Abstract
The paper presents the hybrid model of the objective clustering inductive technology based on complex using of the self-organizing SOTA and the density DBSCAN clustering algorithms. The inductive methods of complex systems analysis were used as the basis to implement the objective clustering inductive technology of gene expression profiles. To estimate the clustering quality for equal power subsets (include the same quantity of pairwise similar objects) the complex multiplicative criterion was calculated as the combination of the Calinski-Harabasz criterion and WB-index. The external clustering quality criterion is calculated as the normalized difference of the internal clustering quality criteria for the equal power subsets. The final decision concerning the determination of the optimal parameters of the clustering algorithm operation is done based on the maximum value of the Harrington desirability function that takes into account both the character of the objects and the clusters distribution in various clustering and the difference between clustering, which are implemented on the equal power subsets. The studied data grouping within the framework of the objective clustering inductive technology was performed in two stages. Firstly, the studied gene expression profiles were grouped with the use DBSCAN clustering algorithm. Then, the obtained set of gene expression profiles was divided into two clusters using SOTA clustering algorithm. This step-by-step procedure of the data clustering crates the conditions to save more useful information for following data processing.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Objective clustering
- Inductive modeling
- Gene expression profiles
- Clustering quality criteria
- SOTA clustering algorithm
- DBSCAN clustering algorithm
1 Introduction
The gene regulatory network creation based on the gene expression profiles is one of the current problems of the modern bioinformatics. The gene regulatory network is a set of genes, which interact with each other to control the specific cells functions. Qualitatively constructed gene regulatory network allows us to study the influence of the corresponding group of genes or the individual genes on functional possibilities of the biology objects in order to correct this process. The gene expression profiles, which are obtained by DNA microarray experiments or by RNA sequences technology are the basis to construct the gene regulatory networks. High dimension of feature space is one of the distinctive peculiarities of the studied profiles. About tens of thousands genes are contained in the gene expression profiles. The creation of gene regulatory network based on the whole dataset of the gene expression profiles is very difficult problem because: it requests large computer resources; it needs large time expenses to process the information; the complexity of the obtained network complicates the interpretation of results. In this context, it is necessary firstly to divide the gene expression profiles into subsets, each of which includes a group of genes that performs similar functions in the studied biological object. Biclustering technology is actual to solve this problem nowadays. Implementation of this technology allows grouping the objects and the features according to their mutual correlation. So, in the paper [14, 17] authors provide a review of a large quantity of biclustering approaches existing in the literature with analysis their advantages and disadvantages. In [7] authors have proposed and implemented the convex biclustering method using gene expression profiles of the lung cancer patient. The authors have shown the efficiency of the proposed method during simulation process. However, it should be noted that one of the significant problem of this technology qualitative implementation is selection of the biclustering level during the objects and the genes grouping. The qualitative validation of the obtained model is another task, which has no solution nowadays. High dimension of the features space promotes to the large quantity of the obtained biclusters. Limitation of their quantity by removing of small biclusters leads to the loss of some useful information. To solve this problem we propose the cluster-bicluster technology the implementation of which involves two stage: clustering of the gene expression profiles at the first step and biclustering of the obtained clusters at the second step. The reproducibility error is one of the current problems of the existing clustering algorithms, in other words, successful clustering results obtained on one dataset do not repeat while using another similar dataset. Reduction of this error can be achieved by careful verification of the obtained model using “fresh information”, which was not used during the model making. A higher degree of coincidence between the clustering results on the similar data corresponds to a higher degree of the obtained model objectivity. This idea is the basis of the objective clustering inductive technology, the main conception of which was presented in [15] and further developed in [16, 18, 19]. The practical implementation of the objective clustering inductive technology is possible using various clustering algorithms. The choice of the clustering algorithm is determined by the structure and character of the studied data. The practical implementation of this technology based on the agglomerative hierarchical and self-organizing SOTA clustering algorithms were presented in [2, 3]. One of the key conditions of successful implementation of this technology is careful determination of the internal, the external and the complex balance clustering quality criteria, which should take into account both the character of the objects grouping within the clusters and the character of the clusters distribution in the features space. This paper presents the research concerning the complex using of the density-based DBSCAN (Density Based Spatial Clustering of Application with Noise)[9] and self-organizing SOTA (Self Organizing Tree Algorithm)[8, 10] clustering algorithms within the framework of the objective clustering inductive technology. The implementation of the proposed step-by-step procedure of the gene expression profiles grouping allows us to save more useful information of following data processing.
