Abstract
Regulation of cell growth and proliferation is crucial for development and function of organs in all animals. Genetic defects in growth control can lead to developmental disorders and cancers. Translationally controlled tumor protein (TCTP) is a family of evolutionarily conserved proteins implicated in cancer. Recent studies have revealed multiple roles of TCTP in diverse cellular events, but TCTP functions in vivo are poorly understood in vertebrate systems. We have used Drosophila melanogaster, the fruit fly, as a model organism for genetic dissection of Tctp function. Our studies have shown that Tctp is essential for organ development by regulating growth signaling. Furthermore, it is required for genome stability by promoting DNA repair and chromatin remodeling in the nucleus. Thus, Tctp acts as a multifaceted cytosolic and nuclear factor for regulating organ growth and genome stability. In this chapter, we describe an overview of our findings on Tctp functions in Drosophila and discuss their implications in cancer.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
8.1 Introduction
TCTP family proteins are widely expressed in eukaryotes. It was first identified in mouse tumor cells in the early growth phase (Bohm et al. 1989; Chitpatima et al. 1988). TCTP is abundantly expressed in rat and human testes, a tissue that undergoes intense mitotic activity (Guillaume et al. 2001). In yeast, TCTP is expressed throughout the cell cycle (Chung et al. 2000), but is greatly upregulated during exponential growth (Norbeck and Blomberg 1997), whereas it is repressed in conditions of growth arrest (Bonnet et al. 2000). Furthermore, extensive analysis of differential gene expression in the tumor cells and revertants revealed that TCTP is upregulated in cancer cells derived from different organs and is among the most downregulated genes during tumor reversion, the process of “quitting the malignant phenotype” (Amson et al. 2012, 2013; Bommer and Thiele 2004; Tuynder et al. 2002, 2004). These studies suggest important roles of TCTP in growth regulation, tumorigenesis, and its reversion process.
In addition, TCTPs have been implicated in several other functions, including histamine release (Nielsen et al. 1998), microtubule association (Gachet et al. 1999; Jeon et al. 2016), and Ca++ regulation (Kim et al. 2000; Sanchez et al. 1997). An important clue to the molecular function of TCTP was provided from its structure. TCTP family proteins turned out to be related to a guanine nucleotide exchange factor (GEF) for Rab proteins (Dong et al. 2009; Thaw et al. 2001), suggesting that TCTPs might be involved in intracellular vesicle trafficking (Predic et al. 2002). Many of these TCTP functions are based on biochemical interactions and functional assays in culture cells but not in animals. Thus, whether TCTP plays such roles in tissues and organs of animals in vivo is an important question to be addressed.
Drosophila is an ideal system for studying gene functions in vivo. We became interested in Tctp initially by chance (hereafter, Drosophila TCTP gene and protein are labeled “Tctp” and “Tctp”, respectively, according to the FlyBase nomenclature). Our work since then has revealed important functions of Tctp in the regulation of growth and genome stability, providing new insights into the roles of mammalian TCTP genes. Firstly, we will begin with a brief background of our Tctp work. Secondly, we will discuss the function of Tctp in TOR signaling for organ growth. Thirdly, we will present a nuclear function of Tctp for DNA repair and genome stability. Lastly, we will discuss possible implications of our works on mammalian TCTP functions and cancer.
8.2 Identification of Drosophila Tctp Function in Organ Growth
Nothing was known about the Tctp gene in Drosophila when we first noticed its putative role in tissue growth. At the time, we had been studying how the adult compound eye develops from the eye imaginal disc, an epithelial primordium for the eye. In the early stages of eye development, establishment of the dorso-ventral (DV) axes of the eye disc is critically important for its growth and patterning (Singh et al. 2005). While searching for genes involved in the growth of the eye disc, we found a strong genetic interaction between a mutation affecting eye growth and a deficiency chromosome uncovering the Tctp locus.
This genetic enhancement of the eye growth phenotype suggested a role of Tctp in organ development. Because there was no known mutation in the Tctp gene, we first generated Tctp mutants by imprecise excision of a P-element inserted in an intron of the Tctp gene (Hsu et al. 2007). Genetic tests suggested one of these mutations to be a null allele. This mutant was lethal as homozygotes, resulting in death during early larval stages. Some Tctp null homozygotes go through embryogenesis probably due to the presence of wild-type maternal Tctp protein deposited from heterozygous mothers, but they all die during early larval stages. Thus, it is clear that Tctp plays important functions for larval stages during which primordia for adult organs undergo active development.
To understand the function of Tctp in organ development, we mainly used the eye and the wing, two appendages that have been studied extensively (Baker 2007; Cohen and Di Nardo 1993). These organs develop from the eye and the wing imaginal disc, respectively. Although development of these organs involves several conserved signaling mechanisms, the ways these signaling pathways are used for axial patterning and differentiation of the eye and the wing are quite different (Baker 2007). Nonetheless, tissue-specific Tctp knockdown using RNA interference (RNAi) in these developing organs by using Gal4–UAS system (Brand and Perrimon 1993) results in similar growth defects. Because organ growth depends not only on cell proliferation but also on the cell size, it is important to determine whether Tctp is involved in the control of cell size, cell number, or both. Since each cell of adult wing has a single hair protruding from the cell surface, the hair density provides an approximate measure of cell size and number. Using the hair density, we showed that both cell number and size are reduced when Tctp is partially depleted by RNAi (Hsu et al. 2007), indicating the requirements of Tctp for cell growth as well as proliferation.
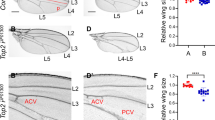
To support the results of Tctp RNAi phenotypes in organ growth, Tctp null mutant was also examined using genetic mosaic clones. Because Tctp null mutant flies die during early stage of development, we utilized methods of making genetic mosaic animals in which patches of homozygous Tctp mutant clones can be generated in heterozygous flies by mitotic recombination (Xu and Rubin 1993) (Fig. 8.1a). The clone size depends on the number of cell division after the first mitotic recombination event. When Tctp mutant clones were examined in wing discs soon after mitotic recombination was induced at first instar larval stage, both +/+ wild-type twin spots and Tctp − /Tctp − mutant clones were very small but showed similar sizes (Fig. 8.1b). On the contrary, when mosaic wings were examined during late third instar stage, Tctp − /Tctp − clones remained small or were eliminated while +/+ twin spot clones had grown much faster, eventually competing out Tctp − /Tctp − mutant cells (Fig. 8.1c). This clonal analysis demonstrated that Tctp is essential for growth of tissues in wing discs. Another interesting point is that Tctp mutant cells cannot compete well with adjacent wild-type cells, eventually being lost, thus indicating the importance of Tctp in cell survival.
