Abstract
In this paper we show how we used multiple large knowledge sources to construct a much smaller knowledge graph that is focussed on single disease (in our case major depression disorder). Such a disease-centric knowledge-graph makes it more convenient for doctors (in our case psychiatric doctors) to explore the relationship among various knowledge resources and to answer realistic clinical queries.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Major Depressive Disorder
- Regular Expression
- Clinical Decision Support
- Knowledge Source
- Knowledge Resource
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Major depressive disorder (MDD) has become a serious problem in modern society. Using antidepressants has been considered the dominant treatment for MDD. However, 30% to 50% of the individuals treated with antidepressants do not show a response. Hence, psychiatric doctors confront the challenge to make clinical decision efficiently by gaining a comprehensive analysis over various knowledge resources about depression. In this paper we propose an approach to constructing a knowledge graph of depression using semantic web technology to integrate those knowledge resources, achieving a high degree of inter-operability. With a single semantic query over integrated knowledge resources, psychiatric doctors can be much more efficient in finding answers to queries which currently require them to explore multiple databases and to make a time-consuming analysis on the results of those searches.
The term “Knowledge Graph” is widely used to refer to a large scale semantic network of entities and concepts plus the semantic relationships among them. Medical knowledge graphs typically cover very wide areas of medical knowledge: all proteines (UniProt), as many drugs as possible (Drugbank), as many drug-drug interactions as are known (Sider), and massively integrated knowledge graphs such as Bio2RDF and LinkedLifeData. Such knowledge graphs are very a-specific in terms of the diseases that they cover, and are often prohibitively large, hampering both efficiency for machines and usability for people. In this abridged paper we propose an approach to the construction of disease-centric knowledge graphs. Our claims are (i) that it is indeed possible to make disease-centric subgraphs and (ii) that realistic clinical queries can still be answered over such disease-specific knowledge graphs without substantial loss of recall.
We illustrate our general idea by integrating various knowledge resources about depression (e.g., clinical trials, antidepressants, medical publications, clinical guidelines, etc.). We call the generated knowledge graph DepressionKG for short. DepressionKG is represented in RDF/NTriple format [2]. DepressionKG provides a data infrastructure to explore the relationship among various knowledge and data-sources about depression. We show how it provides support for clinical question answering and knowledge browsing.
2 Challenges
In order to integrate various knowledge resources of depression, we had to confront the following challenges: (i) Heterogeneity: Different knowledge resources are generated by multiple creators. We have to achieve semantic inter-operability among those knowledge resources; (ii) Free text processing. Some of our knowledge resources contained a lot of free text. We have to use a natural language processing tool with a medical terminology to extract semantic relations from free text; (iii) Partiality, inconsistency, and incorrectness. Knowledge resources often contain some partial, inconsistent, noisy or even erroneous data. We have to develop efficient methods to deal with this; (iv) Expressive Representation of Medical Knowledge. The formal languages of Knowledge Graphs (RDF, RFD Schema, OWL) are a decidable fragment of first order logic. Such languages are usually not expressive enough for medical knowledge representation.
In this paper we present some methods for dealing with the first two challenges when constructing a knowledge graph of depression, while leaving the third and fourth challenges for future work. We will show how DepressionKG can be used in some realistic scenarios for clinical decision support. We have implemented a system of DepressionKG aimed at psychiatric doctors with no background knowledge in knowledge graphs.
3 Knowledge Resources and Integration
Following commonly used technology, we will construct our knowledge graph as an RDF graph. A summary of DepressionKG is shown in the table below. This shows that the resulting knowledge graph is only of moderate size (8M triples), whereas many of the original knowledge graphs are many times larger than this (10M–100M triples). In Sect. 4 we will illustrate that we can still answer a diversity of clinically relevant questions with such a small disease-centric knowledge graph. Our 8M triples are dominated by SNOMED. We decided to include all of SNOMED (instead of only those parts of the hierarchies relevant to depression) because some of the clinical use-cases presented to us by our psychiatric experts concern comorbidities (e.g., Alzheimer disease is a frequent comorbidity with MDD), and restricting SNOMED would hamper such comorbidity queries.
Knowledge resource | Number of data item | Number of triple |
|---|---|---|
ClinicalTrial | 10,190 trials | 1,606,446 |
PubMed on depression | 46,060 papers | 1,059,398 |
Medical guidelines | 1 guideline | 1,830 |
DrugBank | 4,770 drugs | 766,920 |
DrugBook | 264 antidepressants | 13,046 |
Wikipedia side effects | 17 antidepressants | 6,608 |
SNOMED CT | 5,045,225 | |
Patient data | 1,000 patients | 200,000 |
Total | 8,699,473 |

We use the following four methods to integrate the various knowledge resources. (i) Direct Entity identification. Some knowledge resources refer to the same entity with identical names, e.g. the PubMed IDs used in both PubMed and the clinical trials. Such entities are obvious links between these knowledge sources; (ii) Direct Concept identification. Numerous knowledge resources can be integrated by using direct concept identification. For example, both a publication in PubMed and a clinical trial are annotated with MeSH terms. This provides us with a way to detect a relationship between a clinical trial and a publication directly; (iii) Semantic Annotation with an NLP tool. We used Xerox’s NLP tool XMedlan [1] for semantically annotating medical text (both concept identification and relation extraction) with medical terminologies such as SNOMED CT; (iv) Semantic Queries with regular expressions. The previous three approaches are offline approaches to integrate knowledge sources. Semantic Queries with regular expressions are an online approach, because such queries find relationships among knowledge resources at query time. Although online methods lead to more latency, they do provide a method to detect a connection among different knowledge resources based on free text.