The aim of the paper is development of the hybrid model of the objective clustering inductive technology of gene expression profiles based on DBSCAN and SOTA clustering algorithms.
2 Problem Statement
Let the initial dataset of the gene expression profiles is a matrix: \(A = \{x_{ij}\},\,i = 1,\dots ,n;\, j = 1,\dots ,m\), where n \( \textendash \) is the quantity of genes observed, m \(\textendash \) is the quantity of the studied objects. The aim of the clustering process is a partition of the genes expression profiles into non empty subsets of pairwise non-intersecting clusters in accordance with the clustering quality criteria taking into account the properties of the studied profiles:
where k \( \textendash \) is the quantity of clusters, \( i,j = 1,\dots ,k \). The objective clustering technology is based on the inductive methods of complex systems analysis, which involves sequential enumeration of the clustering within a given range in order to select from them the best variants. Let W \( \textendash \) is a set of all admissible clustering for given set A. The clustering is the best (an optimal) in terms of clustering quality criteria QC(K) is the following condition is performed:
The clustering \(K_{opt}\subseteq W\) is the objective if the difference of the objects and clusters distribution in different clustering for equal power subsets is minimal:
The architecture of the objective clustering inductive technology is presented in Fig. 1. Implementation of the technology involves the following steps:
-
1.
Problem statement. Clustering aim formation according to the stated task. Studied data preprocessing and their formation as a matrix.
-
2.
Determination of the affinity function of the studied data. Division of the initial dataset into two equal power subsets A and B using chosen affinity function. The equal power subsets include the same quantity of the pairwise similar objects.
-
3.
Choice of the clustering algorithm. Setup of its initial parameters. These parameters are changed during the algorithm operation to obtain the different variants of the studied data clustering.
-
4.
Data clustering on subsets A and B concurrently and clusters formation within the range of the algorithm’s parameters change. If the clusters quantity in various clustering differs, it is necessary to change the setup of the algorithm or to use another admissible clustering algorithm and to repeat the step 5.
-
5.
Calculation of the internal \( QC_{int} \), the external \( QC_{ext} \) and the complex balance \( QC_{bal} \) clustering quality criteria for the current clustering on equal power subsets A and B.
-
6.
Analysis of the complex balance clustering quality criterion values. In case of absence of this criterion extremums or if their values are less than admissible standards, choose another clustering algorithm and repeat the steps 4–7 of this procedure.
-
7.
Fixation of the objective clustering in correspondents with the maximum values of the complex balance clustering quality criterion.
The idea of the algorithm to divide the initial dataset of the objects \( \varOmega \) into two equal power subsets \( \varOmega ^{A} \) and \( \varOmega ^{B} \) is stated in [15] and further developed in [18]. Implementation of this algorithm involves the following steps:
-
1.
Calculation of \( \dfrac{n\times (n-1)}{2} \) pairwise distances between the gene expression profiles of the initial data. The result of this step is a triangular matrix of the distances.
-
2.
Allocation of the pairs of objects \( X_{s} \) and \( X_{p} \), the distance between which is minimal:
$$\begin{aligned} d(X_{s},X_{p}) = \min _{i,j} d(X_{i},X_{j}); \end{aligned}$$ -
3.
Distribution of the object \( X_{s} \) to subset \( \varOmega ^{A} \), and the object \( X_{p} \) to subset \( \varOmega ^{B} \).
-
4.
Repetition of the steps 2 and 3 for the remaining objects. If the quantity of objects is odd, the last object is distributed to the both subsets.
The example of the objects and the clusters distribution in the objective clustering inductive technology is shown in the Fig. 2. Obviously, that the best clustering corresponds to the higher density of the objects distribution relative to the mass centers of the clusters where these objects are and less density of the clusters’ mass centers distribution in the feature space. Moreover, it is necessary that the difference of the clustering results which are obtained on the equal power subsets was minimal. Thus, to implement this technology it is necessary to determine the gene expression profiles proximity metric, the internal, the external and the complex balance clustering quality criteria.
