Abstract
Recent advances in medical treatment and the development of new mechanical devices have greatly improved the prognosis for heart disease patients. However, heart disease, particularly heart failure, is still a major health issue with continuously increasing numbers of affected patients. Because adult heart muscle has a low regenerative capacity, cardiac function declines with age after cardiac injury. A potential approach to solve this problem is regenerative medicine, aiming at the remuscularization of damaged hearts. Studies conducted in small animals and humans revealed that transplanting various types of cells into failing hearts resulted in the repair of injured hearts and improved cardiac function, but the effects were modest, and further improvement is needed before the method can be widely applied in the clinic. Moreover, true muscle regeneration or cardiac differentiation from so far clinically tested adult stem cells seems to be a rare event, and the beneficial effects of these cell-based therapies are likely due to paracrine factors secreted by the transplanted cells. To regenerate cardiac muscle, it is important to first understand the mechanism of cardiac cell fate determination. Several groups including ours recently found that somatic cells can be directly reprogrammed into cardiomyocyte-like cells using combinations of master regulators. The cardiac reprogramming approach is applicable not only in vitro but also in vivo. It can repair injured hearts and improve cardiac function. Thus, this new technology may open an avenue for regenerative therapy for heart disease.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
6.1 Introduction
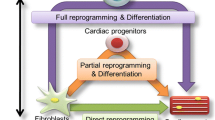
Cardiomyocytes are terminally differentiated cells and are generally considered to have little regenerative potential. Therefore, after cardiomyocytes are damaged, the damaged areas are replaced with fibroblasts and fibrous tissue. Since fibroblasts have no contractile activity, cardiac function decreases, resulting in arrhythmia and heart failure. Cardiac regenerative medicine has arisen as a promising novel therapeutic field to overcome the naturally limited regenerative potential of the heart. The approaches for developing cardiac regeneration techniques are currently focused on cell transplantation using cardiomyocytes induced ex vivo from stem cells, such as embryonic and induced pluripotent stem cells (iPSCs) (Chong et al. 2014; Lalit et al. 2014; Shiba et al. 2012). These approaches, however, have multiple challenges, including tumorigenicity derived from residual pluripotent stem cells, long-term cell engraftment, and high costs, requiring further improvements (Okano et al. 2013; Yang et al. 2014). In contrast to cell transplantation, we have been developing a novel myocardial regeneration technique for direct reprogramming of cardiac non-myocytes (fibroblasts) into cardiomyocytes (Ieda et al. 2010; Inagawa and Ieda 2013; Sadahiro et al. 2015; Wada et al. 2013; Yamakawa et al. 2015). If pre-existing non-cardiomyocytes can be converted to cardiomyocytes within patient’s heart, this new approach would be an attractive approach for pharmacologically induced remuscularization of the failing heart.
6.2 Discovery of Master Regulators for Direct Cardiac Reprogramming
In 1987, MyoD was identified as the single master gene for skeletal muscle differentiation (Davis et al. 1987). Similar investigational efforts failed to identify a single master gene for myocardial differentiation. The discovery of the four “Yamanaka factors” in 2006 suggested that the co-transfection of multiple transcription factors could potentially enable the reprogramming of terminally differentiated cells (Takahashi et al. 2007a, b; Takahashi and Yamanaka 2006). Thus, we performed a similar trial and identified three transcription factors (Gata4, Mef2c, and Tbx5, hereafter referred to as GMT) crucial for direct cardiac reprogramming in mice. In order to identify the transcription factors, we isolated cardiomyocytes and cardiac fibroblasts (CFs) using fluorescence-activated cell sorting (FACS: flow cytometry) and screened 14 genes that are specifically expressed in embryonic cardiomyocytes and play a critical role in cardiac formation as candidate factors (Ieda et al. 2009, 2010). Next, we established a genetically modified mouse that specifically expressed green fluorescent protein (GFP) only in differentiated cardiomyocytes (αMHC-GFP transgenic mouse) and screened gene candidates by quantitatively evaluating GFP expression. When all 14 genes were transduced to the CFs via retrovirus vectors, GFP-positive cells were detected at a level of approximately 1.7% a week after infection with the virus. Based on further GFP quantitative evaluations after removing each of the 14 genes, we identified three transcription factors (GMT) that are minimally required for cardiomyocyte generation. In addition, we identified a higher reprogramming efficiency with GMT (17% of GFP+ cells and 5% of cTnT+ cells). We named the cells induced from CFs that express these three transcription factors induced cardiomyocytes (iCMs).
We also determined whether iCMs have a similar morphology and function as native cardiomyocytes. Immunocytochemistry and microarray methods were applied to analyze expression at the protein and gene levels. The immunocytochemistry results demonstrated that iCMs express the cardiomyocyte-specific proteins α-actinin, cardiac troponin T (cTnT), and atrial natriuretic factor (ANF) and possess cardiomyocyte-specific sarcomeric structures. The microarray results demonstrated that gene expression changed from a CF pattern to a cardiomyocyte-like pattern, and the epigenetic status of histone methylation and DNA methylation was similar to that of cardiomyocytes. Functionally, Ca2+ imaging demonstrated similar intracellular Ca2+ kinetics as mouse neonatal cardiomyocytes. Furthermore, we confirmed spontaneous beating in some iCMs.
Subsequently, we demonstrated the direct reprogramming of CFs to a cardiomyocyte fate. Mesp1 and Isl1 are specifically expressed in cardiac progenitor cells (CPCs) (Laugwitz et al. 2005; Saga et al. 1999). We generated genetically modified mice that expressed yellow fluorescent protein (YFP) in all cells derived from CPCs by crossing Mesp1-Cre mice or Isl1-Cre mice with YFP-flox mice (Mesp1-YFP mice or Isl1-YFP mice). The iCMs derived from the fibroblasts in these mice did not express YFP, providing proof of the direct conversion to iCMs without a step involving CPCs.