Suppression of Tctp mutant defects by CycE and P35. (a–c) Growth defects of Tctp mutant clones in wing disc. (a) Tctp − mutant clones are generated by mitotic recombination in Tctp − /+ heterozygote cells. Tctp − clones and wild-type twin spots (+/+) are marked by the absence or presence of GFP expression. (b) At 24 h after clone induction, both Tctp − and +/+ twin spot clones are small but similar in size. (c) By 60 h after clone induction, +/+ clones grow large but most Tctp − clones are eliminated. (d–f) Partial suppression of Tctp − mutant defects by CycE or P35. (d) Clones are generated by the MARCM method. The presence of Gal80 in Tctp − /+ heterozygous wing cells represses Gal4-dependent GFP expression. The presence of Tub-Gal4 and UAS-GFP are not shown for simplicity. After recombination, Tctp mutant cells can express GFP due to loss of Gal80 whereas +/+ twin spot cannot. (e) Tctp − clones are very small or not formed. (f) Expression of CycE or P35 in mutant clones partially suppresses growth defects, resulting in the formation of Tctp − clones
8.3 Role of Tctp in TOR Signaling
Clonal analysis of Tctp null mutation demonstrated that Tctp is required for organ growth. We tested whether the growth defects of Tctp mutant cells are due to abnormalities in cell proliferation and/or cell survival. A genetic technique called MARCM (Mosaic Analysis with a Recessive Cell Marker) (Lee and Luo 2001) provides a powerful in vivo tool for expressing a gene within mutant clones. Using this technique, we overexpressed either Cyclin E (CycE) or P35 caspase inhibitor in Tctp mutant clones. Notably, small sizes of Tctp null mutant clones were enlarged by expressing cyclin E or P35 in the MARCM mutant clones (Fig. 8.1d–f), indicating that loss of Tctp not only affects cell proliferation but also impairs cell survival (Hsu et al. 2007).
Genetic analysis of Tctp mutation and knockdown indicates that Tctp is involved in the regulation of both cell proliferation and cell size. Because the Target of Rapamycin (TOR) signaling regulates both cell growth and proliferation (Laplante and Sabatini 2012b), it was conceivable that Tctp might function in the TOR pathway to control organ growth. A central component of this pathway is TOR protein kinase that phosphorylates S6 kinase (S6k) and Thor (Drosophila 4EBP) to promote cell growth (Miron et al. 2001; Oldham et al. 2000). TOR kinase is activated by parallel inputs from growth factor signaling and nutrient conditions (Jewell et al. 2013). Upon insulin receptor (InR) signaling, the GTPase Activating Protein (GAP) activity of the Tuberous sclerosis complex (Tsc1/Tsc2) is inhibited, thus activating Rheb (Ras-related human protein enriched in brain) GTPase and its associated effector TOR (Dong and Pan 2004; Garami et al. 2003; Saucedo et al. 2003; Stocker et al. 2003; Zhang et al. 2000, 2003). Indeed, there was striking genetic interaction between Tctp and the TOR pathway genes, including upstream genes like Tsc1/2, Rheb, and InR as well as a downstream gene S6k. For instance, tissue growth induced by overexpression of InR or Rheb can be suppressed by Tctp knockdown (Hsu et al. 2007). Genetic relationships between Tctp and these TOR components suggest that Tctp acts at a step close to Rheb.
In addition to the genetic interaction of Tctp with Rheb, we noted that yeast TCTP/DSS4 (mammalian MSS4) was initially identified as a genetic suppressor of sec4 gene encoding a Rab GTPase (Burton et al. 1993; Moya et al. 1993). Since TCTP is structurally similar to the Mss4 GTPase regulator (Thaw et al. 2001), Tctp might function in TOR signaling by interacting with Rheb GTPase. Biochemical evidence indicated that Tctp can physically interact with Rheb, and it has a guanine nucleotide exchange (GEF) activity for Rheb. The glutamate residue at the 12th position of TCTP was implicated in binding to Sec4 Rab GTPase (Thaw et al. 2001). We showed that the E to V mutation (TctpE12V) abolished the GEF activity. This residue is also critical for the in vivo function of Tctp because, unlike wild-type Tctp, TctpE12V fails to rescue the growth defects of Tctp RNAi (Hsu et al. 2007). Taken together, multiple pieces of evidence suggest that Tctp acts through Rheb in order to activate TOR signaling for organ growth (Fig. 8.2a).
14-3-3 promotes Tctp–Rheb interaction. (a) Tor kinase in TORC1 is activated by Rheb GTPase. Tor phosphorylates S6k and Thor/4EBP. Phosphorylated S6k promotes protein synthesis for cell growth. Phosphorylated Thor/4EBP cannot inhibit the function of eIF4, thereby increasing protein synthesis. Increased translation leads to expression of cell cycle regulators. Rheb activity is inhibited by TSC1/2 while facilitated by Tctp. (b) In wild-type condition, 14-3-3 isoforms directly interact with Tctp and Rheb, resulting in normal eye size. Tctp RNAi causes a reduction in the eye size. Knockdown of either 14-3-3ε or 14-3-3ζ has no effect, but it strongly enhances the Tctp RNAi eye phenotype. Knockdown of both forms of 14-3-3 abolishes the Tctp–Rheb interaction and disrupts eye disc development causing the headless phenotype. It is unknown whether homo- or hetero-dimerization of 14-3-3 isoform directly links Tctp and Rheb
The proposed Tctp function as a GEF toward Rheb had been questioned by Wang et al. (2008) and Rehmann et al. (2008) based on the observations that mammalian TCTP could not bind to Rheb and did not reproducibly affect mTORC1 signaling. The cause of these discrepancies on the function of TCTP is not yet clear but is likely to be due to differences in cell cultures and assay conditions. In fact, Dong et al. (2009) demonstrated that human Tctp not only binds to Rheb but also accelerates GDP release from hRheb. Additionally, they showed that hTCTP can prolong the activation of mTOR signaling in amino acid-depleted cells whereas hTCTPE12V mutant form cannot. hTCTP also acts upstream to Rheb for the activation of S6k phosphorylation. All of these results were consistent with the Tctp–Rheb relationships shown in Drosophila. In addition, analysis of the structure model of the hRheb–hTCTP complex showed that hTCTP binding to hRheb opens the nucleotide binding site to facilitate the dissociation of GDP. Moreover, key residues involved in the hTCTP–hRheb interaction were experimentally validated (Dong et al. 2009), supporting the function of TCTP as a GEF for Rheb.
The binding between TCTP and Rheb has also been shown in Arabidopsis (Brioudes et al. 2010), indicating that this interaction is conserved in invertebrates, vertebrates, and plant systems. We have shown that human TCTP can fully rescue the growth defects in Tctp-depleted Drosophila organs (Hsu et al. 2007). Remarkably, Drosophila Tctp can also restore the defects in Arabidopsis TCTP mutants (Brioudes et al. 2010). These studies suggest strong structural and functional conservation of TCTP family genes among plants, invertebrate animals, and humans.
