Abstract
In this paper, we present a new method to cluster DNA sequence. The proposed method is based on using the Power Spectrum and the Wavelet Neural Network (WNN). The satisfying performance of the Wavelet Neural Networks (WNN) depends on an appropriate determination of the WNN structure. Our approach uses the Least Trimmed Square (LTS) to select the wavelet candidates from the Multi Library of the Wavelet Neural Networks (MLWNN) for constructing the WNN. The LTS has been able to optimize the wavelet neural network. The LTS algorithm is to find the regressors, which provide the most significant contribution to the approximation of error reduction. This wavelet can reduce the approximation error.
In this study, the DNA sequence is coded by using a binary format. The Fourier transform is applied to attain respective Power Spectra (PS) by using the binary indicator sequence. The PS is applied to construct the mathematical moments which be used to build the vectors of real numbers, which are applied to compare easily the sequences with different lengths. Our aim is to construct classifier method that gives highly accurate results. This classifier permits to classify the DNA sequence of organisms. The classification results are compared to other classifiers. The experimental results have shown that the WNN-PS model outperformed the other classifier in terms of both the running time and clustering. In this paper, our approach consists of three phases. The first one, which is called transformation, is composed of three sub steps; binary codification of DNA sequences, Fourier Transform and Power Spectrum Signal Processing. The second section is the approximation; it is empowered by the use of Multi Library Wavelet Neural Networks (MLWNN). Finally, the third section, which is called the classification of the DNA sequences. The Euclidean distances is used to classify the signatures of the DNA sequences.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Various approaches are used for clustering the DNA sequences such as the WNN, which is applied to construct a classification system. Cathy H. et al. used an artificial neural network to classify the DNA sequences [10]. Moreover, Agnieska et al. are proposed a method to classify the mitochondrial DNA Sequences. This approach joins the WNN and a Self-Organizing map method. The feature vector sequences constructed by using the WNN [11]. Xiu Wen et al. used a Wavelet packet analysis to extract features of DNA sequences, which are applied to recognize the types of other sequences [12]. C. Wu et al. applied the neural network to classify the nucleic acid sequence. This classifier used three-layer and feed-forward networks that employ back-propagation learning algorithm [13]. Since a DNA sequence can be converted into a sequence of digital signals, the feature vector can be built in time or frequency domains. However, most traditional methods, such as k-tuple and DMK,… models build their feature vectors only in the time domain, i.e., they use direct word sequences [14,15,16,17,18,19].
The construction of the neural networks structure suffers from some deficiencies: the local minima, the lack of efficient constructive methods, and the convergent efficiency, when using ANNs. As a result, the researchers discovered that the WNN, is a new class of neural networks which joins the wavelet transform approach. The WNN were presented by Benveniste and Zhang. This approach is used to approximate the complex functions with a high rate of convergence [1]. This model has recently attracted extensive attention for its ability to effectively identify nonlinear dynamic systems with incomplete information [1,2,3,4,5]. The satisfying performance of the WNN depends on an appropriate determination of the WNN structure. To solve this task many methods are proposed to optimize the WNN parameters. These methods are applied for training the WNN such as the least-square which is used to train the WNN when outliers are present. These training methods are applied to reduce some function costs and improve performed the approximation quality of the wavelet neural network. On the other hand, the WNN has often been used on a small dimension [6]. The reason is that the complexity of the network structure will exponentially increase with the input dimension. The WNN structure has been studied by several researchers. Moreover, the research effort has been made to deal with this problem over the last decades [6,7,8,9]. The application of WNN is usually limited to problem of small dimension. The number of wavelet functions in hidden layer increases with the dimension. Therefore, building and saving WNN of large dimension are of prohibitive cost. Many methods are used to reduce the size of the wavelet neural networks to solve large dimensional task. In this study, we use the Least Trimmed Square (LTS) method to select a little subset of wavelet candidates from MLWNN constructing the WNN structure in order to build a method to classify a collection containing a dataset of DNA sequences. This method is used to optimize an important number of inputs of DNA sequences. The Beta wavelet function is used to build the WNN. This wavelet makes the WNN training very efficient a reason of adjustable parameters of this function.
This paper contains five sections: in Sect. 2, we present our proposed approach. Section 2.6 presents the wavelet theory used to construct the WNN of our method. Section 3 shows the simulation results of our approach and Sect. 4 ends up with the conclusion.