Lastly, we also determined whether non-cardiac fibroblasts can be reprogrammed into iCMs by GMT. Mouse tail-tip fibroblasts (TTFs) were reprogrammed using GMT retrovirus vectors. The TTFs were successfully able to differentiate into αMHC-GFP+ cardiomyocyte-like cells. However, the reprogramming efficiency was low, as TTF-derived iCMs accounted for only ~2.5% according to a quantification cTnT+ cells, approximately half of the number of CF-derived iCMs. In addition, although TTF-derived iCMs possessed some cardiomyocyte functions such as Ca2+ regulation, they expressed higher levels of H3K27me3, a histone marker for gene suppression, in the promoters of cardiac genes than CF-derived iCMs and neonatal cardiomyocytes. These lines of evidence suggest that although various cells can be reprogrammed into iCMs, the induction rate varies depending on the original cell type. We confirmed cardiomyocyte conversion 1 week after the GMT infection by cTNT staining; few of the converted cells started to beat 4–5 weeks later. These results indicate that iCMs represent a heterogeneous cell population and that only 0.01–0.1% of CFs were reprogrammed completely into functional (i.e., beating) cardiomyocytes, warranting further improvement. Nevertheless, we identified the master regulators for cardiomyocytes that are minimally required for cardiac induction from other somatic cell types.
6.3 New Cardiac Reprogramming Factors and Stoichiometry of Transcription Factors for Efficient Reprogramming
Although GMT was identified to be a combination of critical transcription factors and enabled the reprogramming of mouse fibroblasts into functional cardiomyocytes, the poor induction rate warranted further improvements (Srivastava and Ieda 2012). Multiple studies on cardiac reprogramming have been performed since our report on the three “GMT transcription factors” and production of iCMs (Fig. 6.1).
Modification of reprogramming factors and fibroblast properties affect cardiac reprogramming efficiency. (a) Addition of Hand2 to Gata4, Mef2c, and Tbx5 (GMT), a polycistronic vector for GMT, and addition of the MyoD-M3 segment to Mef2c promoted cardiac reprogramming. (b) Cardiac reprogramming was inefficient in adult CFs and TTFs
Song et al. explored the optimal combination of core cardiac transcription factors necessary for efficient reprogramming of adult TTFs into functional cardiomyocytes (Song et al. 2012). Six cardiac transcription factors were screened using αMHC-GFP reporter mice to determine the core factors involved in cardiac reprogramming. When Hand2, a basic helix-loop-helix (bHLH) transcription factor, was added to GMT (GHMT—Gata4, Hand2, Mef2c, and Tbx5), adult CFs and TTFs were reprogrammed into functional cardiomyocyte-like cells more efficiently compared to other combinations. GMT alone resulted in only approximately 3% of the original fibroblasts becoming positive for both αMHC-GFP and cTnT. In contrast, GHMT induced approximately 9% of the adult fibroblasts to adopt a cTnT+ and αMHC-GFP+ phenotype. In addition, 5% of adult CF-derived iCMs and 1.8% of adult TTF-derived iCMs possessed sarcomere-like structures. Microarray and quantitative RT-PCR (qPCR) analyses of gene expression patterns demonstrated the upregulation of a broad range of cardiac genes, indicative of a differentiated cardiac-like phenotype, and concomitant suppression of non-myocyte genes, including Fsp1 (fibroblast-specific protein 1), in fibroblasts transduced with GHMT. Furthermore, when adult CFs or TTFs transduced with GHMT were continuously cultured for more than 5 weeks, Ca2+ transients, action potentials (APs), and spontaneous contractions were observed in the cell subsets, indicating that induced cardiomyocyte-like cells possessed similar functions as native cardiomyocytes (Nam et al. 2014). This study also demonstrated that a limited 10-day exogenous GHMT expression was sufficient to reprogram the fibroblasts toward a cardiomyocyte fate. This result suggests that GHMT may play a critical role in the onset of reprogramming, but thereafter, the reprogramming can continue to progress without exogenous GHMT expression. Based on these study results, the addition of Hand2 to GMT is considered to improve the efficiency of reprogramming of fibroblasts into cardiomyocytes compared to GMT alone (Nam et al. 2014; Song et al. 2012).
Protze et al. used a different approach to identify the optimal combination of transcription factors (Protze et al. 2012). They did not search a pool of transcription factor candidates for a single crucial gene but directly screened all triplet combinations of ten candidate factors combined with a qPCR assay to determine multiple cardiac-specific genes (MYH6, Myl2, Actc1, Nkx2.5, and SCN5A). Through this screening method, the combination of Mef2c, Myocd, and Tbx5 (MMT) was found to upregulate a broader spectrum of cardiac genes compared to other combinations. When neonatal CFs were transduced with MMT or GMT via lentivirus vectors, the expression of proteins involved in cardiac contractility and sodium and potassium ion channels were observed, and 0.08% of the CFs exhibited spontaneous contractions as well as action potential. These results indicate that the transduction of MMT as well as GMT can directly reprogram neonatal CFs into functional cardiomyocytes.
Addis et al. reported that Nkx2.5 is another important cardiac reprogramming factor that can improve the functionality of iCMs (Addis and Epstein 2013; Addis et al. 2013). They analyzed combinations of reprogramming factors using calcium activity in iCMs as a functional measure of cardiomyocytes. They constructed a GCaMP5 reporter lentivirus that allows for the real-time detection of calcium flux in live cells driven by the cardiomyocyte-specific troponin T (TNNT2) promoter. They transduced the GCaMP5 reporter virus containing the reprogramming factors into mouse fibroblasts. The results indicated that the addition of Nkx2.5 to GMT produced more functional cardiomyocytes, with 0.7 ± 0.3% of cells exhibiting GCaMP activity at 14 days post-induction, which was 22.5-fold higher than that with GMT alone (0.03 ± 0.02%). They also showed that the addition of Nkx2.5 to GHMT further increased the number of functional iCMs up to 1.6 ± 0.3% of the cells expressing GCaMP5, which is a 52-fold increase over GMT alone. Nkx2.5 did not increase the number of iCMs by promoting cell proliferation; instead, Nkx2.5 and Hand2 augmented the expression of cardiac genes related to excitation–contraction coupling, such as phospholamban (Pln) and calsequestrin (Casq2).