3 Criteria to Estimate the Gene Expression Sequences Proximity and Clustering Quality
It is obvious that the qualitative clustering corresponds to the high division ability of different clusters and high density of the objects concentration inside the clusters. Thus, it is necessary firstly to determine the proximity metric of the gene expression profiles. The [4] presents the results of the research concerning comparison of the three well know metrics efficiency to estimate the proximity level of numeric vectors: Manhattan, Euclidean and Correlation distances. Evaluation of the effectiveness of the studied metrics was performed using the model data representing the gene expression profiles of the objects in two different clusters. Centers of the corresponding clusters are calculated by the formula:
where \( N_{S} \) is the quantity of gene expression profiles in cluster S, \( x_{i}^{S} \) is \( i\textendash \)th sequence in cluster S. The research technique consists the next steps:
-
calculation of the average distance \( d_{int} \) from the profiles to the clusters’ centers, where these profiles are:
$$\begin{aligned} d_{int}(X^{S,P},C_{S,P})=\dfrac{1}{N}(\sum _{i=1}^{N_{S}}d(x_{i}^{S},C_{S})+\sum _{j=1}^{N_{P}}d(x_{j}^{P},C_{P})); \end{aligned}$$ -
calculation the average distance \( d_{ext} \) from the profiles to the centers of the neighbouring clusters:
$$\begin{aligned} d_{ext}(X^{S,P},C_{S,P})=\dfrac{1}{N}(\sum _{i=1}^{N_{S}}d(x_{i}^{S},C_{P})+\sum _{j=1}^{N_{P}}d(x_{j}^{P},C_{S})); \end{aligned}$$ -
calculation the relative coefficient:
$$\begin{aligned} d_{rel}(X^{S,P},C_{S,P})=\dfrac{d_{ext}(X^{S,P},C_{S,P})}{d_{int}(X^{S,P},C_{S,P})}; \end{aligned}$$
It is obvious the higher value of the relative coefficient corresponds to the higher separating ability of the used proximity metric. In order to estimate the effectiveness of the metrics we used the data of the lung cancer patients of the database Array Express [5], which includes the gene expression profiles of 96 patients, 10 of which were healthy and 86 patients were divided by the degree of the health severity into three groups (Well, Moderate and Poor). Each of the profiles includes 7129 genes expressions. Data preprocessing in order of gene expression matrix formation was carried out accordingly to the technique, which is presented in [1]. To choose the metrics of the gene expression profiles similarity class of the health patient (10 profiles) and class of patients with Poor state of health (21 profiles) were used. The results of the relative criteria values distribution while using different metrics to estimate the level of the gene expression profiles similarity are shown in Fig. 3. The analysis of the Fig. 3 allows us to conclude that in case of the gene expression profiles the correlation metric has higher separating ability in comparison with Euclid and Manhattan metrics because the values of the relative criterion which is calculated basing on the correlation distance, are higher in comparison with the use of Euclid and Manhattan distances.