2 Proposed Approach
This paper presents a new approach based on the wavelet neural network and Power Spectrum. The WNN is constructed by using the Multi-Library Wavelet Neural Networks (MLWNN). The WNN structure is solved by using the LTS method. The power spectrum is used to construct mathematical moments to solve the DNA sequence lengths. Our approach is divided into two stages: approximation of the input signal sequence and clustering of feature extraction of the DNA sequences using the WNN and the Euclidean distances is used to classify the feature extraction of the DNA sequences.
2.1 Fourier Transform and Power Spectrum Signal Processing
The proposed approach uses a natural representation of genomic data by binary indicator sequences of each nucleotide (adenine (A), cytosine (C), guanine (G), and thymine (T)). Afterwards, the discrete Fourier transform is used to these indicator sequences to calculate spectra of the nucleotides [11,12,13,14,15,16,17,18,19,20]. For example, if x[n] = [TT A A …], we obtain: x[n] = [000100011000 1000. ..]. The indicator sequence is manipulated with mathematical methods. The sequence of complex numbers, called f(x) (1), is obtained by using the discrete Fourier Transform:
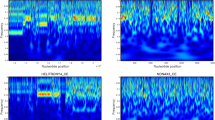
The Power Spectrum is applied to compute the Se[k] (2) for frequencies k = 0, 1, 2, …, N-1 is defined as,
Se [k] has been plotted (Fig. 1).
2.2 Wavelet Neural Network
The wavelet neural network is defined by the combination of the wavelet transform and the artificial neuron networks [33, 34]. It is composed of three layers. The salaries of the weighted outputs are added. Each neuron is connected to the other following layer. The WNN (Fig. 2) is defined by pondering a set of wavelets dilated and translated from one wavelet candidate with weight values to approximate a given signal f. The response of the WNN is:
where (x1, x2,…, xNi) is the vector of the input, Nw is the number of wavelets and y is the output of the network. The output can have a component refine in relation to the variables of coefficients a k (k = 0, 1… Ni) (Fig. 2). The wavelet mother is selected from the MLWNN, which is defined by dilation (ai) which controls the scaling parameter and translation (bi) which controls the position of a single function (Ψ(x)). A WNN is used to approximate an unknown function:
where \( f \) is the regression function and \( \varepsilon \) is the error term.
2.3 Multi Library Wavelet Neural Network (MLWNN)
Many methods are used to construct the Wavelet Neural Network. Zhang applied two stages to construct the Wavelet neural Network [2, 3]. First, the discretely dilated and translated version of the wavelet mother function Ψ is used to build the MLWNN [21, 22].
where L is the number of wavelets in W and xk is the sampled input. Then the best M wavelet mother function is selected based on the training sets from the wavelet library W, in order to construct the regression:
where M \( \le \) L and I is a subset wavelet from the wavelet library.
Secondly, the minimized cost function:
The gradient algorithms used to train the WNN, like least mean squares to reduce the mean-squared error:
where \( j(w) \) is the output of the Wavelet neural networks. The time-frequency locality property of the wavelet is used to give a signal \( f \) , a candidate library \( w \) of wavelet basis can be constructed.
2.4 Wavelet Network Construction Using the LTS Method
The set of training data \( TN = \left\{ {x_{1} ,x_{2} , \ldots ,x_{k} ,f(x_{k} )} \right\}_{k = 1}^{N} \) is used to adjust the weights and the WNN parameters, and the output of the three layers of the WNN in Fig. 2 can be expressed via (7). The model selection is used to select the wavelet candidates from the Multi Library Wavelet Neural Networks (MLWNN). These wavelet mothers are used to construct the wavelet neural network structure [37, 38]. In this study, the Least Trimmed Squares estimator (LTS) is proposed to select a little subset of wavelet candidates from the MLWNN. These wavelet candidates are applied to construct the hidden layer of the WNN [30,31,32, 36]. Furthermore, the Gradient Algorithm is proposed to optimize the wavelet neural networks parameter. The residual (or error) ei at the ith ouput of the WNN due to the ith example is defined by:
The Least Trimmed Square estimator is used to select the WNN weights that minimize the total sum of trimmed squared errors:
The Gradient Algorithm used to optimize the parameters (ai,bi,wi) of the WNN.