Hirai et al. reported that fusion of the MyoD transactivation domain to the pluripotency transcription factor Oct4 facilitated the transcriptional activity of Oct4, resulting in the highly efficient production of iPSCs (Hirai et al. 2010). They then applied this strategy to iCMs and showed that fusion of the MyoD transactivation domain to the cardiac reprograming factor Mef2c could greatly promote the direct reprogramming of fibroblasts into cardiomyocyte-like cells (Hirai et al. 2013). They fused the MyoD domain to Mef2c, Gata4, Hand2, and Tbx5 and transduced these four genes in various combinations into mouse fibroblasts. Transduction of the chimeric Mef2c with the wild-type forms of other three genes produced a much higher number of beating iCMs than with the other combinations of reprogramming factors. The induction efficiency of beating iCMs using chimeric Mef2c was 3.5%, which was 15-fold greater than when using the wild-type GHMT. Although the MyoD domain effectively increased the efficiency of iCM generation when fused at the carboxy terminus of Mef2c, it was not effective when fused to the other reprogramming factors Gata4, Hand2, and Tbx5. These results suggest that an optimal balance of the transcriptional activities of reprogramming factors is critical for successful cardiac induction; however, the exact molecular basis for this remains elusive.
Wang et al. also reported that a precise stoichiometry of GMT is critical for the efficiency and quality of iCM generation (Wang et al. 2014). They generated all possible combinations of G, M, and T with identical 2A sequences in a single transgene and transduced these viral vectors into mouse fibroblasts. They demonstrated that each combination of G, M, and T gave rise to distinct G, M, and T protein expression levels and that the MGT vector that expressed a higher protein level of Mef2c and lower levels of Gata4 and Tbx5 significantly enhanced reprogramming efficiency compared to the other GMT variants. Finally, the MGT vector resulted in more than a tenfold increase in the number of beating iCM loci than the separate GMT vectors, and the molecular characterization revealed that the optimal stoichiometry of G, M, and T correlated with the high expression of cardiac genes (Muraoka and Ieda 2015).
Given that a precise stoichiometry and the transcriptional activity of reprogramming factors are critical for reprogramming, it is conceivable that not all laboratories can reproduce cardiac reprogramming due to subtle differences in experimental conditions (Carey et al. 2011; Polo et al. 2012; Qian et al. 2013). Chen et al. reported that they could not produce functional iCMs using lentiviral GMT vectors from adult mouse fibroblasts (Chen et al. 2012). Although they were able to generate partially reprogrammed iCMs expressing some cardiac markers, the cells did not beat spontaneously or exhibit APs, suggesting that they were not fully reprogrammed iCMs.
In contrast to iCM generation, the induction of iPSCs has been widely reproduced by many laboratories. The iPSCs can expand indefinitely under standardized culture conditions, and the iPSC colonies are easily detected in the culture dish. In contrast, iCMs do not proliferate because they are post-mitotic cells, which may hinder the detection of low numbers of beating iCMs (Yoshida and Yamanaka 2012b). Collectively, the available data highlight that cardiac reprogramming needs further optimization of the reprogramming factors and that defined culture conditions are necessary to establish a routine procedure that can be performed in a wide range of laboratories, as discussed below.
6.4 Identification of MicroRNAs for Cardiac Reprogramming
MicroRNAs (miRNAs) are ∼22-nucleotide RNAs that modulate gene expression by inhibiting mRNA translation and promoting mRNA degradation (Liu and Olson 2010). Previous studies have revealed that MEF2 and SRF regulate the expression of two bicistronic muscle miRNA clusters encoding miR-133a-1/miR-1-2 and miR-133a-2/miR-1-1 in the embryonic and adult heart (Zhao et al. 2005). Loss-of-function mutation studies of these miRNAs in mice revealed that miR-1 and miR-133a regulate the gene expression programs required for normal cardiac growth and function (Zhao et al. 2007). For example, the absence of miR-133a expression results in the ectopic expression of smooth muscle genes and aberrant cardiomyocyte proliferation in the developing heart, leading to embryonic lethality in half of the mutant mice (Liu et al. 2008). These abnormal phenotypes can be attributed, at least in part, to the elevated expression of SRF and cyclin D2, which are direct targets of miR-133a. These results demonstrated that miRNAs and transcription factors orchestrate a complex network involved in normal heart development and cardiac cell fate determination.
There are four known muscle-specific miRNAs, miR-1, 133, 208, and 499, which regulate various stages of cardiac differentiation and development. Jayawardena et al. reported that a combination of these four miRNAs can reprogram mouse neonatal CFs into cardiomyocyte-like cells in vitro (Jayawardena et al. 2012). Their study demonstrated that a transduction of miR-1, 133, 208, and 499 resulted in 5% of adult fibroblasts from αMHC-CFP mice possessing an αMHC-CFP+ phenotype. In addition, the induced cells expressed cardiac-specific proteins and sarcomeric structures, indicative of iCMs. Furthermore, a JAK inhibitor improved the induction rate and quality of the miRNA-mediated reprogramming, leading to high induction rate of 1–2% of the initial fibroblast population into iCMs with spontaneous contractions. Although induction by miRNAs alone may not be sufficient to reprogram the fibroblasts from other origins such as tail-tip fibroblasts into cardiomyocytes, these results demonstrated miRNAs can reprogram at least CFs into functional cardiomyocytes. However, the molecular mechanisms of cardiac reprogramming by these miRNAs were not clarified.
We identified the molecular mechanisms and direct target of miR-133a in cardiac reprogramming (Muraoka and Ieda 2014; Muraoka et al. 2014). We also analyzed the cardiac reprogramming efficiency when using the miR-1, 133, 208, and 499 miRNAs in mouse embryonic fibroblasts (MEFs). We found that the miRNAs alone were not sufficient for cardiac reprogramming in MEFs, but the addition of miR-133a to GMT greatly increased the reprogramming efficiency compared with other miRNAs. Compared to GMT alone, miR-133a overexpression with GMT generated sevenfold more cTnT+ cells and beating iCMs from MEFs and shortened the duration to induce beating cells from 30 to 10 days. Microarray analyses revealed that more than 100 genes were downregulated by miR-133a, many of which were fibroblast-related genes, such as Col1a1, Col1a2, Fn1, and Postn. While the expression levels of Ccnd2, Cdc42, Hand2, RhoA, and Srf, which have been shown previously to be the direct targets of miR-133a, were not significantly altered, the expression of Snai1, a master regulator of epithelial-to-mesenchymal transformation, was significantly downregulated by miR-133a overexpression. Snai1 is a putative direct target of miR-133a and contains two conserved miR-133a-binding sites within its 3′-UTR as shown by bioinformatics analyses. Luciferase assays confirmed that miR-133a binds to both sites. The expression levels of Snai1 mRNA and protein were suppressed by miR-133a transduction, suggesting that Snai1 is a new direct target of miR-133a. To investigate the possible contribution of Snai1 to cardiac reprogramming, we suppressed Snai1 expression with siRNA in GMT-transduced MEFs. Snai1 knockdown suppressed fibroblast genes, upregulated cardiac gene expression, and promoted cardiac reprogramming, recapitulating the effects of miR-133a overexpression. In contrast, overexpression of Snai1 in GMT/miR-133a-transduced cells suppressed reprogramming by maintaining fibroblast signatures. MiR-133a-mediated Snai1 repression was also critical for cardiac reprogramming in adult mouse and human CFs. These results suggest that miR-133a-induced Snai1 and fibroblast gene suppressions are critical for cardiac reprogramming. Given that Snai1 suppression is also important for iPSC generation, which requires a mesenchymal-to-epithelial transition before reprogramming, Snai1 is a common target for cellular reprogramming from fibroblasts (Li et al. 2010; Unternaehrer et al. 2014). Moreover, the balance between master regulators of original cells (fibroblasts) and target cells (iCMs or iPSCs) may determine the success of the cell fate switch in general. Further studies might identify other new targets of the miRNAs involved in cardiac reprogramming, as miRNAs have numerous targets related to signal transduction, transcription factors, and epigenetic regulation. Nonetheless, this is the first study demonstrating a molecular circuit and mechanism of cardiac reprogramming (Fig. 6.2).