3.1 Internal and External Clustering Quality Criteria
As it was noted herein before, it is obvious that the qualitative clustering corresponds to the high division ability of different clusters and high density of the objects concentration inside the clusters. Thus, the internal clustering quality criterion should be complex and takes into account both the objects distribution inside different clusters and the clusters distribution in the features space. The first component of the complex internal criterion is calculated as average distance from the objects to the mass centers of the clusters, where these objects are:
The second component of this criterion, which takes into account the singularity of the clusters distribution in the feature space, is calculated as an average distance between the mass centers of the clusters:
where K \( \textendash \) is the quantity of clusters, N \( \textendash \) is the general quantity of objects, \( N_{s} \) \( \textendash \) is the quantity of the objects in cluster s, \( x_{i}^{s} \) \( \textendash \) is the i-th vector in S cluster, \( C_{i} \), \( C_{j} \) and \( C_{s} \) \( \textendash \) are the mass centers of the clusters i, j and S concurrently,  – is the metric used to estimate the proximity level of the studied vectors. Various combinations of these components allow obtaining the clustering quality criteria for studied data subsets. During the simulation process the following internal criteria to estimate the data grouping quality were used:
– is the metric used to estimate the proximity level of the studied vectors. Various combinations of these components allow obtaining the clustering quality criteria for studied data subsets. During the simulation process the following internal criteria to estimate the data grouping quality were used:
-
Calinski-Harabasz [6]:
$$\begin{aligned} Q_{CH}=\dfrac{QCB(N-K)}{QCW(K-1)}; \end{aligned}$$ -
WB index [20]:
$$\begin{aligned} Q_{WB}=\dfrac{KQCW}{QCB}; \end{aligned}$$ -
Hartigan [12]:
$$\begin{aligned} Q_{H}=\log _{2}(\dfrac{QCB}{QCW}). \end{aligned}$$
In order to obtain more complete information about the effectiveness of these criteria operation the complex multiplicative criteria were calculated as follow:
The external clustering quality criterion is calculated as the normalized difference of the internal clustering quality criteria for the equal power subsets A and B:
To estimate the effectiveness of the internal and the external clustering quality criteria within the framework of the objective clustering inductive technology the gene expression profiles of the lung cancer patients were used [5]. Firstly, the data were divided into two equal power subsets with the use of the algorithm that had been presented in [15, 18]. Then, each of the subsets was sequentially divided into clusters from Kmin = 2 to Kmax = 5. In case of two-cluster structure in first cluster there were the gene expression profiles of the healthy patients (NORM) and gene expression of the patients with good state of health (WELL), second cluster included the gene expression of the patients with poor (POOR) and moderate (MODERATE) states. In case of three-cluster structure the first cluster contained the data of the healthy patients, the second \( \textendash \) the data of the patients with good state, the third cluster included the gene expression of the patients with poor and moderate states. In case of four-cluster structure the first cluster contained the data of the healthy patients, the second \( \textendash \) the data of the patients with good state, the third cluster included the gene expression of the patients with poor state and the fourth cluster contained the gene expression of the patients with moderate state. To obtain the five-cluster structure the gene expression profiles of the patients with moderate state were divided into two groups randomly. Objective clustering in this case corresponds to four-cluster structure. To estimate the proximity level of the appropriate vectors the correlation metric was used. Figure 4 shows the charts of the internal clustering quality criteria for equal power subsets A and B versus the clusters quantity. Figure 5 presents the charts of the complex multiplicative internal criteria versus the clusters quantity. Figure 6 shows the charts of the external clustering quality criteria, which were calculated based on the internal criteria versus the clusters quantity. Analysis of the charts which are shown in Fig. 4 and Fig. 5 allows us to conclude that the internal clustering quality criteria give the same results in terms of the local extremums existence. They have local extremums, which corresponds to the objects division into 4 clusters, however, it should be noted that in case of the \( QC_{H} \), \( QC_{CX2} \), \( QC_{CX3} \) and \( QC_{CX4} \) criteria use, the clustering, which correspond to the objects division into 4 and 5 clusters are badly distinguished. Analysis of the external criteria values, which are shown in Fig. 6, allows concluding that in terms of the clustering objectivity (proximity level of the results, which have been obtained on equal power subsets A and B) the \( QC_{H} \) Hartigan criterion and the \( QC_{CX2} \), \( QC_{CX3} \) complex criteria are ineffective, because they have not a local minimums corresponding to the objects division into 4 clusters (the objective clustering). The \( QC_{CX1} \) and \( QC_{CX4} \) criteria are the most informative to select the objective clustering, however, the \( QC_{CX1} \) criterion is more preferable because it has more expressed local minimum, which corresponds to four clusters existence in the obtained clustering.