2.5 Approximation of DNA Sequence Signal
The classification of DNA sequences is an NP-complete problem; the alignment is outside the range of two sequence of DNA, the problem rapidly becomes very complex because the space of alignment becomes very high. The recent advance of the sequence technology has brought about a consequent number of DNA sequences that can be analyzed. This analysis is used to determine the structure of the sequences in homogeneous groups using a criterion to be determined. In this paper, the Power Spectrum is used to process the signal of the DNA sequence. These signals are used by the wavelet neural networks (WNN) to extract the signatures of DNA sequences, which are used to match the DNA test with all the sequences in the training set [17,18,19,20,21,22,23,24,25,26,27,28,29]. Initially, the signatures of DNA sequences developed by the 1D wavelet network during the learning stage gave the wavelet coefficients which are used to adapt the DNA sequences test with all the sequences in the training set. Then, the DNA test sequence is transmitted onto the wavelet neural networks of the learning DNA sequences and the coefficients specific to this sequence are computed. Finally, the coefficients of the learning DNA sequences compared to the coefficients of the DNA test sequences by computing the Correlation Coefficient. In this stage, the Euclidean distances is used to classify the signatures of the DNA sequences [27].
The Euclidean distances of different DNA sequences are measured and applied as a measure of similarity for these DNA sequences. The pairwise Euclidean distances of DNA sequences are used to generate a similarity matrix, which can be used to classify the DNA sequence.
2.6 Learning Wavelet Network
In this section, we show how the library wavelet is used to learn a wavelet neural network [15, 16, 26, 27].
-
Learning approach
-
Step 1: The data set of DNA sequence is divided into two groups: training and testing dataset. These groups are applied to train and test the wavelet neural network.
-
Step 2: Conversion of DNA sequence to a genomic signal using a binary indicator and Power Spectrum Signal Processing
-
Step 3: The discretely dilated and translated versions are used to construct the library W. The training data are proposed to create this library wavelet, apply the Least Trimmed Square (LTS) algorithm to select the optimal mother wavelet function (10)(11) and choose, from the library, the N wavelet candidate that best matches an output vector.
-
Step3.1: Initializing of the mother wavelet function library
-
Step 3.2: Randomly initialize \( {\texttt{w}}_{{{\texttt{jk}}}} \) and \( {\texttt{V}}_{{{\texttt{ij}}}} \) .
-
Step 3.3: For k = 1,…,m
-
Calculate the predicted output \( \hat{y}_{i} \) via (3).
-
Compute the residuals \( e_{ik} = y_{i} - \hat{y}_{i} \) via (9).
The algorithm is stopped when the criteria diverged, then stop; otherwise, go to the next step.
-
Find the arranged values \( e^{2}_{ik} \le \ldots \le e^{2}_{im} \). Choosing the N best mother wavelet function to initialize the WNN.
-
Step 4: The values of \( \mathop w\nolimits_{ij}^{opt} ,\mathop a\nolimits_{i}^{opt} and\mathop b\nolimits_{i}^{opt} \) are computed using the Gradient algorithm go to step 3.3.
-
-
Clustering using the Euclidean distances
-
Step 1: Generate a similarity matrix, which can be used to classify the DNA sequence \( (\mathop w\nolimits_{ij}^{opt}, \mathop a\nolimits_{i}^{opt} and\mathop b\nolimits_{i}^{opt}). \)
-
Step 2: Use the similarity matrix to classify to the DNA sequence.
-
To construct a phylogenetic tree of these sequences and Generate the classes of the DNA sequences. (The phylogenetic trees constructed from a similarity matrix reflect groups(classes) information, hierarchical similarity and evolutionary relationships of the DNA sequences) (Figure 3).
3 Results and Discussion
This paper used three datasets HOG100, HOG200, and HOG300 selected from microbial organisms [23]. In this study, different experiments are used to evaluate the performance of our approach. The data set of DNA sequences are divided into test and train data. The published empirical and synthetic datasets are selected to perform the clustering comparative analysis [23] (Table 1).
3.1 Classification Results
Experiment results were performed to prove the effectiveness of our proposed approach. Evaluation metrics namely Precision, Recall and accuracy are used to compare our approach with other competitive methods. The classification accuracy Ai of an individual program i depends on the number of samples correctly classified (true positives plus true negatives) and is evaluated by the formula:
where t is the number of sample cases correctly classified, and n is the total number of sample cases.
Table 2 and Fig. 4 show that WNN-PS (our method) is better than other models(WFV, K-tuple and DMK) in terms of the classification results and optimal settings. The number of classes obtained by our approach is little less than in the other methods. The accuracy proves the efficiency of our method. The accuracy is increased using WNN and LTS method. The LTS is applied to optimize the WNN structure.