Mechanisms of cardiac reprogramming. (a) Addition of miR-133, a TGFβ inhibitor, and a ROCK inhibitor increased cardiac reprogramming by repressing pro-fibrotic signaling, while TGFβ reduced it by maintaining fibroblast signatures. These interventions changed the generation of partially reprogrammed iCMs at the early stage of cardiac reprogramming. (b) Cultivation of the reprogrammed cells in serum-free medium with FGF2, FGF10, and VEGF promoted the conversion of partially reprogrammed iCMs into fully reprogrammed iCMs at the late stage of cardiac reprogramming. FGF2, FGF10, and VEGF increased the expression of cardiac reprogramming factors (Hand2, Nkx2.5, and Gata6) via the PI3K/Akt and p38MAPK pathways. The defined conditions increased the expression of Gata4 and enabled cardiac reprogramming with only MT. Activation of Akt1 promoted cardiac reprogramming and maturation of iCMs with alteration of mTOR and FoxO expression levels
6.5 Inhibition of Pro-fibrotic Signaling with Small Molecules Enhances Cardiac Reprogramming
Several small molecules have been utilized to enhance the differentiation of pluripotent stem and progenitor cells to cardiomyocytes and to promote the reprogramming of fibroblasts into iPSCs (Burridge et al. 2014; Kattman et al. 2011; Li et al. 2010). Ifkovits et al. reported that the inhibition of transforming growth factor β (TGFβ) signaling by small molecules increased the direct conversion of mouse fibroblasts to iCMs (Ifkovits et al. 2014). They overexpressed GHMT plus Nkx2.5 (GHMNT) with the calcium indicator GCaMP5, driven by the cTnT promoter, to quantify iCM yield in MEFs. They screened five small molecules, including a G9a histone methyltransferase inhibitor (BIX01294), canonical Wnt signaling activator (CHIR99021), Wnt inhibitor (XAV939), TGFβ/Activin/Nodal inhibitor (SB431542), and BMP inhibitor (DMH1), which were reported to promote directed differentiation from pluripotent stem cells to cardiomyocytes and iPSC reprogramming. Among them, only SB431542 promoted cardiac induction by GHMNT, and LY364947, a specific inhibitor of TGFβ, also showed similar effects. In contrast, addition of TGFβ to the culture media greatly reduced cardiac induction, suggesting that inhibition of TGFβ signaling increased the efficiency of iCM generation.
These results were confirmed and analyzed in more detail by Zhao et al. (2015). They performed RNA sequencing to identify the genes that were regulated by GHMT overexpression at day 7. Surprisingly, not only cardiac gene but also pro-fibrotic gene expression was activated during GHMT-mediated cardiac reprogramming at day 7. The expression of fibrotic genes was reduced 12 days post-GHMT infection, suggesting that transient activation of pro-fibrotic signaling at the early stage of reprogramming may inhibit the conversion of fibroblasts into cardiomyocytes. TGFβ signaling is an important pathway involved in controlling fibrotic events, and the expression levels of TGFβ signaling components, including phosphorylated Smad transcription factors, Smad2 and Smad3, and Tgfb2 and Tgfbr1, were all upregulated by GHMT transduction during the early stage of cardiac reprogramming. They showed that stimulation of pro-fibrotic signaling by TGFβ1 supplementation attenuated cardiac reprogramming by GHMT or GHMT plus miR-1 and 133 (GHMT2m). The addition of miR-1 and 133 to GHMT significantly decreased the expression of pro-fibrotic genes concomitant with the activation of cardiac genes, which was consistent with our previous data (Muraoka et al. 2014). In addition to TGFβ signaling, Rho triggers the formation of stress fibers and stimulates pro-fibrotic events via activation of its downstream effector, Rho-associated kinase (ROCK). Treatment with the ROCK inhibitor Y-27632 decreased the expression of the fibrotic genes Fn-EDA and aSMA and promoted cardiac reprogramming with GHMT and GHMT2m. Treatment of MEFs with A83-01, a selective inhibitor of TGFβ signaling, also decreased the phosphorylation of Smad2 and inhibited the expression of Fn-EDA, Col1a1, Col3a1, and SMA in GHMT- and GHMT2m-infected cells. Although A83-01 alone did not induce cardiac reprogramming in MEFs, the addition of A83-01 to GHMT or GHMT2m transduction greatly increased the cardiac reprogramming efficiency, with up to 60% of the MEFs reprogrammed into functional iCMs. The electrophysiological analysis revealed that APs were recorded from single spontaneously beating cells on day 9 of reprogramming. The APs of iCMs mimic those of fetal or nodal cardiomyocytes, as they show a high rate of spontaneous firing, short AP durations, and slow upstroke velocity, suggesting that they were an immature myocyte form. The frequency of cell contraction and spontaneous calcium transients in the iCMs were modulated by the administration of isoproterenol, a β-adrenergic agonist, and nifedipine, a blocker of L-type calcium channels, suggesting that functional excitation–contraction coupling machinery and β-adrenergic signaling components were developed in the iCMs. Inhibition of TGFβ signaling by A83-01 also enhanced the reprogramming of adult cardiac and dermal fibroblasts into functional cardiomyocytes, with frequencies of 2.5% and 4%, respectively. These results suggest that the inhibition of pro-fibrotic signaling by small molecules promotes cardiac reprogramming, but the manipulation of other molecules and signaling pathways will be necessary to further improve cardiac reprogramming in adult fibroblasts.
