis. Native T1 has been demonstrated to correlate with histology in diffuse fibrosis [43] whereas ECV has been validated with biopsy in patients with severe aortic stenosis and hypertrophic cardiomyopathy [44, 45]. Cardiac T1 mapping is still far from routine clinical use. It is primarily used as a means to observe group differences, or to provide optimal contrast for specific tissues to the radiologist. A truly quantitative approach would allow comparisons between patients, scanners, and sites, as well as across time for a given patient.
Recommendations
A consensus statement on T1 mapping was recently issued by the Society for Cardiovascular Magnetic Resonance (SCMR) and the CMR Working Group of the European Society of Cardiology [15]. Regular updates by these societies are expected. A number of recommendations for performing and standardizing T1 mapping have been provided in that statement, related to terminology, scan type, scan planning and acquisition, site preparation, quality control visualization and analysis, and technical development. It is important that each site be aware of the above recommendations, to enable better standardization of T1 mapping across sites.
T1 mapping including ECV quantification is on its way to becoming an important imaging biomarker in cardiology. It has significant diagnostic and prognostic value. It overcomes limitations of signal-intensity based methods and provides unique information in diseases affecting the myocardial tissue. Current limitations include the variations and lack of standardization among sites, scanners, sequences and evaluation procedures. Even though many diseases (e.g., amyloidosis, myocarditis, Fabry’s disease) have been explored by T1 mapping, data from large scale multicenter trials is lacking, and standards for data acquisition and reference values for normal and diseased myocardium will have to be established before T1 mapping can be more broadly applied. The field continues to evolve in a complex interplay between engineering progress and demonstration of clinical utility. Limitations of any existing technique have to be broadly acknowledged to make room for improved techniques that do not necessarily agree with past clinical literature.
References
Levitt MH. Spin dynamics: basics of nuclear magnetic resonance. Chichester, UK: Wiley; 2001.
Tofts P. Quantitative MRI of the brain: measuring changes caused by disease. Chichester: Wiley; 2003.
Frank JS. The myocardial interstitium: its structure and its role in ionic exchange. J Cell Biol. 1974;60(3):586–601.
Rienks M, Papageorgiou A-P, Frangogiannis NG, Heymans S. Myocardial extracellular matrix: an ever-changing and diverse entity. Circ Res. 2014;114(5):872–88.
Scholz TD, Fleagle SR, Burns TL, Skorton DJ. Nuclear magnetic resonance relaxometry of the normal heart: relationship between collagen content and relaxation times of the four chambers. Magn Reson Imaging. 1989;7(6):643–8.
Hinojar R, Foote L, Ucar EA, Jackson T, Jabbour A, Chung-Yao Y, et al. Native T1 in discrimination of acute and convalescent stages in patients with clinical diagnosis of myocarditis: a proposed diagnostic algorithm using CMR. JACC Cardiovasc Imaging. 2015;8(1):37–46.
Kali A, Avinash K, Eui-Young C, Behzad S, Kim YJ, Bi X, et al. Native T1 mapping by 3-T CMR imaging for characterization of chronic myocardial infarctions. JACC Cardiovasc Imaging. 2015;8(9):1019–30.
Kim RJ, Fieno DS, Parrish TB, Harris K, Chen EL, Simonetti O, et al. Relationship of MRI delayed contrast enhancement to irreversible injury, infarct age, and contractile function. Circulation. 1999;100(19):1992–2002.
Pennell DJ, Sechtem UP, Higgins CB, Manning WJ, Pohost GM, Rademakers FE, et al. Clinical indications for cardiovascular magnetic resonance (CMR): consensus panel report. Eur Heart J. 2004;25(21):1940–65.
Wesbey GE, Higgins CB, McNamara MT, Engelstad BL, Lipton MJ, Sievers R, et al. Effect of gadolinium-DTPA on the magnetic relaxation times of normal and infarcted myocardium. Radiology. 1984;153(1):165–9.