3.2 Running Time
Tables 2 and 3 show that the WNN can produce very good the prediction accuracy. The results of our approach WNN-PS tested on datasets show that accuracy outperforms the other techniques in terms of percentage of the correct species identification. Tables 2 and 3 show the distribution of the good classifications by class as well as the rate of global classification for all the DNA sequences of the validation phase. The WNN-PS(our approach) is faster than the other methods. This speed is due to the use of the Least Trimmed Square (LTS) algorithm; this method is a robust estimator.
4 Conclusions
In this study, we have used the LTS method to select a subset of wavelet function from the Library Wavelet Neural Network Model. This subset wavelet is applied to build Wavelet Neural Network (WNN). The WNN is used to approximate function f (x) of a DNA sequence signal. Firstly, the binary codification and Power Spectrum are used to process the DNA sequence signal. Secondly, the Library Wavelet is constructed. The LTS method is used to select the best wavelet from library. These wavelets are applied to construct the WNN. Thirdly, the Euclidean distances of signatures of DNA are used to classify the similar DNA sequences according to some criteria. This clustering aims at distributing DNA sequences characterized by p variables X1, X2…Xp in a number m of subgroups which are homogeneous as much as possible while every group is well differentiated from the others. The proposed approach helps to classify DNA sequences of organisms into many classes. These clusters can be used to extract significant biological knowledge.
References
Zhang, Q., Benveniste, A.: Wavelet networks. IEEE Trans. Neural Networks 3(6), 889–898 (1992)
Zhang, J., Walter, G., Miao, Y., et al.: Wavelet neural networks for function learning. IEEE Trans. Signal Process. 43(6), 1485–1497 (1995)
Zhang, Q.: Using wavelet network in nonparametric estimation. IEEE Trans. Signal Process. 8, 227–236 (1997)
Pati, Y.C., Krishnaprasad, P.S.: Analysis and synthesis of feed-forward neural networks using discrete affine wavelet transformations. IEEE Trans. Neural Networks 4, 73–85 (1993)
Billings, S.A., Wei, H.L.: A new class of wavelet networks for nonlinear system identification. IEEE Trans. Neural Networks 16, 862–874 (2005)
Xu, J.H., Ho, D.W.C.: A basis selection algorithm for wavelet neural networks. Neurocomputing 48, 681–689 (2002)
Mallat, S.G., Zhifeng, Z.: Matching pursuits with time-frequency dictionaries. IEEE Trans. Signal Process. 41, 3397–3415 (1993)
Chen, S., Wigger, J.: Fast orthogonal least squares algorithm for efficient subset model selection. IEEE Trans. Signal Process. 43, 1713–1715 (1995)
Han, M., Yin, J.: The hidden neurons selection of the wavelet networks using support vector machines and ridge regression. Neurocomputing 72, 471–479 (2008)
Wu, C.H.: Artificial neural networks for molecular sequence analysis. Comput. Chem. 21(4), 231–256 (1997)
Jach, E.A., Marín, J.M.: Classification of genomic sequences via wavelet variance and a self-organizing map with an application to mitochondrial DNA. Stat. Appl. Genet. Mol. Biol. 9, 1544–6115 (2010)
Zhao, J., Yang, X.W., Li, J.P., Tang, Y.Y.: DNA sequences classification based on wavelet packet analysis. In: Tang, Y.Y., Yuen, P.C., Li, C.-H., Wickerhauser, V. (eds.) WAA 2001. LNCS, vol. 2251, pp. 424–429. Springer, Heidelberg (2001). doi:10.1007/3-540-45333-4_53
Wu, C., Berry, M., Fung, Y.-S., McLarty, J.: Neural Networks for Molecular Sequence Classification. In: Proceedings of the International Conference on Intelligent Systems for Molecular Biology, pp. 429–437 (1993)
Vinga, S., Almeida, J.: Alignment-free sequence comparison-a review. Bioinformatics 19, 513–523 (2003)
Wei, D., Jiang, Q.: A DNA sequence distance measure approach for phylogenetic tree construction. In: Proceedings of the IEEE Fifth International Conference on Bio-Inspired Computing: Theories and Applications (BIC-TA), pp. 204–212, IEEE (2010)