6.6 PI3K/Akt and p38MAPK Pathways Enhanced Cardiac Reprogramming Under Defined Culture Conditions
Our previous results demonstrated that transduction with GMT activated cardiac reporters and protein expression in ~20% of fibroblasts after 1 week; however, only 0.1% of the starting fibroblasts were fully reprogrammed into functional iCMs after 4 weeks under conventional serum-based culture conditions, suggesting that most cells remained partially reprogrammed or immature iCMs with the original method (Ieda et al. 2010). As discussed above, inhibition of fibroblast signatures by miR-133 and small molecules promoted cardiac reprogramming at the early stage of reprogramming; however, the molecular mechanisms underlying the conversion of partially reprogrammed cells into functional iCMs at the later stage remained unclear (Sadahiro et al. 2015). Moreover, the use of undefined serum-containing medium in the original method was associated with considerable batch-to-batch variation in cardiac reprogramming, leading to the variable and low reprogramming efficiencies in previous studies (Chen et al. 2012; Srivastava and Ieda 2012; Yoshida and Yamanaka 2012b). Recently, we were the first to describe the defined culture conditions that increased cardiac reprogramming by 100-fold compared with the conventional serum-based conditions (Yamakawa et al. 2015). We screened eight cardiogenic compounds and found that a combination of fibroblast growth factor (FGF) 2, FGF10, and vascular endothelial growth factor (VEGF), termed FFV, greatly improved the quality of cardiac reprogramming in mouse fibroblasts under serum-free culture conditions. FFV did not increase the generation of partially reprogrammed iCMs, while this treatment activated multiple cardiac transcriptional regulators, including Gata4/6, Hand2, and Nkx2.5, and converted partially reprogrammed iCMs into functional iCMs through the p38 mitogen-activated protein kinase (MAPK) and phosphoinositol 3-kinase (PI3K)/AKT pathways. Moreover, FFV enabled cardiac reprogramming with only Mef2c and Tbx5 without the need for Gata4, which may enable pharmacological reprogramming in the future. Thus, our results demonstrated that the FGF- and VEGF-mediated PI3K/Akt and p38MAPK signaling pathways are critical for the late stage of cardiac reprogramming, which has been a major hurdle for successful reprogramming. Identifying the efficient, reproducible, and defined culture conditions at least for mouse cells could allow for the standardization of the cardiac reprogramming procedure and lead to further improvements in the protocol in the future.
Consistent with our results, Zhou et al. reported that Akt dramatically enhanced cardiac reprogramming in three different types of fibroblasts (mouse embryo, adult cardiac, and tail tip) (Zhou et al. 2015). They used a retroviral expression library and screened 192 protein kinases that might augment the generation of functional iCMs by GHMT. They found that Akt1 overexpression induced Akt1 phosphorylation and increased the expression of cardiac genes, whereas a kinase-dead form of Akt1 abrogated the stimulatory activity on GHMT, suggesting that activation of Akt signaling is critical for cardiac reprogramming. Approximately 50% of the reprogrammed MEFs displayed spontaneous beating after 3 weeks of induction by Akt plus GHMT, while ~1% of the adult CFs and TTFs could be reprogrammed into beating iCMs with the same treatment, suggesting that some epigenetic barriers still exist in adult fibroblasts. Nevertheless, the iCMs generated by Akt plus GHMT displayed a more mature phenotype compared with those generated without Akt and that were polynucleated, hypertrophic, and responsive to β-adrenoceptor pharmacologic modulation. Mechanistically, insulin-like growth factor 1 (IGF1) and PI3K acted upstream of Akt, whereas the mitochondrial target of rapamycin complex 1 (mTORC1) and forkhead box o3 (Foxo3a) acted downstream of Akt to promote cardiac reprogramming. These findings provide new insights into the molecular mechanisms of cardiac reprogramming and might be valuable for future research on human cardiac reprogramming (Fig. 6.2).
6.7 Discovery of Human Cardiac Reprogramming Factors
We determined whether cardiomyocytes can be induced from human fibroblasts by direct reprogramming (Wada et al. 2013). First, we transduced GMT into human CFs in vitro; however, there was insufficient induction of cardiomyocytes. Thus, we explored new human cardiac reprogramming factors. When Mesp1 and Myocd, which are cardiomyocyte or CPC-specific transcription regulators, were added to GMT (GMT + Mesp1 + Myocd, hereafter referred to as GMTMM), the cardiac induction rate was improved. Microarray analysis demonstrated that the GMTMM-mediated human iCMs had increased expression levels of cardiomyocyte-specific genes and possessed sarcomeric structures. In addition, under co-culture conditions with primary cultured rat cardiomyocytes, the human iCMs demonstrated physical cardiac functions, indicating that the five factors of GMTMM can induce the reprogramming of human CFs to cardiomyocyte-like cells. We also demonstrated that the addition of miR-133a to GMTMM increased cardiac reprogramming in human fibroblasts via Snai1 repression. The induction of the cardiac markers α-actinin and cTnT increased from ~2% to ~27% by the addition of miR-133a (Muraoka et al. 2014).
Nam et al. also reported that the mouse reprogramming factors GHMT were ineffective in activating cardiac gene expression in human fibroblasts and that Myocd was required for human cardiac induction (Nam et al. 2013). Furthermore, the addition of miR-1 and miR-133a improved the myocardial conversion of human fibroblasts and eliminated the requirement for Mef2c in cardiac induction from human neonatal foreskin fibroblasts, adult CFs, and dermal fibroblasts. The induced human cardiomyocyte-like cells expressed multiple cardiac genes and developed sarcomere-like structures. Although the efficiency of inducing cTnT-expressing cells from human fibroblasts was 10–20%, only a small subset of the cells could exhibit spontaneous contractility after 11 weeks of culture.