Arheden H, Saeed M, Higgins CB, Gao DW, Bremerich J, Wyttenbach R, et al. Measurement of the distribution volume of gadopentetate dimeglumine at echo-planar MR imaging to quantify myocardial infarction: comparison with 99mTc-DTPA autoradiography in rats. Radiology. 1999;211(3):698–708.
Fontana M, White SK, Banypersad SM, Sado DM, Maestrini V, Flett AS, et al. Comparison of T1 mapping techniques for ECV quantification. histological validation and reproducibility of ShMOLLI versus multibreath-hold T1 quantification equilibrium contrast CMR. J Cardiovasc Magn Reson. 2012;14:88.
Treibel TA, Fontana M, Maestrini V, Castelletti S, Rosmini S, Simpson J, et al. Automatic measurement of the myocardial interstitium: synthetic extracellular volume quantification without hematocrit sampling. JACC Cardiovasc Imaging. 2016;9(1):54–63.
Schelbert EB, Piehler KM, Zareba KM, Moon JC, Martin U, Messroghli DR, et al. Myocardial fibrosis quantified by extracellular volume is associated with subsequent hospitalization for heart failure, death, or both across the spectrum of ejection fraction and heart failure stage. J Am Heart Assoc. 2015;4(12):e002613.
Moon JC, Messroghli DR. Myocardial T1 mapping and extracellular volume quantification: a Society for Cardiovascular Magnetic Resonance (SCMR) and CMR Working Group of the European Society of Cardiology consensus statement. J Cardiovasc Magn Reson. 2013;15:92. https://doi.org/10.1186/1532-429X-15-92.
Kellman P, Wilson JR, Xue H, Patricia Bandettini W, Shanbhag SM, Druey KM, et al. Extracellular volume fraction mapping in the myocardium, part 2: initial clinical experience. J Cardiovasc Magn Reson. 2012a;14:64.
Barral JK, Gudmundson E, Stikov N, Etezadi-Amoli M, Stoica P, Nishimura DG. A Robust methodology for in vivo T1 mapping. Magn Reson Med. 2010;64(4):1057–67.
Donoho DL. An invitation to reproducible computational research. Biostatistics. 2010;11(3):385–8.
Tannús A, Alberto T, Michael G. Adiabatic pulses. NMR Biomed. 1997;10(8):423–34.
Gold GE, Eric H, Jeff S, Graham W, Jean B, Christopher B. Musculoskeletal MRI at 3.0 T: relaxation times and image contrast. Am J Roentgenol. 2004;183(2):343–51.
Bevington PR, Keith Robinson D, Morris Blair J, John Mallinckrodt A, Susan M. Data reduction and error analysis for the physical sciences. Comput Phys. 1993;7(4):415.
Lustig M, Michael L, David D, Pauly JM. Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007;58(6):1182–95.
Weingärtner S, Akçakaya M, Roujol S, Basha T, Stehning C, Kissinger KV, et al. Free-breathing post-contrast three-dimensional T1 mapping: volumetric assessment of myocardial T1 values. Magn Reson Med. 2015;73(1):214–22.
Brix G, Schad LR, Deimling M, Lorenz WJ. Fast and precise T1 imaging using a TOMROP sequence. Magn Reson Imaging. 1990;8(4):351–6.
Look DC, Locker DR. Nuclear spin-lattice relaxation measurements by tone-burst modulation. Phys Rev Lett. 1968;20(21):1222.
Deichmann R, Haase A. Quantification of T1 values by SNAPSHOT-FLASH NMR imaging. J Magn Reson. 1992;96(3):608–12.
Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified look-locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52(1):141–6.
Piechnik SK, Ferreira VM, Dall’Armellina E, Cochlin LE, Andreas G, Stefan N, et al. Shortened modified look-locker inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson. 2010;12(1):69.
Kellman P, Peter K, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16(1):2.
Slavin GS. On the use of the ‘look-locker correction’ for calculating T1 values from MOLLI. J Cardiovasc Magn Reson. 2014;16(Suppl 1):P55.
Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T(1) mapping. Magn Reson Med. 2014;71(6):2082–95.
Captur G, Gaby C, Peter G, Peter K, Heslinga FG, Katy K, et al. A T1 and ECV phantom for global T1 mapping quality assurance: the T1 mapping and ECV standardisation in CMR (T1MES) program. J Cardiovasc Magn Reson. 2016;18(Suppl 1):W14.
Teixeira T, Hafyane T, Stikov N, Akdeniz C, Greiser A, Friedrich MG. Comparison of different cardiovascular magnetic resonance sequences for native myocardial T1 mapping at 3T. J Cardiovasc Magn Reson. 2016;18(1):65.
Robson MD, Piechnik SK, Tunnicliffe EM, Neubauer S. T1 measurements in the human myocardium: the effects of magnetization transfer on the SASHA and MOLLI sequences. Magn Reson Med. 2013;70(3):664–70.
Schelbert EB, Messroghli DR. State of the art: clinical applications of cardiac T1 mapping. Radiology. 2016;278(3):658–76.
Cerqueira MD, Weissman NJ, Dilsizian V, Jacobs AK. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the cardiac imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105(4):539–42. http://circ.ahajournals.org/content/105/4/539.short.
Avendi MR, Kheradvar A, Jafarkhani H. Fully automatic segmentation of heart chambers in cardiac MRI using deep learning. J Cardiovasc Magn Reson. 2016;18(Suppl 1):P351.
Luo G, An R, Wang K, Dong S, Zhang H. A deep learning network for right ventricle segmentation in short: axis MRI. In 2016 Computing in Cardiology Conference (CinC). 2016. https://doi.org/10.22489/cinc.2016.139-406.
Kellman P, Wilson JR, Xue H, Ugander M, Arai AE. Extracellular volume fraction mapping in the myocardium, part 1: evaluation of an automated method. J Cardiovasc Magn Reson. 2012b;14:63.
Keenan K, Katy K, Stupic KF, Boss MA, Russek SE. Standardized phantoms for quantitative cardiac MRI. J Cardiovasc Magn Reson. 2015;17(Suppl 1):W36.
Stikov N, Boudreau M, Levesque IR, Tardif CL, Barral JK, Bruce Pike G. On the accuracy of T1 mapping: searching for common ground. Magn Reson Med. 2015;73(2):514–22.
Kellman P, Arai AE, Xue H. T1 and extracellular volume mapping in the heart: estimation of error maps and the influence of noise on precision. J Cardiovasc Magn Reson. 2013;15:56.
Bull S, White SK, Piechnik SK, Flett AS, Ferreira VM, Loudon M, et al. Human non-contrast T1 values and correlation with histology in diffuse fibrosis. Heart. 2013;99(13):932–7.
Flett AS, Hayward MP, Ashworth MT, Hansen MS, Taylor AM, Elliott PM, et al. Equilibrium contrast cardiovascular magnetic resonance for the measurement of diffuse myocardial fibrosis: preliminary validation in humans. Circulation. 2010;122(2):138–44.
White SK, Sado DM, Fontana M, Banypersad SM, Maestrini V, Flett AS, et al. T1 mapping for myocardial extracellular volume measurement by CMR: bolus only versus primed infusion technique. JACC Cardiovasc Imaging. 2013;6(9):955–62.
Acknowledgements
The authors would like to thank Pascale Beliveau and Tarik Hafyane for their help with preparing the figures, and Reeve Ingle for insightful discussions.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Barral, J.K., Friedrich, M.G., Stikov, N. (2018). Fundamentals of Cardiac T1 Mapping. In: Yang, P. (eds) T1-Mapping in Myocardial Disease. Springer, Cham. https://doi.org/10.1007/978-3-319-91110-6_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-91110-6_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-91109-0
Online ISBN: 978-3-319-91110-6
eBook Packages: MedicineMedicine (R0)