8.4 Regulation of Tctp Function by 14-3-3
Our study described above suggests that Tctp facilitates the Rheb function in vivo for TOR signaling. Because growth signaling must be precisely controlled for normal development, it is an intriguing question how Tctp function is regulated during organogenesis. In an effort to identify factors that modulate the function of Tctp, we have performed a genetic screen using the Gal4-UAS system (Brand and Perrimon 1993) for targeted knockdown of gene(s). This screen was based on the fact that Tctp RNAi in the eye disc using eyeless (ey)-Gal4 driver results in a reduction of the eye size. We screened a library of UAS-dsRNA (RNAi for short) lines to identify specific RNAi lines that either enhance or suppress the Tctp RNAi eye phenotype.
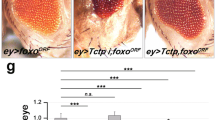
From this screen, we found many RNAi lines that modify the Tctp RNAi eye phenotype. Tctp modifier genes identified in this screen belong to diverse categories, including the ones that are involved in growth signaling, cell death, cytoskeleton, and transcription. Interestingly, 14-3-3 RNAi was found as an enhancer of Tctp RNAi. 14-3-3 is a family of highly conserved proteins expressed in all eukaryotic cells. 14-3-3 proteins play important roles as adaptors in multiple signaling pathways (Morrison 2009). 14-3-3 genes have also been implicated in oncogenesis as well as tumor suppression (Aghazadeh and Papadopoulos 2016; Aitken et al. 2002; Zhao et al. 2011). Mammals have several isoforms of 14-3-3 that have redundant functions as well as cell-type specific roles (Aghazadeh and Papadopoulos 2016; Obsilova et al. 2008). Drosophila has two genes, 14-3-3ε and 14-3-3ζ (Skoulakis and Davis 1996). Although silencing of either isoform of 14-3-3 in the eye disc does not affect the eye growth, it synergistically enhances the effects of Tctp RNAi, resulting in much smaller eyes than the size of Tctp RNAi eyes (Fig. 8.2b, Le et al. 2016). Strong genetic interaction between Tctp and 14-3-3 isoforms raised a possibility that 14-3-3 might be involved in regulating the Tctp function in TOR signaling.
Previous studies in mammalian cells have also found that 14-3-3 proteins participate in TORC1 signaling by interacting with TSC2 and PRAS40. Both TSC2 and PRAS40 are negative regulators of Rheb and TOR kinase, respectively (Fonseca et al. 2007; Inoki et al. 2002; Li et al. 2002). Upon phosphorylation of TSC2 by Akt, 14-3-3 binds to phosphorylated TSC2 to inhibit the GAP function of TSC2. PRAS40 directly binds to TOR kinase to inhibit its kinase activity. Binding of 14-3-3 to PRAS40 leads to its dissociation from the TORC1 complex, thereby activating the TORC1 activity (Jewell et al. 2013; Morrison 2009). Hence, 14-3-3 proteins promote TOR signaling by inhibiting these negative regulators of TOR signaling. The serine and threonine phosphorylation sites of Akt (S924 and T1518) are conserved in Drosophila and mammalian TSC2 proteins. Interestingly, however, a mutated TSC2 with both substitutions of S924 and T1518 with unphosphorylatable alanine is fully functional to inhibit TOR signaling in Drosophila (Dong and Pan 2004). Thus, this phosphorylation-dependent 14-3-3 binding is probably not essential for the TSC2 function in Drosophila. Moreover, PRAS40 in Drosophila regulates fertility but is not required for growth of the fly, despite its importance in TOR signaling in mammalian cells (Pallares-Cartes et al. 2012). It has not been tested in vivo whether loss of 14-3-3 impairs TOR signaling in mammals. Thus, further studies are necessary to determine the importance of 14-3-3 interaction with TSC2 and PRAS40 in vivo.
Due to the fact that TSC2 phosphorylation is dispensable in Drosophila, we figured that genetic interaction between 14-3-3 and Tctp is probably independent of 14-3-3 binding to TSC2. Instead, we found that both 14-3-3 isoforms could directly interact with Tctp (Le et al. 2016), raising the possibility that 14-3-3 might promote TOR signaling through a new mechanism by binding to Tctp. Furthermore, 14-3-3 isoforms can physically interact with Rheb protein as well. Because Tctp and Rheb act together, these proteins seem to function together with 14-3-3 isoforms. One possibility is that 14-3-3 isoforms might be involved in facilitating the interaction between Tctp and Rheb. In testing this possibility, we found that knockdown of both 14-3-3 isoforms abolished the interaction between Tctp and Rheb to an undetectable level in co-immunoprecipitation assays using Drosophila S2 cells. However, depletion of either 14-3-3ε or 14-3-3ζ isoform did not significantly affect the Tctp–Rheb interaction, which is correlated with the observation that knockdown of a single 14-3-3 isoform causes noticeable defects in neither eye nor wing. These results indicate that 14-3-3 isoforms are critical for the interaction between Tctp and Rheb, although the two isoforms share redundant roles.
To test the functional redundancy of the 14-3-3 isoforms, we examined the phenotypes of depleting either one or both of the 14-3-3 isoforms in specific tissues. In striking contrast to the single knockdown of either isoform of 14-3-3, knockdown of both 14-3-3 isoforms using ey-Gal4 results in pupal lethality. Ey-Gal4 drives the expression of Gal4 in the primordia for both eye and head during the early stage of development. Examination of dead pupae showed relatively normal body parts, but there was specific loss of entire head and eye tissues to which Gal4 expression was targeted (Fig. 8.2b). Knockdown of both 14-3-3 isoforms in developing wing discs also resulted in severe loss of cell proliferation and induction of specific cell death in the targeted wing area (Le et al. 2016). These tests clearly indicate that two isoforms of 14-3-3 can function redundantly, thereby compensating the loss of one isoform. However, based on the findings that 14-3-3ε or 14-3-3ζ null mutations are semi- or fully lethal as homozygotes, respectively (Acevedo et al. 2007), we presume that these two 14-3-3 isoforms cannot be entirely redundant. Thus, 14-3-3ε and 14-3-3ζ isoforms seem to be partially redundant in certain conditions or tissues, while they also have unique functions. Nevertheless, knockdown of both 14-3-3 isoforms critically impairs normal growth of imaginal discs.