3.2 Complex Balance Clustering Quality Criterion
It is obvious that the objective clustering corresponds to the minimum values of the internal and the external clustering quality criteria. However, it is possible that the extremums of these criteria correspond to different clustering. Thus, it is necessary to determine the complex balance clustering quality criterion that takes into account both the character of the objects and the clusters distribution in various clustering and the difference between clustering, which are implemented on the two equal power subsets. To calculate the complex balance clustering quality criterion the Harrington desirability function [11] was used. The implementation of this function involves transformation of the scales of the internal and the external criteria into reaction scale the values of which are changed linearly within the range from −2 to 5. Then the private desirabilities of the appropriate criteria are calculated by the formula:
The chart of the Harrington desirability function versus the reaction index Y is shown in Fig. 7. The transformation of the criteria scales into the reaction scales were performed by linear equation:
The parameters a and b are determined empirically. The general desirability index value is calculated as geometric average of the private desirabilities indexes:
In case of the objective clustering inductive technology the general Harrington desirability index was used as the complex balance criterion:
The largest value of the complex balance criterion corresponds to the best parameters of the clustering algorithm operation.
4 Implementation of SOTA Clustering Algorithm Within the Framework of the Objective Clustering Inductive Technology
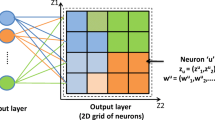
The SOTA clustering algorithm (Self-Organizing Tree Algorithm) [8] represents a type of self-organizing neural networks based on the Kohonen maps and the Fritzke algorithm of the spatial cell structure growing [10]. Opposed to the Kohonen maps that reflect a set of high dimensional input data on the elements of the two-dimensional array of small dimension, the SOTA algorithm generates a binary topological tree. The Fritzke algorithm performs self-organization of output nodes of the network in such a way that the quantity of the nodes increases in the field of the higher density of objects concentration and decreases in the field of the lower density. Two parameter are determined the effectiveness of the SOTA clustering algorithm operation: weight coefficient of the sister’s cell (scell) and maximum divergence coefficient value. The weight coefficients of the parent’s and winner’ cells are determined automatically: \( pcell = scell\cdot 5 \); \( wcell = pcell\cdot 2 \). This ratio is recommended by the algorithm’s authors. The block-scheme of the objective clustering model based on the SOTA clustering algorithm is shown in Fig. 8. Implementation of this model involves the following steps:
-
1.
Presentation of the studied data as a matrix \( n\times m \), where n \( \textendash \) is the quantity of the studied profiles or the quantity of the rows and m \( \textendash \) is the quantity of the objects or the quantity of the columns.
-
2.
Division of the initial dataset into two equal power subsets.
-
3.
Setup of the SOTA clustering algorithm. Setting of the initial value of scell weight coefficient, the interval and the step of its change.
-
4.
Data clustering on the equal power subsets A and B concurrently. Clusters formation and the internal, the external and the balance clustering quality criteria calculation within a range of the interval of the algorithm’s parameter change.
-
5.
Fixation of the optimal scell parameter corresponding to the maximum value of the balance criterion.
-
6.
Setting of the initial value of the maximum divergence parameter, the interval and the step of its change. Repetition of the step 4 of this algorithm. Fixation of the optimal maximum divergence parameter.
-
7.
Full data clustering by the SOTA clustering algorithm using the optimal parameters of the algorithm operation.
5 Implementation of DBSCAN Clustering Algorithm Within the Framework of the Objective Clustering Inductive Technology
DBSCAN clustering algorithm (Density Based Spatial Clustering of Application with Noise Algorithm) [9] initially needs two parameters: EPS-neighborhood of points (EPS) and the least quantity of the points within EPS-neighborhood (MinPts). Choice of these parameters determines the character of the studied objects grouping during the algorithm operation. In [9] authors proposed the technology based on the sorted 4-dist graph. However, the implementation of this technology does not allow determination of EPS and MinPts values exactly and this fact influences the quality of the algorithm operation. To determine EPS and MinPts values we propose to use the objective clustering inductive technology. The structural scheme of the objective clustering model based on DBSCAN clustering algorithm is presented in Fig. 9. Implementation of this model involves the following steps:
-
1.
The matrix of the studied data formation. The matrix contains n rows or studied profiles and m columns or the objects.
-
2.
Division of the initial dataset into two equal power subsets.
-
3.
The distance matrix between studied profiles for both subsets is calculated using correlation distance. This distance matrix is the input matrix for the next step of the algorithm operation.
-
4.
Setup of DBSCAN clustering algorithm, choice of the intervals and steps of EPS and MinPts change.
-
5.
Fixation of MinPts value (MinPts = 3). Initialization of EPS = EPSmin.