Shi, L., Huang, H.: DNA sequences analysis based on classifications of nucleotide bases. In: Luo, J. (ed.) Affective Computing and Intelligent Interaction. AISC, vol. 137, pp. 379–384. Springer, Heidelberg (2012). doi:10.1007/978-3-642-27866-2_45
Bauer, M., Schuster, S.M., Sayood, K.: The average mutual information profile as a genomic signature. BMC Bioinform. 9, 48 (2008)
Qi, J., Wang, B., Hao, B.I.: Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach, vol. 58, pp 1–11 (2004)
Bonham-Carter, O., et al.: Alignment-free genetic sequence comparisons: a review of recent approaches by word analysis. Brief. Bioinform. 15(6), 890–905 (2013)
Bao, J.P., Yuan, R.Y.: A wavelet-based feature vector model for DNA clustering. Genet. Mol. Res. 14, 19163–19172 (2015)
Amar, C.B., Bellil, W., Alimi, M.A.: Beta function and its derivatives: a new wavelet family. Trans. Syst. Signals Devices 1, 275–293 (2006)
Bellil, W., Othmani, M., Amar, C.B.: Initialization by selection for multi-library wavelet neural network training. In: Conference: Artificial Neural Networks and Intelligent information Processing (ANNIIP), Angers, France (2007)
Mejdoub, M., Amar, C.B.: Classification improvement of local feature vectors over the KNN algorithm. Multimedia Tools Appl. 64(1), 197–218 (2013)
Zaied, M., Said, S., Jemai, O., Amar, C.: A novel approach for face recognition based on fast learning algorithm and wavelet network theory. Int. J. Wavelets Multiresolut. Inf. Process. 19, 923–945 (2011). World Scientific
Said, S., Amor, B.B., Zaied, M., Amar, C.B., Daoudi, M.: Fast and efficient 3D face recognition using wavelet networks. In: 16th IEEE International Conference on Image Processing, Cairo, Egypt, pp. 4153–4156 (2009)
Jemai, O., Zaied, M., Amar, C.B.: Fast learning algorithm of wavelet network based on fast wavelet transform. Int. J. Pattern Recogn. Artif. Intell. 25(8), 1297–1319 (2011)
Jemai, O., Zaied, M., Amar, C.B., Alimi, A.: Pyramidal hybrid approach: wavelet network with OLS algorithm- based image classification. Int. J. Wavelets Multiresolut. Inf. Process. 9, 111–130 (2011). World Scientific Publishing Company
Ejbali, R., Benayed, Y., Zaied, M., Alimi, A.: Wavelet networks for phonemes recognition. International Conference on Systems and Information Processing (2009)
Ejbali, R., Zaied, M., Amar, C.B.: Multi-input Multi-output Beta wavelet network modeling of acoustic units for speech recognition. Int. J. Adv. Comput. Sci. Appl. (IJACSA), The Science and Information Organization(SAI), vol. 3 (2012)
Ejbali, R., Zaied, M., Amar, C.B.: Wavelet network for recognition system of arabic word. Int. J. Speech Technol. 13, 163–174 (2010). Springer edition
Bouchrika, T., Zaied, M., Jemai, O., Amar, C.B.: Ordering computers by hand gestures recognition Based on wavelet networks. In: International Conference on Communications, Computing and Control Applications, Marseilles, France, pp. 36–41 (2012)
Mejdoub, M., Fonteles, L., Amar, C.B., Antonini, M.: Embedded lattices tree: an efficient indexing scheme for content based retrieval on image databases. J. Vis. Commun. Image Represent. 20(2), 145–156 (2009)
Dammak, M., Mejdoub, M., Zaied, M., Amar, C.B.: Feature vector approximation based on wavelet network. In: Proceedings of the 4th International Conference on Agents and Artificial Intelligence (ICAART 2012), vol. 1, pp. 394–399
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Dakhli, A., Bellil, W., Ben Amar, C. (2017). DNA Sequence Classification Using Power Spectrum and Wavelet Neural Network. In: Abraham, A., Haqiq, A., Alimi, A., Mezzour, G., Rokbani, N., Muda, A. (eds) Proceedings of the 16th International Conference on Hybrid Intelligent Systems (HIS 2016). HIS 2016. Advances in Intelligent Systems and Computing, vol 552. Springer, Cham. https://doi.org/10.1007/978-3-319-52941-7_39
Download citation
DOI: https://doi.org/10.1007/978-3-319-52941-7_39
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52940-0
Online ISBN: 978-3-319-52941-7
eBook Packages: EngineeringEngineering (R0)