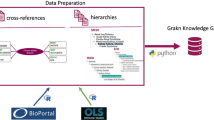
The figure shows the connectivity of DepressionKG. An arrow denotes a direct concept connection via a property, and a dashed arrow denotes a concept identification in a medical terminology by using an NLP tool. The figure shows that our set of knowledge resources is well integrated.
4 Use Cases
In this section, we will discuss several use cases how the knowledge graph on depression can be used by psychiatric doctors for clinical decision support through SPARQL queries over the knowledge graphs. Because of space constraints, we only give the code of the SPARQL query for one example.
Case 1. Patient A, female, aged 20. She has suffered from MDD for three years. In the past, she took the SSRI antidepressant Paroxetine, however, gained a lot of weight. She wants an antidepressant which has the effect of weight loss. This can be answered with a SPARQL query over a single knowledge source, combining semantic properties and a regular expression on the textual description of symptoms From the result of this query, we learn that taking Bupropion may lead to weight loss, and taking Fluoxetine may lead to a modest weight loss.
Case 2. Patient B, a female adolescent with MDD. She failed to respond to first-line treatment with Fluoxetine. The doctor wants to know the details of any clinical trial which investigates the effect of Fluoxetine and the publications of those trials. The following query searches over two knowledge resources ClincalTrial and PubMed in the knowledge graph:

This query finds three relevant trials, one with two publications, the others with one publication.
Case 3. Patient C, an adult male, suffers from mood disorder and hopes to try a clinical trial on depression. His clinical doctor wants to find an on-going trial which uses a drug intervention with target “neurotransmitter transporter activity”. This requires a search that covers both DrugBank and ClinicalTrial. From DrugBank we find the drug target, and from ClinicalTrial we find which trial has an intervention with the required drugs. This query returns 25 clinical trials whose starting date is in 2016, and which meet the specified condition.
Case 4. Patient D, male, aged 45, has complained a lot that the antidepressant Clomipramine has lead to fatigue. Indeed fatigue is a very common side effect of Clomipramine. The psychiatric doctor wants to know if there exists any other antidepression drug of the same class where fatigue is a rare or uncommon side effect. In this example, we use the predicate skos:narrower and skos:broader to search over the SNOMED concept hierarchy to find two sibling concepts (i.e. two antidepressants of the same class). The answer for this search is “Dosulepin”.
5 Implementation, Discussion and Conclusion
We have implemented the DepressionKG system with a graphical user interface, so that psychiatric doctors can use the system to search for the knowledge they need and to explore the relationships among various knowledge resources for clinical decision support. The DepressionKG system supports knowledge browsing and querying, and will be evaluated in Beijing Anding Hospital, one of the biggest psychiatric hospitals in China, for experiments in the Smart Ward project. The objective of the Smart Ward project is to develop a knowledge-based platform for monitoring and analyzing the status of patients and for supporting clinical decision making in a psychiatric ward.
In this paper, we have proposed an approach to making a knowledge graph of depression, and we have shown how various knowledge resources concerning depression can be integrated for semantic inter-operability. We have provided several use cases for such a knowledge graph of depression. From those use cases, we can see that by using a knowledge graph with its semantic search, it is rather convenient for us to detect relationship which cover multiple knowledge resources.
References
Ait-Mokhtar, S., Bruijn, B.D., Hagege, C., Rupi, P.: Intermediary-stage ie components, D3.5, Technical report, EURECA Project (2014)
Cyganiak, R., Wood, D., Lanthaler, M.: RDF 1.1 concepts and abstract syntax (2014)
Acknowledgments
This work is partially supported by the Dutch national project COMMIT, the international cooperation project No. 61420106005 funded by National Natural Science Foundation of China, and the NWO-funded Project Re-Search. The fourth author is funded by the China Scholarship Council.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Huang, Z., Yang, J., van Harmelen, F., Hu, Q. (2017). Constructing Disease-Centric Knowledge Graphs: A Case Study for Depression (short Version). In: ten Teije, A., Popow, C., Holmes, J., Sacchi, L. (eds) Artificial Intelligence in Medicine. AIME 2017. Lecture Notes in Computer Science(), vol 10259. Springer, Cham. https://doi.org/10.1007/978-3-319-59758-4_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-59758-4_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-59757-7
Online ISBN: 978-3-319-59758-4
eBook Packages: Computer ScienceComputer Science (R0)