-
6.
Data clustering on the two subsets A and B using DBSCAN algorithm in range from EPSmin to EPSmax. Clustering fixation at each step.
-
7.
The internal, the external and the complex balance clustering quality criteria is calculated at each step of the algorithm operation.
-
8.
Analysis of the balance criterion values. Fixation of the optimal value EPS, which corresponds to the maximum value of the balance clustering quality criterion.
-
9.
Data clustering on the two equal power subsets A and B in the range from MinPtsmin to MinPtsmax. Clustering fixation at each step.
-
10.
Repetition of the steps 7 and 8 of this algorithm for MinPts values. Fixation of EPS and MinPts optimal values which correspond to the maximum of the complex balance clustering quality criterion.
-
11.
Studied data clustering using obtained parameters of DBSCAN algorithm operation.
6 Experiment, Results and Discussion
To estimate the effectiveness of the algorithm’s operation within the framework of the proposed technology the genes expressions of the lung cancer patients [5] were used. Firstly, the data were divided into two equal power subsets with the use of the algorithm that was presented herein before The simulation was carried out using software R [13]. Figure 10 shows the charts of the internal, the external and the complex balance criteria versus EPS-neighborhood values for gene expression profiles of the lung cancer patient. Two thousand profiles were studied during the experiment. Firstly, these profiles were divided into two equal power subsets using correlation metric. Then the dissimilarity matrices for all pairs of the studied objects of the both subsets using correlation distance was calculated. These dissimilarity matrices were used as the input data for next steps of DBSCAN algorithm operation. Three values of EPS-neighborhood were selected based on the maximum values of the complex balance criterion which is shown in Fig. 10: \( EPS_{1} \) = 0,13; \( EPS_{2} \) = 0,17; \( EPS_{3} \) = 0,44. Figure 11 shows the charts of the complex balance criteria for selected EPS versus MinPts values. The analysis of the charts allows concluding that the best clustering in terms of maximum value of the complex balance clustering quality criterion is achieved using the following parameters of DBSCAN algorithm: (a) EPS = 0,13, MinPts = 3; (b) EPS = 0,17, MinPts = 8; (c) EPS = 0,44, MinPts = 6. However, the detail analysis of the obtained results has shown what in the first and in the second cases there were differ clusters quantity in the obtained clustering. Only in case of EPS = 0,44 and MinPts = 6 both clustering contained the same quantity of clusters. The initial dataset contained 2000 gene expression profiles. In this case the studied data were divided in such a way: the first cluster contained 1663 profiles, in the second cluster there were 16 profiles, there were 321 profiles in the third cluster. The objects in the third were identified as the noise component. The results of the simulation have shown also that the largest quantity of the gene expression profiles are concentrated in the first cluster. This fact can be explained by the fact that these genes define the main processes, which are carried out in biological organisms, therefore they have more correlation between each other to compare with genes in other clusters or genes, which are identified as the noise. The results of the internal criteria for the equal power subsets A and B, the external and the balance criteria versus the weight parameter of the sisters cell using SOTA clustering algorithm are presented in Fig. 12. The maximum divergence value in this case E = 0,001 was taken. As it can be seen from Fig. 12, the internal clustering quality criteria \( CX\_1 \) and \( CX\_2 \), which have been calculated on equal power subsets A and B do not allow determining the optimal scell value corresponding the objective clustering of the studied data. The external clustering quality criterion CQE has several local minimums corresponding to the successful grouping of the studied vectors. However, the analysis of the complex balance criterion values, which takes into account both the internal and the external criteria, allows us to conclude that the best clustering corresponds to the scell = 0,001. In this case the 6659 profiles were divided into two clusters. The first cluster contained 4276 profiles and the second – 2383 ones. Variation of the maximum divergence value in the range from 0,001 to 1 has not changed the obtained results. The obtained results create the conditions to create the step-by-step technology of gene expression profiles grouping at early stage of the gene regulatory network construction. Objective clustering based on DBSCAN algorithm allows us to select the genes with higher level of their mutual correlation. The noise component also is removed at this step. Then at the second step of the profiles grouping the selected profiles are divided into two group using SOTA clustering algorithm. At the third step of the gene expression profiles grouping the biclustering technology is implemented on the obtained clusters. To our mind the implementation of the proposed technology allows saving more useful information to follow create the gene regulatory network.