Fu et al. reported that GMT plus Esrrg, Mesp1, Myocd, and Zfpm2 induced global cardiac gene expression and a phenotypic shift to a cardiac fate in human fibroblasts derived from human ESCs (Fu et al. 2013). While most cells were partially reprogrammed, a subset of human iCMs had sarcomere structures, calcium transients, and APs. They demonstrated that the epigenetic status of human iCMs resembled that of hESC-derived cardiomyocytes in terms of DNA and histone methylation status and that the iCMs were stably reprogrammed to a cardiac state without the need for the continuous expression of reprogramming factors.
Islas et al. reported that the transient overexpression of Ets2 and Mesp1, followed by activin A and BMP2 treatment, could reprogram human dermal fibroblasts into cardiac progenitor-like cells (Islas et al. 2012). The induced cardiac progenitor-like cells differentiated into immature cardiomyocytes that expressed several cardiac genes and exhibited sarcomeric structures and Ca2+ activities in a prolonged culture. Given that the induced progenitor-like cells formed colonies in a culture dish and expressed several cardiac progenitor genes, the route of cardiac induction by Ets2 and Mesp1 seemed to be different from that with the direct cardiac reprogramming performed by our group and other groups (Fu et al. 2013; Nam et al. 2013; Wada et al. 2013). These findings represent an important initial step toward potential therapeutic applications of the direct reprogramming approach in clinical situations. However, cardiac reprogramming of human fibroblasts was much slower and less efficient than that in mouse fibroblasts, and future studies are needed to optimize the necessary reprogramming factors and culture conditions for human cardiomyocyte induction and functional maturation.
6.8 Systemic Approach to Identify the Transcription Factors Required for Direct Reprogramming
Thus far, identification of the key transcription factors required for reprogramming has been performed by a process of exhaustive testing of large sets of plausible transcription factors (Han et al. 2014; Huang et al. 2011, 2014; Ieda et al. 2010; Riddell et al. 2014; Sekiya and Suzuki 2011; Vierbuchen et al. 2010; Zhou et al. 2008). As there are roughly 2000 different transcription factors, it is technically challenging to test all possible combinations of factors necessary for cardiac reprogramming. Recently, the Daley and Collins groups developed a network biology platform, CellNet, which can compare gene regulatory networks in engineered cell populations with those in in vivo counterparts and identify systematically the factors that can improve cellular reprogramming (Fig. 6.3) (Cahan et al. 2014; Morris et al. 2014). In the case of cardiac reprogramming, Cahan et al. used CellNet to analyze the gene regulatory network of the GMT-induced aMHC-GFP+ population derived from mouse fibroblasts, of which the vast majority of the cells were partially reprogrammed iCMs (Cahan et al. 2014). They found that the aMHC-GFP+ cells were exclusively classified as cardiomyocytes; however, multiple cardiac transcription factors, including Gata6, Tbx20, Hand2, and Nkx2.5, were incompletely activated in the GMT-mediated aMHC-GFP+ population. As discussed, at least some of the candidate factors, such as Hand2 and Nkx2.5, could improve the cardiac reprogramming by GMT, suggesting that this approach can be valuable for screening cardiac reprogramming factors (Addis et al. 2013; Song et al. 2012).
Strategies to identify reprogramming factors. (a) Candidate approach for identification of reprogramming factors. Candidate factors defined empirically or experimentally are screened in in vitro experiments. (b) CellNet accurately assessed the fidelity of cellular engineering and identified the transcription factors that might be needed to enhance reprogramming. (c) The Mogrify algorithm for predicting transcription factors for cell conversion. The cell type ontology tree, gene expression data, and network analyses are used to predict the reprogramming factors necessary to induce cell conversion
More recently, Rackham et al. reported a predictive system (Mogrify) that combines human gene expression data with regulatory network information to predict the reprogramming factors necessary to induce cell conversion (Rackham et al. 2016). They applied Mogrify to 1173 human cell types and 1134 tissues and defined a cellular reprogramming atlas. To assess the predictive power of Mogrify, they first determined how Mogrify performed against previously published human cell conversions. Mogrify predicted NANOG, OCT4, and SOX2 as the top three transcription factors for iPSC conversion, a combination that was experimentally validated. For the conversion of human dermal fibroblasts into cardiomyocytes, Mogrify’s predicted list included Gata4, Hand2, Mef2c, Nkx2.5, and Tbx5. Mogrify correctly predicted the transcription factors used in known transdifferentiations, and the average recovery rate of published transcription factors was 84%. Thus, these predictive computational programs can be useful to identify new transcription factors that could improve the efficiency of human cardiac reprogramming.
6.9 Cardiac Regeneration by In Vivo Direct Cardiac Reprogramming
For use in regenerative medicine, it would be ideal if cardiomyocytes could be reprogrammed from the endogenous CFs in situ. Based on our in vitro results, we determined whether the direct reprogramming approach could also be used to reprogram endogenous CFs into cardiomyocytes by applying cardiac reprogramming factors in vivo (Inagawa et al. 2012). We used a myocardial infarction (MI) mouse model, which was generated by coronary artery ligation. Retrovirus vectors were directly injected to transfer the transcription factors to the CFs on the day of coronary artery ligation. After 2 weeks, the hearts were removed, and the induction of cardiomyocytes was evaluated. In the negative control group that received a control retrovirus vector injection, there was no induction of cardiomyocytes from fibroblasts. In contrast, the group that received the GMT retrovirus vector injection exhibited induction of cardiomyocytes from fibroblasts. However, the majority of the inducted cells were immature cardiomyocyte-like cells, suggesting that all three transcription factors might not be transferred into the cells simultaneously. Thus, we constructed a single polycistronic GMT to ensure simultaneous delivery of the three factors. As a result, multiple cardiac genes were expressed, and the induction rate of sarcomere+ cardiomyocytes increased by twofold.
Similar outcomes were reported by multiple research groups. Qian et al. directly injected a GMT retrovirus into the mouse heart after coronary ligation and reported that 35% of the cardiomyocytes from the border/infarct zone were iCMs newly derived from endogenous CFs (Qian et al. 2012). Approximately 50% of the iCMs exhibited functional characteristics of adult ventricular cardiomyocytes, including an organized sarcomeric structure, cell contraction, electrophysiologic properties, and functional coupling to other cardiomyocytes. In order to determine whether iCMs were derived from CFs, genetic lineage tracing with a fibroblast-specific gene was applied using the transgenic mice (Postn-Cre and Fsp1-Cre with reporter mice) after direct injection with the GMT retrovirus into the heart. The post-MI myocardium contained iCMs, which expressed Postn-Cre- or Fsp1-Cre-induced reporter expression, confirming that these iCMs were derived from CFs and not from native cardiomyocytes. In addition, the functional evaluation after MI revealed that GMT retrovirus injection significantly improved cardiac function and suppressed fibrosis at least up to 3 months after the cardiac infarction.