The levels of pS6k and pThor are convenient readouts of TOR signaling, as they are phosphorylation targets of TOR kinase. Interestingly, assays in S2 cells have shown that single knockdown of either of the 14-3-3 isoforms causes considerable reduction of pS6k and pThor (4EBP) levels, indicating that each 14-3-3 isoform is required for full TOR signaling (Le et al. 2016). Thus, TOR signaling in S2 cells might be more sensitive to a reduction of 14-3-3 isoforms than it is for developing organ tissues, although the molecular basis for this difference is unknown. Another important question is how 14-3-3 isoforms promote the interaction between Tctp and Rheb. 14-3-3 proteins are known to function as a homo- or a hetero-dimer (Acevedo et al. 2007; Yaffe 2002). Thus, it is plausible that dimerization of 14-3-3 proteins bound to Tctp and Rheb might bring them together to facilitate their direct interaction. Alternatively, 14-3-3 may be involved in modifying the structure and/or subcellular localization of Tctp and Rheb so that they can form a complex to activate Rheb activity. Additional studies are necessary to pinpoint the precise function of 14-3-3 in the formation of Tctp–Rheb complex.
8.5 Effects of 14-3-3 Isoforms and Tctp on Cyclin E
CycE, as a key regulator of the G1-S transition in cell cycle, plays an essential role in cell proliferation. TOR signaling regulates not only cell growth through activation of S6k and 4EBP but also regulates proliferation by promoting cell cycle. Consistent with genetic and physical interaction of 14-3-3 and Tctp, loss of function clones of 14-3-3ε or 14-3-3ζ null mutation results in a partial reduction of CycE level in the eye imaginal disc (Le et al. 2016). However, the partial CycE reduction by depleting one 14-3-3 isoform is insufficient to disrupt proliferation or differentiation of retinal cells of eye disc. We have shown that reduced organ size caused by Tctp mutation or RNAi can be rescued by CycE overexpression (Hsu et al. 2007). Therefore, it is likely that defects in organ growth caused by double knockdown of Tctp and one of 14-3-3 isoforms might be due to loss of CycE. As expected, organ growth defects caused by reducing both Tctp and 14-3-3 (or knockdown of both 14-3-3 isoforms) were strongly suppressed by CycE (Le et al. 2016).
In Drosophila, TOR kinase activates cell proliferation by regulating the level of CycE (Zhang et al. 2000), although it is not clearly defined how activated TOR leads to CycE expression. Phosphorylation of S6k and 4EBP by activated mTOR signaling results in increased protein synthesis, leading to the promotion of cell cycle (Jewell et al. 2013; Laplante and Sabatini 2012b). In mammalian cells, the eIF4E pathway activated by phosphorylation of 4EBP results in increased translation of mRNAs for cell cycle regulators such as CycD1 (Hashemolhosseini et al. 1998; Laplante and Sabatini 2012a; Rosenwald et al. 1995), thus allowing more CycE–Cdk2 complex to promote cell proliferation. Drosophila CycD is not only involved in cell cycle progression but also cell growth through an independent pathway (Datar et al. 2006). It is currently unknown whether the regulation of TOR-dependent CycE level in Drosophila is mediated by CycD. Interestingly, suppression of human 14-3-3ε inhibits proliferation of cancer cells and tumor growth. 14-3-3ε inhibition suppresses CycE expression while inducing the cell cycle inhibitor p27kip1 at the G1 stage, consistent with the antitumor effect of 14-3-3ε inhibition (Gong et al. 2014). This study supports the role of Drosophila 14-3-3 in cell cycle progression during organ development. It would be interesting to see whether the effects of human14-3-3ε inhibition are at least in part mediated by TCTP/Rheb-dependent TOR signaling.
8.6 Tctp Function in DNA Damage Control
TCTP is known to be expressed in the cytosol and the nucleus of normal as well as cancer cells. Although the roles of TCTP in the nucleus have not been extensively studied, it has been reported that TCTP can function as a transcriptional regulator to induce oct4 genes for the maintenance of stem cell fate (Cheng et al. 2012; Koziol et al. 2007). These findings suggest that TCTP can have important nuclear functions distinct from its known functions in the cytosol.
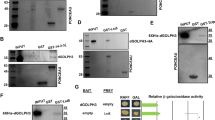
In Drosophila, first clue to the potential nuclear function of Tctp was provided by the finding that Tctp directly interacts with Ataxia Telangiectasia Mutated (ATM). Previously, we have used a phage display method to identify specific peptide ligands that bind to Tctp. Screening of aptamer libraries helped identify Tctp-binding peptide sequences. ATM was one of the proteins that contain Tctp-binding aptamer sequences and was confirmed to bind Tctp (Hong and Choi 2013). ATM is a serine–threonine protein kinase pivotal for repairing DNA damages caused by double strand break (DSB). Importantly, Tctp was found to form nuclear foci colocalized with ATM upon γ-ray irradiation, which is consistent with its function in DNA damage response. Mammalian ATM forms a protein complex with other factors such as Mre11 and Rad50 for DNA repair. Drosophila Tctp was also found to be associated with Mre11 and Rad50, suggesting that Tctp is a member of the functional DNA repair complex. In fact, Tctp mutants were defective in various DNA damage responses, showing frequent aberrant chromosomes, abnormal G2/M checkpoint response, and higher cell death upon ionizing irradiation. Consistent with the physical interaction between Tctp and ATM, defective DNA damage responses in atm mutants including abnormal chromosomes were strongly enhanced by reducing the level of Tctp. Likewise, growth defects by Tctp RNAi were strongly enhanced by reduced dosages of atm and other known DNA damage control genes. Together with the physical interaction between Tctp and ATM, these genetic interactions support the direct involvement of Tctp in DNA damage control in vivo.
Upon sensing DSBs, ATM kinase phosphorylates H2Av (H2AX in mammals), a variant form of histone 2A. The generation of phosphorylated H2Av (γH2Av) is an initial step for recruiting DNA repair proteins. Thus, γH2Av is a biomarker for DSBs and the sites of damage repair foci (Kuo and Yang 2008). Larval salivary gland cells undergo endocycling without mitosis and have a high level of γH2Av. In Tctp mutants, levels of γH2Av in salivary glands were diminished compared with the wild-type level. The lower level of γH2Av was rescued by adding a wild-type Tctp gene in the mutant background. Furthermore, the level of γH2Av induction upon γ-irradiation was also significantly lower in Tctp mutant wing discs than wild-type discs. These results provide in vivo evidence that Tctp is required for repairing both endogenous and exogenous DNA damages.
An important question is how the interaction between Tctp and ATM leads to DSB repair. Since Tctp mutations result in a reduction of the γH2Av level, Tctp might be involved in the promotion of ATM kinase activity. In vitro assays showed that addition of Tctp increased the ATM kinase activity toward the H2Av substrate in a dose-dependent manner. Thus, Tctp is directly involved in enhancing the ATM kinase activity, facilitating the DSB repair process. It is worth noting that defective Tctp with E12V substitution could not activate ATM kinase activity, indicating that the E12 residue is critical for the function of Tctp in TOR signaling as well as for the interaction with ATM. Remarkably, human TCTP is also associated with ATM and several proteins involved in DSB repair such as Ku70/80, DNA-binding subunits of DNA-dependent protein kinase. Knockdown of TCTP impairs its ability to repair DSBs in irradiated human cells (Zhang et al. 2012). Thus, Drosophila and human TCTP proteins show strong conservation in their nuclear functions for DNA damage response (Fig. 8.3).