7 Conclusion
The paper presents the model of the objective clustering inductive technology of gene expression profiles based on DBSCAN and SOTA clustering algorithm. The implementation of this technology involves the concurrent data clustering on the two equal power subsets which include the same quantity of the pairwise similar objects. The correlation metric was used as the proximity metric of the gene expression profiles. The internal clustering quality criteria take into account both the character of objects distribution within clusters relative to the mass center of the appropriate cluster and the character of the clusters distribution in the features space. The external clustering quality criteria were calculated as a normalized difference of the internal clustering quality criteria, which were calculated on the equal power subsets A and B. The simulation process involved sequential evaluation of the internal and external criteria for clustering during increase the clusters quantity from Kmin to Kmax. The objective clustering corresponded to the global minimum of the external clustering quality criterion. The gene expression sequences of the patients of the database Array Express, which were investigated on the lung cancer, were used as the experimental data. The quantity of the clusters was changed from 2 to 5 during clustering process. The objective clustering corresponded to the four-cluster structure. The results of the simulation have shown that the complex multiplicative criterion, which is the combination of the WB-index and Calinski-Harabasz criterion is the most effective to determine the objective clustering. This criterion has the clearly expressed minimum corresponding to the four-cluster structure both in case of estimation of the character of the objects and the clusters distribution in the equal power subsets and in case of estimation of the result of clustering difference on these subsets. The external criterion was calculated as the normalized difference of the internal clustering quality criteria which are calculated on the two equal power subsets. The general Harrington desirability index based on the internal and external criteria was used as the complex balance clustering quality criterion. Determination of optimal parameters of the used algorithm operation has been performed based on the maximum value of the complex balance clustering quality criterion during the algorithm operation. The results of the simulation have shown the high efficiency of the proposed technology. In case of DBSCAN clustering algorithm using the noise component in terms of density of the objects distribution was selected during algorithm operation. Implementation of the proposed technology also allows us to group the gene expression sequences based on the similarity of their profiles. The gene expression sequences with high correlation coefficient were distributed into one cluster. This fact allows us to select the groups of the gene expression sequences, which determine the main processes in the biological organisms in order to study and to correct these processes. In case of SOTA clustering algorithm using the studied gene expression profiles were divided into two groups. This fact create the conditions to create the step-by-step technology of gene expression profiles grouping at early stage of the gene regulatory network construction. Objective clustering based on DBSCAN algorithm allows selecting the genes with higher level of their mutual correlation. Then the selected profiles are divided into two group using SOTA clustering algorithm. The further perspective of the authors’ research is the development of the hybrid technology of the step-by-step gene expression profiles grouping based on the complex use of the objective clustering and biclustering technologies.
References
Babichev, S., Kornelyuk, A., Lytvynenko, V., Osypenko, V.: Computational analysis of gene expression profiles of lung cancer. Biopolymers Cell 32(1), 70–79 (2016). http://biopolymers.org.ua/content/32/1/070/
Babichev, S., Taif, M.A., Lytvynenko, V.: Inductive model of data clustering based on the agglomerative hierarchical algorithm. In: Proceeding of the 2016 IEEE First International Conference on Data Stream Mining and Processing (DSMP), pp. 19–22 (2016). http://ieeexplore.ieee.org/document/7583499/