Song et al. generated an ischemic cardiac disease mouse model and reported that GHMT retrovirus injection reprogrammed endogenous CFs into functional cardiomyocyte-like cells (Song et al. 2012). They also used transgenic mice with fibroblast lineage tracing and demonstrated that 2–6% of the cardiomyocytes from the border/infarct zone were newly induced cardiomyocyte-like cells. The newly differentiated cardiomyocyte-like cells possessed similar characteristics as the endogenous cardiomyocytes, such as a clear sarcomeric structure and functional properties. Their study also determined whether the newly induced cardiomyocyte-like cells might have arisen from the fusion of native cardiomyocytes with non-cardiomyocytes using mice with an inducible αMHC-MerCreMer transgene and Rosa26-LacZ reporter. The results demonstrated that cardiomyocyte-like cells were newly differentiated and independent of cell fusion. Lastly, GHMT-transduced mice had a twofold higher cardiac ejection fraction compared to the control mice. Furthermore, the damaged scar area was reduced by 50% at 12 weeks post-myocardial infarction.
Mathison et al. reported that in a rat cardiac infarction model, intramyocardial treatment with the proangiogenic VGEF together with GMT transduction enhanced the recovery of cardiac function and reduced the fibrosis area compared to GMT transduction alone (Mathison et al. 2012). This beneficial effect of VGEF treatment suggests that VEGF-mediated neovascularization may at least partially contribute to improved differentiation and survival of newly iCMs in the damaged myocardium, resulting in improved recovery after MI.
Jayawardena et al. used transgenic mice (Fsp1-Cre) with reporter mice and genetic tracing of fibroblast origin and demonstrated that direct injection of lentiviral miR-1, 133, 208, and 499 into the mouse heart after MI reprogrammed endogenous CFs into cardiomyocyte-like cells (Jayawardena et al. 2014). They used Fsp1-Cre mice for the linage tracing of non-myocytes and found that 12% of cardiomyocytes in the border/infarct area were newly generated iCMs. Serial cardiac echo mapping revealed that there was a progressive improvement in ventricular function following miRNA treatment, which began 1 month post-surgery and was enhanced at 3 months, similar to the period required for reprogramming using transcription factors. Thus, our group and other groups demonstrated that cardiac reprogramming can be achieved in vivo by efficiently transferring cardiac reprogramming factors into CFs (Fig. 6.4). Furthermore, in vivo cardiac reprogramming reduced scar size and improved cardiac function after MI, suggesting that in vivo cardiac reprogramming might be a promising approach for regenerative medicine. Given that endogenous CFs can be converted into more fully reprogrammed functional iCMs using in vivo reprogramming than using in vitro conditions, undefined factors in the microenvironment may improve the quality of cardiac reprogramming, which will be investigated in future studies (Yoshida and Yamanaka 2012a).
Conclusions
The heart consists of various types of cells, and cardiomyocytes account for only 30% of the total cells in the heart. Because of the low regenerative capability of adult cardiomyocytes, once they are damaged, they will be replaced by fibroblasts and fibrous tissue, resulting in impaired cardiac function and arrhythmia. Cardiac regenerative medicine has traditionally focused on procedures in which cardiomyocytes are prepared ex vivo and transplanted into the damaged heart. However, if a procedure for the direct reprogramming of endogenous CFs into cardiomyocytes using reprogramming factors is developed, it could become a promising therapeutic approach.
To date, our group and other groups have reported cardiac reprogramming in vitro and in vivo. However, the current reprogramming efficiency is not sufficient, especially in human cardiac reprogramming, and further improvement of the protocol and a better understanding of the molecular mechanisms are needed. In addition, the protocol using retrovirus vectors may potentially alter cellular function due to insertional mutagenesis by the integration of transgenes, requiring further investigation for safety concerns. Nevertheless, since our first discovery of cardiac reprogramming in 2010, there has been enormous progress in this direct reprogramming field as discussed in this chapter. We believe that future research can overcome these challenges, leading to the practical use of direct cardiac reprogramming in the regeneration of the failing heart.
Direct cardiac reprogramming converts endogenous CFs directly into cardiomyocytes by defined factors in vivo. The cardiac reprogramming factors identified in in vitro experiments can be applied to in vivo reprogramming. The fibrous tissue that mainly consists of CFs and extracellular matrix can be repaired by cardiac reprogramming.