Tctp–ATM interaction for DNA repair in mammals and fly. Relationships between TCTP and ATM in DNA damage response in humans and fly. In humans, ATM inhibits TCTP expression in nonirradiated cells. DNA damage by γ-ray irradiation leads to an increase in the TCTP level through ATM. TCTP forms a protein complex with Filamin A, P53, and Ku70/80 for DNA repair. It is unknown whether TCTP directly binds to ATM. In Drosophila, γ-irradiation has little effect on the Tctp level. Tctp directly binds to ATM and promotes its kinase activity. Tctp shows genetic interaction with P53 and Ku70/78, but it is unknown whether this interaction is direct
We have mentioned earlier that growth defects of Tctp mutant cell clones can be partially suppressed by providing CycE as well as the P35 cell death inhibitor, suggesting that Tctp is also required for preventing cell death. This is an important point because mammalian TCTP proteins are anti-apoptotic (Telerman and Amson 2009). One of the mechanisms of TCTP’s anti-apoptotic function is related to its ability to bind Bcl-2 family proteins Bcl-xL and MCL1 that inhibit the pro-apoptotic activity of Bax, located in the outer mitochondrial membrane (Liu et al. 2005; Susini et al. 2008). A recent study has shown that TCTP contains a BH3-like domain that recognizes the BH3 domain of Bcl-xL to activate the anti-apoptotic function of Bcl-xL (Thebault et al. 2016). Interestingly, Drosophila Tctp also has a putative BH3 domain with conserved hydrophobic residues. In addition to the TCTP interaction with anti-apoptotic mitochondrial proteins, mammalian TCTPs are known to inhibit p53 tumor suppressor-induced cell death upon DNA damage (Amson et al. 2011; Kloc et al. 2012). TCTP not only represses transcription of p53 but it also destabilizes p53 in order to inhibit apoptosis (Rho et al. 2011). We have shown that reduced eye caused by Tctp RNAi can be suppressed by overexpression of dominant-negative p53 (Hong and Choi 2013). This suggests that cell death resulting from Tctp knockdown is due in part to p53 upregulation or activation and that the relationship between p53 and TCTP in mammalian systems might also be conserved in Drosophila.
8.7 Tctp in Chromatin Remodeling and Genome Stability
A closer examination of the nuclear localization of Tctp in the salivary gland indicates that it is associated with most interband regions of polytene chromosomes. Furthermore, from a yeast two-hybrid screen, Tctp was found to interact with Brahma (Brm), the Drosophila homolog of yeast Swi/SNF chromatin remodeling factor (Hong and Choi 2016). These findings raised the possibility that Tctp may have a regulatory role at the level of chromatin. A series of genetic and biochemical tests revealed that Tctp is required for inhibiting the ATPase activity of Brm and therefore antagonizing the Brm function in developing organs. Consistent with the role of the Brm remodeler complex in transcriptional gene regulation, loss of Tctp increases RNA polymerase II activity, enhancing transcription in a number of genes.
Interestingly, a reduction in the Tctp level leads to dramatic increases in the transcriptional expression of retrotransposons inserted in the pericentromeric regions, situated near the centromere and highly modified by heterochromatin marks (Hong and Choi 2016). Silencing of retrotransposons by heterochromatin marks is important for the maintenance of genome stability (Larson et al. 2012; Peng and Karpen 2007, 2009; Shi et al. 2008). In addition to its role for inhibiting transposon expression, Tctp is necessary to maintain the stability of rDNA genes and other repeated sequences. Methylation of histone 3 at lysine 9 (H3K9me2/3) by SU(VAR)3–9 histone methyl transferase (HMT) is a critical mark for HP1a-dependent heterochromatin formation and gene silencing. Our data demonstrate that Tctp is required for transcription of su(var)3–9, hence affecting the levels of H3K9 methylation and HP1a protein (Fig. 8.4).
A model for Tctp function in chromatin remodeling and genome stability. Tctp keeps genome stability through two different compaction mechanisms: (1) Heterochromatin formation by inducing transcription of su(var)3–9, thus increasing H3K9 methylation at the initial step of chromatin compaction and (2) maintenance of the proper level of chromatin opening by negatively modulating the Brm chromatin remodeler
Position effect variegation (PEV) is a genetic phenomenon caused by repression of the genes abnormally transposed near heterochromatic regions. Tctp mutations suppress various PEV phenotypes, consistent with its role in promoting the gene silencing effects of heterochromatin. Tctp and Brm also show the opposing relationship in their effects on PEV. These observations support that the antagonistic relationship between Tctp and Brm contributes to the regulation of the chromatin boundary between euchromatin and heterochromatin (Hong and Choi 2016).
8.8 Concluding Remarks
Tctp is an evolutionarily conserved protein. Most of structural features of mammalian TCTP family proteins are shared in invertebrate systems like Drosophila. TCTP family proteins also seem to be conserved in their in vivo functions, based on the successful complementation of mutant phenotypes by TCTP transgenes from other species. Despite the structural and functional conservation, however, there are several differences between the Drosophila Tctp and vertebrate TCTP proteins.
Firstly, TCTP is known as a histamine releasing factor (HRF) that is secreted to promote immune responses (MacDonald et al. 1995). TCTP homologs are secreted from the parasite Plasmodium falciparum that causes malaria and found in the plasma of infected hosts (MacDonald et al. 2001). Evidence suggests that TCTP is secreted via exosomal secretory pathway (Amzallag et al. 2004; Lespagnol et al. 2008). It is an intriguing question whether secretion is a general property of TCTP family proteins. If Drosophila Tctp can be secreted to act at a distance, phenotypes of Tctp mutant clones in genetically mosaic tissues might be rescued by Tctp protein secreted from the adjacent wild-type cells. Thus far, we have not noticed any obvious sign of non-cell autonomous function of Tctp in developing Drosophila tissues. It has been shown that truncation of the N terminal sequence and dimerization of TCTP is necessary for its cytokine-like extracellular activity (Kim et al. 2009). A critical cysteine residue of human TCTP involved in the dimerization is also conserved in Drosophila Tctp. Therefore, although the function of Tctp in organ growth seems to be cell-autonomous, we cannot exclude the possibility that Tctp can act as a secreted factor in Drosophila.