Babichev, S., Taif, M.A., Lytvynenko, V., Korobchynskyi, M., Taif, M.A.: Objective clustering inductive technology of gene expression sequences features. In: Proceeding of the 13th International Conference Beyond Databases, Architectures and Structures. Communication in Computer and Information Science, Ustron, Poland, pp. 359–372 (2017). https://springerlink.bibliotecabuap.elogim.com/content/pdf/10.1007/978-3-319-58274-0_29.pdf
Babichev, S., Taif, M.A., Lytvynenko, V., Osypenko, V.: Criterial analysis of gene expression sequences to create the objective clustering inductive technology. In: Proceeding of the 2017 IEEE 37th International Conference on Electronics and Nanotechnology (ELNANO), pp. 244–249 (2017). http://apps.webofknowledge.com/full_record.do?product=WOS&search_mode=GeneralSearch&qid=3&SID=U2bB7H8kqTrSyZ2eAKs&page=1&doc=2
Beer, D., Kardia, S., et al.: Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat. Med. 8(8), 216–224 (2002). https://www.ncbi.nlm.nih.gov/pubmed/12118244
Calinski, T., Harabasz, J.: A dendrite method for cluster analysis. Commun. Stat. 3, 1–27 (1974)
Chi, E., Allen, G., Baraniuk, R.: Convex biclustering. Biometrics 73, 10–19 (2016). http://onlinelibrary.wiley.com/doi/10.1111/biom.12540/full
Dorazo, J., Corazo, J.: Phylogenetic reconstruction using an unsupervised growing neural network that adopts the topology of a phylogenetic tree. J. Mol. Evol. 44(2), 226–259 (1997). https://www.ncbi.nlm.nih.gov/pubmed/9069183
Ester, M., Kriegel, H., Sander, J., Xu, X.: A density-based algorithm for discovering clusters in large spatial datasets with noise. In: Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, Portland, pp. 226–231 (1996). http://dl.acm.org/citation.cfm?id=3001507
Fritzke, B.: Growing cell structures a self-organizing network for unsupervised and supervised learning. Neural Netw. 7(9), 1441–1460 (1994). http://www.sciencedirect.com/science/article/pii/0893608094900914
Harrington, J.: The desirability function. Ind. Qual. Control 21(10), 494–498 (1965). http://asq.org/qic/display-item/?item=4860
Hartigan, J.: Clustering Algorithms. Wiley, New York (1975). http://dl.acm.org/citation.cfm?id=540298
Ihaka, R., Gentleman, R.: R: a linguage for data analysis and graphics. J. Comput. Graph. Stat. 5(3), 299–314 (1996). http://www.tandfonline.com/doi/abs/10.1080/10618600.1996.10474713
Kaiser, S.: Biclustering: Methods, Software and Application (2011). https://edoc.ub.uni-muenchen.de/13073/
Madala, H., Ivakhnenko, A.: Inductive Learning Algorithms for Complex Systems Modeling, pp. 26–51. CRC Press, Boca Raton (1994). http://www.gmdh.net/articles/theory/ch2.pdf
Osypenko, V.V., Reshetjuk, V.M.: The methodology of inductive system analysis as a tool of engineering researches analytical planning. Agric. Forest Eng. 58, 67–71 (2011). http://annals-wuls.sggw.pl/?q=node/234
Pontes, B., Giraldez, R., Aguilar-Ruiz, J.S.: Biclustering on expression data: a review. J. Biomed. Inf. 57, 163–180 (2015). https://www.ncbi.nlm.nih.gov/pubmed/26160444
Sarycheva, L.: Objective cluster analysis of data based on the group method of data handling. Problems of Control and Automatics 2, 86–104 (2008)
Stepashko, V.: Elements of the Inductive Modeling Theory, State and Prospects of Informatics Development in Ukraine, pp. 471–486. Scientific Thought, Kiev (2010). Monograph/Team of autors
Zhao, Q., Xu, M., Frnti, P.: Sum-of-squares based cluster validity index and significance analysis. In: Proceeding of International Conference on Adaptive and Natural Computing Algorithms, pp. 313–322 (2009). https://springerlink.bibliotecabuap.elogim.com/chapter/10.1007/978-3-642-04921-7_32
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG
About this paper
Cite this paper
Babichev, S., Lytvynenko, V., Skvor, J., Fiser, J. (2018). Model of the Objective Clustering Inductive Technology of Gene Expression Profiles Based on SOTA and DBSCAN Clustering Algorithms. In: Shakhovska, N., Stepashko, V. (eds) Advances in Intelligent Systems and Computing II. CSIT 2017. Advances in Intelligent Systems and Computing, vol 689. Springer, Cham. https://doi.org/10.1007/978-3-319-70581-1_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-70581-1_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-70580-4
Online ISBN: 978-3-319-70581-1
eBook Packages: EngineeringEngineering (R0)