References
Addis RC, Epstein JA (2013) Induced regeneration – the progress and promise of direct reprogramming for heart repair. Nat Med 19:829–836
Addis RC, Ifkovits JL, Pinto F, Kellam LD, Esteso P, Rentschler S, Christoforou N, Epstein JA, Gearhart JD (2013) Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol 60:97–106
Burridge PW, Matsa E, Shukla P, Lin ZC, Churko JM, Ebert AD, Lan F, Diecke S, Huber B, Mordwinkin NM et al (2014) Chemically defined generation of human cardiomyocytes. Nat Methods 11:855–860
Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ (2014) CellNet: network biology applied to stem cell engineering. Cell 158:903–915
Carey BW, Markoulaki S, Hanna JH, Faddah DA, Buganim Y, Kim J, Ganz K, Steine EJ, Cassady JP, Creyghton MP et al (2011) Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9:588–598
Chen JX, Krane M, Deutsch MA, Wang L, Rav-Acha M, Gregoire S, Engels MC, Rajarajan K, Karra R, Abel ED et al (2012) Inefficient reprogramming of fibroblasts into cardiomyocytes using gata4, mef2c, and tbx5. Circ Res 111:50–55
Chong JJ, Yang X, Don CW, Minami E, Liu YW, Weyers JJ, Mahoney WM, Van Biber B, Cook SM, Palpant NJ et al (2014) Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature 510:273–277
Davis RL, Weintraub H, Lassar AB (1987) Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987–1000
Fu JD, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D (2013) Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep 1:235–247
Han JK, Chang SH, Cho HJ, Choi SB, Ahn HS, Lee J, Jeong H, Youn SW, Lee HJ, Kwon YW et al (2014) Direct conversion of adult skin fibroblasts to endothelial cells by defined factors. Circulation 130:1168–1178
Hirai H, Tani T, Kikyo N (2010) Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int J Dev Biol 54:1589–1596
Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N (2013) Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res 100:105–113
Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L (2011) Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475(7356):386–389
Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y et al (2014) Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell Stem Cell 14:370–384
Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D (2009) Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell 16:233–244
Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142:375–386
Ifkovits JL, Addis RC, Epstein JA, Gearhart JD (2014) Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One 9:e89678
Inagawa K, Ieda M (2013) Direct reprogramming of mouse fibroblasts into cardiac myocytes. J Cardiovasc Transl Res 6:37–45
Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, Umei T, Wada R, Katsumata Y, Kaneda R, Nakade K et al (2012) Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ Res 111:1147–1156
Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D et al (2012) Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci U S A 109:13016–13021
Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ (2012) MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110:1465–1473
Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson C, Pratt RE, Rosenberg PB, Mirotsou M, Dzau VJ (2014) MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res 116(3):418–424
Kattman SJ, Witty AD, Gagliardi M, Dubois NC, Niapour M, Hotta A, Ellis J, Keller G (2011) Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell 8:228–240
Lalit PA, Hei DJ, Raval AN, Kamp TJ (2014) Induced pluripotent stem cells for post-myocardial infarction repair: remarkable opportunities and challenges. Circ Res 114:1328–1345
Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M et al (2005) Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 433:647–653
Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q et al (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7:51–63
Liu N, Olson EN (2010) MicroRNA regulatory networks in cardiovascular development. Dev Cell 18:510–525
Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K et al (2008) Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell 3:587–590
Mathison M, Gersch RP, Nasser A, Lilo S, Korman M, Fourman M, Hackett N, Shroyer K, Yang J, Ma Y et al (2012) In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc 1:e005652
Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ (2014) Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell 158:889–902
Muraoka N, Ieda M (2014) Direct reprogramming of fibroblasts into myocytes to reverse fibrosis. Annu Rev Physiol 76:21–37
Muraoka N, Ieda M (2015) Stoichiometry of transcription factors is critical for cardiac reprogramming. Circ Res 116:216–218
Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K et al (2014) MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J 33:1565–1581
Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, Dimaio JM, Baker LA, Bassel-Duby R et al (2013) Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A 110:5588–5593
Nam YJ, Lubczyk C, Bhakta M, Zang T, Fernandez-Perez A, McAnally J, Bassel-Duby R, Olson EN, Munshi NV (2014) Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development 141:4267–4278
Okano H, Nakamura M, Yoshida K, Okada Y, Tsuji O, Nori S, Ikeda E, Yamanaka S, Miura K (2013) Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res 112:523–533
Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J et al (2012) A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 151:1617–1632
Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U (2012) A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol 53:323–332
Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D (2012) In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 485:593–598
Qian L, Berry EC, Fu JD, Ieda M, Srivastava D (2013) Reprogramming of mouse fibroblasts into cardiomyocyte-like cells in vitro. Nat Protoc 8:1204–1215
Rackham OJ, Firas J, Fang H, Oates ME, Holmes ML, Knaupp AS, Consortium F, Suzuki H, Nefzger CM, Daub CO et al (2016) A predictive computational framework for direct reprogramming between human cell types. Nat Genet 48(3):331–335
Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, Mandal PK, Ebina W, Volchkov P, Yuan GC, Orkin SH et al (2014) Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 157:549–564
Sadahiro T, Yamanaka S, Ieda M (2015) Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ Res 116:1378–1391
Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T (1999) MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development 126:3437–3447
Sekiya S, Suzuki A (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475:390–393
Shiba Y, Fernandes S, Zhu WZ, Filice D, Muskheli V, Kim J, Palpant NJ, Gantz J, Moyes KW, Reinecke H et al (2012) Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature 489:322–325
Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG et al (2012) Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485:599–604
Srivastava D, Ieda M (2012) Critical factors for cardiac reprogramming. Circ Res 111:5–8
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Takahashi K, Okita K, Nakagawa M, Yamanaka S (2007a) Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2:3081–3089
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007b) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872
Unternaehrer JJ, Zhao R, Kim K, Cesana M, Powers JT, Ratanasirintrawoot S, Onder T, Shibue T, Weinberg RA, Daley GQ (2014) The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep 3:691–698
Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463:1035–1041
Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K et al (2013) Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A 110:12667–12672
Wang L, Liu Z, Yin C, Asfour H, Chen OM, Li Y, Bursac N, Liu J, Qian L (2014) Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res 116(2):237–244
Yamakawa H, Muraoka N, Miyamoto K, Sadahiro T, Isomi M, Haginiwa S, Kojima H, Umei T, Akiyama M, Kuishi Y et al (2015) Fibroblast growth factors and vascular endothelial growth factor promote cardiac reprogramming under defined conditions. Stem Cell Rep 5:1128–1142
Yang X, Pabon L, Murry CE (2014) Engineering adolescence: maturation of human pluripotent stem cell-derived cardiomyocytes. Circ Res 114:511–523
Yoshida Y, Yamanaka S (2012a) An emerging strategy of gene therapy for cardiac disease. Circ Res 111:1108–1110
Yoshida Y, Yamanaka S (2012b) Labor pains of new technology: direct cardiac reprogramming. Circ Res 111:3–4
Zhao Y, Samal E, Srivastava D (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436:214–220
Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129:303–317
Zhao Y, Londono P, Cao Y, Sharpe EJ, Proenza C, O'Rourke R, Jones KL, Jeong MY, Walker LA, Buttrick PM et al (2015) High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat Commun 6:8243
Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA (2008) In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 455:627–632
Zhou H, Dickson ME, Kim MS, Bassel-Duby R, Olson EN (2015) Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci U S A 112:11864–11869
Acknowledgments
M.I. was supported by research grants from JST CREST, AMED PRIME, JSPS, Keio University Program for the Advancement of Next Generation Research Projects, Banyu Life Science, Senshin Medical Research Foundation, and Takeda Science Foundation.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants performed by any of the authors.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Haginiwa, S., Ieda, M. (2017). Direct Cardiac Reprogramming. In: Ieda, M., Zimmermann, WH. (eds) Cardiac Regeneration. Cardiac and Vascular Biology, vol 4. Springer, Cham. https://doi.org/10.1007/978-3-319-56106-6_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-56106-6_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-56104-2
Online ISBN: 978-3-319-56106-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)