Secondly, one of the most unexpected findings from our Drosophila Tctp studies is the apparent lack of gain-of-function Tctp phenotypes. Human TCTP is upregulated in various cancer cells (Tuynder et al. 2002) and has been implicated in tumorigenesis (Bae et al. 2015; Jung et al. 2011; Niforou et al. 2008). However, despite the critical requirements of Drosophila Tctp for organ growth, ectopic or overexpression of Tctp in various tissues does not induce tumorous overgrowth (Hong and Choi 2013). Thus, Tctp functions seem to be permissive rather than instructive. It is unclear why Tctp overexpression does not induce overgrowth in Drosophila. One possibility is that upregulation of TCTP observed in various cancer cells might be a secondary consequence rather than the primary cause of cancers. Because TCTP is necessary for cell proliferation and survival, knockdown of TCTP might block the growth of cancer cells, possibly explaining the phenomenon of tumor reversion. Alternatively, although Drosophila Tctp overexpression may not be sufficient to induce tumorous growth in normal tissues, it might trigger overgrowth in the presence of an additional factor(s) or under certain cellular conditions. If that is the case, systematic searches for such factors would be critical to understand the mechanism for TCTP-induced tumorigenicity. Studies with transgenic TCTP mice have shown that overexpression of TCTP causes hypertension but with normal appearance at any age (Kim et al. 2008). Therefore, mammalian TCTP may also require additional factors to induce cancer, and it is an interesting possibility that upregulation of TCTP might predispose normal cells to be transformed when combined with an additional factor.
Thirdly, the role of Tctp in DNA damage repair seems to be conserved, but important differences have also been noticed (Fig. 8.3). Both Drosophila Tctp and human TCTP are involved in two distinct mechanisms of DNA repair: homologous recombination (HR) and nonhomologous end-joining (NHEJ). In human cells, depletion of ATM in nonirradiated cells results in an increase of TCTP level. Further, low dose γ-rays upregulate TCTP to protect against DNA damage, and this upregulation depends on ATM (Zhang et al. 2012). Thus, ATM seems to be involved in regulating the level of TCTP expression and/or stability in nonirradiated cells or cells exposed to low dose γ-irradiation. In contrast, the level of Drosophila Tctp is not influenced by irradiation or ATM function (Hong and Choi 2013). Instead, Tctp directly promotes the ATM kinase activity to phosphorylate H2Av. Therefore, loss of Tctp results in a reduction of γH2Av level. This is also in contrast to TCTP-depleted human fibroblast cells in which the number of γH2AX nuclear foci and the level of γH2AX remains high. Human fibroblast cells might have a mechanism to compensate the loss of TCTP to maintain the γH2AX levels. It is also possible that upregulated human TCTP by ATM might be able to activate ATM kinase activity as in the case for Drosophila. Future studies are necessary to understand the basis for these variations in the functional relationship between TCTP and ATM in different organisms.
There is profound evidence that many cancers are associated with genome instability. Because loss of TCTP function impairs DNA repair, it may lead to genome instability causing cancer. Therefore, not only upregulation of TCTP but also its loss may be associated with cancer through distinct mechanisms. Furthermore, Drosophila Tctp is critical for genome stability by regulating global gene expression and chromatin modification. It is an important question to be addressed in the future whether mammalian TCTP might play similar roles in the regulation of chromatin modification and genome stability.
References
Acevedo SF, Tsigkari KK, Grammenoudi S, Skoulakis EM (2007) In vivo functional specificity and homeostasis of Drosophila 14-3-3 proteins. Genetics 177:239–253
Aghazadeh Y, Papadopoulos V (2016) The role of the 14-3-3 protein family in health, disease, and drug development. Drug Discov Today 21:278–287
Aitken A, Baxter H, Dubois T, Clokie S, Mackie S, Mitchell K, Peden A, Zemlickova E (2002) Specificity of 14-3-3 isoform dimer interactions and phosphorylation. Biochem Soc Trans 30:351–360
Amson R, Kubiak JZ, Van Montagu M, Telerman A (2011) Could TCTP contribute to Armin Braun's paradigm of tumor reversion in plants? Cell Cycle 10:1
Amson R, Pece S, Lespagnol A, Vyas R, Mazzarol G, Tosoni D, Colaluca I, Viale G, Rodrigues-Ferreira S, Wynendaele J et al (2012) Reciprocal repression between P53 and TCTP. Nat Med 18:91–99
Amson R, Pece S, Marine JC, Di Fiore PP, Telerman A (2013) TPT1/TCTP-regulated pathways in phenotypic reprogramming. Trends Cell Biol 23:37–46
Amzallag N, Passer BJ, Allanic D, Segura E, Thery C, Goud B, Amson R, Telerman A (2004) TSAP6 facilitates the secretion of translationally controlled tumor protein/histamine-releasing factor via a nonclassical pathway. J Biol Chem 279:46104–46112
Bae SY, Kim HJ, Lee KJ, Lee K (2015) Translationally controlled tumor protein induces epithelial to mesenchymal transition and promotes cell migration, invasion and metastasis. Sci Rep 5:8061
Baker NE (2007) Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev 17:287–293
Bohm H, Benndorf R, Gaestel M, Gross B, Nurnberg P, Kraft R, Otto A, Bielka H (1989) The growth-related protein P23 of the Ehrlich ascites tumor: translational control, cloning and primary structure. Biochem Int 19:277–286
Bommer UA, Thiele BJ (2004) The translationally controlled tumour protein (TCTP). Int J Biochem Cell Biol 36:379–385
Bonnet C, Perret E, Dumont X, Picard A, Caput D, Lenaers G (2000) Identification and transcription control of fission yeast genes repressed by an ammonium starvation growth arrest. Yeast 16:23–33
Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401–415
Brioudes F, Thierry AM, Chambrier P, Mollereau B, Bendahmane M (2010) Translationally controlled tumor protein is a conserved mitotic growth integrator in animals and plants. Proc Natl Acad Sci U S A 107:16384–16389
Burton J, Roberts D, Montaldi M, Novick P, De Camilli P (1993) A mammalian guanine-nucleotide-releasing protein enhances function of yeast secretory protein Sec4. Nature 361:464–467
Cheng X, Li J, Deng J, Li Z, Meng S, Wang H (2012) Translationally controlled tumor protein (TCTP) downregulates Oct4 expression in mouse pluripotent cells. BMB Rep 45:20–25
Chitpatima ST, Makrides S, Bandyopadhyay R, Brawerman G (1988) Nucleotide sequence of a major messenger RNA for a 21 kilodalton polypeptide that is under translational control in mouse tumor cells. Nucleic Acids Res 16:2350
Chung S, Kim M, Choi W, Chung J, Lee K (2000) Expression of translationally controlled tumor protein mRNA in human colon cancer. Cancer Lett 156:185–190
Cohen SM, Di Nardo S (1993) Wingless: from embryo to adult. Trends Genet 9:189–192
Datar SA, Galloni M, de la Cruz A, Marti M, Edgar BA, Frei C (2006) Mammalian cyclin D1/Cdk4 complexes induce cell growth in Drosophila. Cell Cycle 5:647–652
Dong J, Pan D (2004) Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev 18:2479–2484
Dong X, Yang B, Li Y, Zhong C, Ding J (2009) Molecular basis of the acceleration of the GDP-GTP exchange of human ras homolog enriched in brain by human translationally controlled tumor protein. J Biol Chem 284:23754–23764
Fonseca BD, Smith EM, Lee VH, MacKintosh C, Proud CG (2007) PRAS40 is a target for mammalian target of rapamycin complex 1 and is required for signaling downstream of this complex. J Biol Chem 282:24514–24524
Gachet Y, Tournier S, Lee M, Lazaris-Karatzas A, Poulton T, Bommer UA (1999) The growth-related, translationally controlled protein P23 has properties of a tubulin binding protein and associates transiently with microtubules during the cell cycle. J Cell Sci 112(Pt 8):1257–1271
Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G (2003) Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11:1457–1466
Gong X, Yan L, Gu H, Mu Y, Tong G, Zhang G (2014) 14-3-3epsilon functions as an oncogene in SGC7901 gastric cancer cells through involvement of cyclin E and p27kip1. Mol Med Rep 10:3145–3150
Guillaume E, Pineau C, Evrard B, Dupaix A, Moertz E, Sanchez JC, Hochstrasser DF, Jegou B (2001) Cellular distribution of translationally controlled tumor protein in rat and human testes. Proteomics 1:880–889
Hashemolhosseini S, Nagamine Y, Morley SJ, Desrivieres S, Mercep L, Ferrari S (1998) Rapamycin inhibition of the G1 to S transition is mediated by effects on cyclin D1 mRNA and protein stability. J Biol Chem 273:14424–14429
Hong ST, Choi KW (2013) TCTP directly regulates ATM activity to control genome stability and organ development in Drosophila melanogaster. Nat Commun 4:2986
Hong ST, Choi KW (2016) Antagonistic roles of Drosophila Tctp and Brahma in chromatin remodelling and stabilizing repeated sequences. Nat Commun 7:12988
Hsu YC, Chern JJ, Cai Y, Liu M, Choi KW (2007) Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445:785–788
Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4:648–657
Jeon HJ, You SY, Park YS, Chang JW, Kim JS, Oh JS (2016) TCTP regulates spindle microtubule dynamics by stabilizing polar microtubules during mouse oocyte meiosis. Biochim Biophys Acta 1863:630–637
Jewell JL, Russell RC, Guan KL (2013) Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol 14:133–139
Jung J, Kim HY, Kim M, Sohn K, Kim M, Lee K (2011) Translationally controlled tumor protein induces human breast epithelial cell transformation through the activation of Src. Oncogene 30:2264–2274
Kim M, Jung Y, Lee K, Kim C (2000) Identification of the calcium binding sites in translationally controlled tumor protein. Arch Pharm Res 23:633–636
Kim MJ, Kwon JS, Suh SH, Suh JK, Jung J, Lee SN, Kim YH, Cho MC, Oh GT, Lee K (2008) Transgenic overexpression of translationally controlled tumor protein induces systemic hypertension via repression of Na+,K+-ATPase. J Mol Cell Cardiol 44:151–159
Kim M, Min HJ, Won HY, Park H, Lee JC, Park HW, Chung J, Hwang ES, Lee K (2009) Dimerization of translationally controlled tumor protein is essential for its cytokine-like activity. PLoS One 4:e6464
Kloc M, Tejpal N, Sidhu J, Ganachari M, Flores-Villanueva P, Jennings NB, Sood AK, Kubiak JZ, Ghobrial RM (2012) Inverse relationship between TCTP/RhoA and p53/cyclin a/actin expression in ovarian cancer cells. Folia Histochem Cytobiol 50:358–367
Koziol MJ, Garrett N, Gurdon JB (2007) Tpt1 activates transcription of oct4 and nanog in transplanted somatic nuclei. Curr Biol 17:801–807
Kuo LJ, Yang LX (2008) Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In Vivo 22:305–309
Laplante M, Sabatini DM (2012a) mTOR signaling. Cold Spring Harb Perspect Biol 4
Laplante M, Sabatini DM (2012b) mTOR signaling in growth control and disease. Cell 149:274–293
Larson K, Yan SJ, Tsurumi A, Liu J, Zhou J, Gaur K, Guo D, Eickbush TH, Li WX (2012) Heterochromatin formation promotes longevity and represses ribosomal RNA synthesis. PLoS Genet 8:e1002473
Le TP, Vuong LT, Kim AR, Hsu YC, Choi KW (2016) 14-3-3 proteins regulate Tctp-Rheb interaction for organ growth in Drosophila. Nat Commun 7:11501
Lee T, Luo L (2001) Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24:251–254
Lespagnol A, Duflaut D, Beekman C, Blanc L, Fiucci G, Marine JC, Vidal M, Amson R, Telerman A (2008) Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ 15:1723–1733
Li Y, Inoki K, Yeung R, Guan KL (2002) Regulation of TSC2 by 14-3-3 binding. J Biol Chem 277:44593–44596
Liu H, Peng HW, Cheng YS, Yuan HS, Yang-Yen HF (2005) Stabilization and enhancement of the antiapoptotic activity of mcl-1 by TCTP. Mol Cell Biol 25:3117–3126
MacDonald SM, Rafnar T, Langdon J, Lichtenstein LM (1995) Molecular identification of an IgE-dependent histamine-releasing factor. Science 269:688–690
MacDonald SM, Bhisutthibhan J, Shapiro TA, Rogerson SJ, Taylor TE, Tembo M, Langdon JM, Meshnick SR (2001) Immune mimicry in malaria: Plasmodium falciparum secretes a functional histamine-releasing factor homolog in vitro and in vivo. Proc Natl Acad Sci U S A 98:10829–10832
Miron M, Verdu J, Lachance PE, Birnbaum MJ, Lasko PF, Sonenberg N (2001) The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat Cell Biol 3:596–601
Morrison DK (2009) The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 19:16–23
Moya M, Roberts D, Novick P (1993) DSS4-1 is a dominant suppressor of sec4-8 that encodes a nucleotide exchange protein that aids Sec4p function. Nature 361:460–463
Nielsen HV, Johnsen AH, Sanchez JC, Hochstrasser DF, Schiotz PO (1998) Identification of a basophil leukocyte interleukin-3-regulated protein that is identical to IgE-dependent histamine-releasing factor. Allergy 53:642–652
Niforou KM, Anagnostopoulos AK, Vougas K, Kittas C, Gorgoulis VG, Tsangaris GT (2008) The proteome profile of the human osteosarcoma U2OS cell line. Cancer Genomics Proteomics 5:63–78
Norbeck J, Blomberg A (1997) Two-dimensional electrophoretic separation of yeast proteins using a non-linear wide range (pH 3-10) immobilized pH gradient in the first dimension; reproducibility and evidence for isoelectric focusing of alkaline (pI > 7) proteins. Yeast 13:1519–1534
Obsilova V, Silhan J, Boura E, Teisinger J, Obsil T (2008) 14-3-3 proteins: a family of versatile molecular regulators. Physiol Res 57(Suppl 3):S11–S21
Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E (2000) Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev 14:2689–2694
Pallares-Cartes C, Cakan-Akdogan G, Teleman AA (2012) Tissue-specific coupling between insulin/IGF and TORC1 signaling via PRAS40 in drosophila. Dev Cell 22:172–182
Peng JC, Karpen GH (2007) H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9:25–35
Peng JC, Karpen GH (2009) Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet 5:e1000435
Predic J, Soskic V, Bradley D, Godovac-Zimmermann J (2002) Monitoring of gene expression by functional proteomics: response of human lung fibroblast cells to stimulation by endothelin-1. Biochemistry 41:1070–1078
Rehmann H, Bruning M, Berghaus C, Schwarten M, Kohler K, Stocker H, Stoll R, Zwartkruis FJ, Wittinghofer A (2008) Biochemical characterisation of TCTP questions its function as a guanine nucleotide exchange factor for Rheb. FEBS Lett 582:3005–3010
Rho SB, Lee JH, Park MS, Byun HJ, Kang S, Seo SS, Kim JY, Park SY (2011) Anti-apoptotic protein TCTP controls the stability of the tumor suppressor p53. FEBS Lett 585:29–35
Rosenwald IB, Kaspar R, Rousseau D, Gehrke L, Leboulch P, Chen JJ, Schmidt EV, Sonenberg N, London IM (1995) Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem 270:21176–21180
Sanchez JC, Schaller D, Ravier F, Golaz O, Jaccoud S, Belet M, Wilkins MR, James R, Deshusses J, Hochstrasser D (1997) Translationally controlled tumor protein: a protein identified in several nontumoral cells including erythrocytes. Electrophoresis 18:150–155
Saucedo LJ, Gao XS, Chiarelli DA, Li L, Pan D, Edgar BA (2003) Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5:566–571
Shi S, Larson K, Guo D, Lim SJ, Dutta P, Yan SJ, Li WX (2008) Drosophila STAT is required for directly maintaining HP1 localization and heterochromatin stability. Nat Cell Biol 10:489–496
Singh A, Lim J, Choi KW (2005) Dorsoventral boundary for organizing growth and planar polarity in the Drosophila eye. In: Mlodzik M (ed) Advances in developmental biology: planar cell polarization during development. Elsevier, Amsterdam, p 59, 172p
Skoulakis EM, Davis RL (1996) Olfactory learning deficits in mutants for leonardo, a Drosophila gene encoding a 14-3-3 protein. Neuron 17:931–944
Stocker H, Radimerski T, Schindelholz B, Wittwer F, Belawat P, Daram P, Breuer S, Thomas G, Hafen E (2003) Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat Cell Biol 5:559–566
Susini L, Besse S, Duflaut D, Lespagnol A, Beekman C, Fiucci G, Atkinson AR, Busso D, Poussin P, Marine JC et al (2008) TCTP protects from apoptotic cell death by antagonizing bax function. Cell Death Differ 15:1211–1220
Telerman A, Amson R (2009) The molecular programme of tumour reversion: the steps beyond malignant transformation. Nat Rev Cancer 9:206–216
Thaw P, Baxter NJ, Hounslow AM, Price C, Waltho JP, Craven CJ (2001) Structure of TCTP reveals unexpected relationship with guanine nucleotide-free chaperones. Nat Struct Biol 8:701–704
Thebault S, Agez M, Chi X, Stojko J, Cura V, Telerman SB, Maillet L, Gautier F, Billas-Massobrio I, Birck C et al (2016) TCTP contains a BH3-like domain, which instead of inhibiting, activates Bcl-xL. Sci Rep 6:19725
Tuynder M, Susini L, Prieur S, Besse S, Fiucci G, Amson R, Telerman A (2002) Biological models and genes of tumor reversion: cellular reprogramming through tpt1/TCTP and SIAH-1. Proc Natl Acad Sci U S A 99:14976–14981
Tuynder M, Fiucci G, Prieur S, Lespagnol A, Geant A, Beaucourt S, Duflaut D, Besse S, Susini L, Cavarelli J et al (2004) Translationally controlled tumor protein is a target of tumor reversion. Proc Natl Acad Sci U S A 101:15364–15369
Wang X, Fonseca BD, Tang H, Liu R, Elia A, Clemens MJ, Bommer UA, Proud CG (2008) Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J Biol Chem 283:30482–30492
Xu T, Rubin GM (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117:1223–1237
Yaffe MB (2002) How do 14-3-3 proteins work?-- gatekeeper phosphorylation and the molecular anvil hypothesis. FEBS Lett 513:53–57
Zhang H, Stallock JP, Ng JC, Reinhard C, Neufeld TP (2000) Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev 14:2712–2724
Zhang Y, Gao XS, Saucedo LJ, Ru BG, Edgar BA, Pan DJ (2003) Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 5:578–581
Zhang J, de Toledo SM, Pandey BN, Guo G, Pain D, Li H, Azzam EI (2012) Role of the translationally controlled tumor protein in DNA damage sensing and repair. Proc Natl Acad Sci U S A 109:E926–E933
Zhao J, Meyerkord CL, Du Y, Khuri FR, Fu H (2011) 14-3-3 proteins as potential therapeutic targets. Semin Cell Dev Biol 22:705–712
Acknowledgements
We thank Kyungok Cho and Jean Jung for comments on the manuscript. This work was supported by NRF-2014K1A1A2042982, NRF-2017R1A2B3007516 (KWC), 2016R1D1A1B03932093 and 35B-2011-1-C00033 (STH) through the National Research Foundation of Korea funded by the Korean Ministry of Education Science & Technology.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Choi, KW., Hong, ST., Le, T.P. (2017). Function of Translationally Controlled Tumor Protein in Organ Growth: Lessons from Drosophila Studies. In: Telerman, A., Amson, R. (eds) TCTP/tpt1 - Remodeling Signaling from Stem Cell to Disease. Results and Problems in Cell Differentiation, vol 64. Springer, Cham. https://doi.org/10.1007/978-3-319-67591-6_8
Download citation
DOI: https://doi.org/10.1007/978-3-319-67591-6_8
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67590-9
Online ISBN: 978-3-319-67591-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)