Abstract
The stressosome is a multi-protein signal integration and transduction hub found in a wide range of bacterial species. The role that the stressosome plays in regulating the transcription of genes involved in the general stress response has been studied most extensively in the Gram-positive model organism Bacillus subtilis. The stressosome receives and relays the signal(s) that initiate a complex phosphorylation-dependent partner switching cascade, resulting in the activation of the alternative sigma factor σB. This sigma factor controls transcription of more than 150 genes involved in the general stress response. X-ray crystal structures of individual components of the stressosome and single-particle cryo-EM reconstructions of stressosome complexes, coupled with biochemical and single cell analyses, have permitted a detailed understanding of the dynamic signalling behaviour that arises from this multi-protein complex. Furthermore, bioinformatics analyses indicate that genetic modules encoding key stressosome proteins are found in a wide range of bacterial species, indicating an evolutionary advantage afforded by stressosome complexes. Interestingly, the genetic modules are associated with a variety of signalling modules encoding secondary messenger regulation systems, as well as classical two-component signal transduction systems, suggesting a diversification in function. In this chapter we review the current research into stressosome systems and discuss the functional implications of the unique structure of these signalling complexes.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
1.1 Introduction
1.1.1 Environmental Sensing and Signalling in Bacteria
Bacteria have colonized almost every possible environmental niche on earth and are subjected to constant fluctuations in their growth conditions (Aertsen and Michiels 2004). Consequently, they have developed sensing and signalling systems that allow adaptive responses through changes in gene expression and cellular behaviour (Hecker and Völker 2001). These signalling systems can alter some cellular behaviours directly, these include motility (Mitchell and Kogure 2006), the modulation of biochemical pathways by allosteric activation/inhibition (Wang et al. 2008); and affect gene expression through the activation of alternative RNA polymerase sigma factors (Paget 2015), transcription factors , and DNA binding two-component signal transduction systems (Stock et al. 2000; Capra and Laub 2012).
Two-component and hybrid two-component signal transduction systems that regulate gene expression comprise a, usually membrane-embedded but occasionally soluble, sensor histidine kinase that senses environmental signals. The kinase is coupled to a cognate response regulator that mediates a cellular response by affecting the transcription of target genes (Gao and Stock 2009). Upon receipt of the appropriate stimulus, the sensor kinase autophosphorylates on an invariant histidine residue. The phosphoryl group is subsequently transferred to a conserved aspartic acid on the cognate response regulator, triggering a conformational change that activates the effector domain of this protein. Hybrid two-component systems combine the function of both the sensory kinase and the response regulator in a single polypeptide, but function in essentially the same way as the classical two-component systems (Capra and Laub 2012). A number of recent reviews of two component signalling are available, but given the dynamic nature of the field, an exhaustive reference list is impossible because of space limitations (Gao and Stock 2009; Krell et al. 2010; Lowe et al. 2012; Bhate et al. 2015).
RNA polymerase sigma factors are essential for the correct initiation of gene transcription through the recognition of promoters (Paget 2015). In addition to the major sigma factors that are required for the transcription of essential housekeeping genes, many bacteria possess alternative sigma factors that control the expression of distinct subsets of genes. The first identification of an alternative sigma factor was in 1979 by Haldenwang and Losick (Haldenwang and Losick 1979), which was at that time erroneously linked to the sporulation process of Bacillus subtilis . Subsequently, it became clear that this particular sigma factor was actually induced by general stress (Moran et al. 1981) and the interest in σB waned somewhat. Today, σB is known as the sigma factor required for the general stress response in B. subtilis and related bacteria, controlling a large regulon of nearly two hundred genes involved in cellular stress (Price et al. 2001; Petersohn et al. 2001). Owing to the potentially large number of genes that alternative sigma factors can regulate, and the resulting heavy metabolic burden on the cell, the activity of sigma factors are regulated by complex signalling cascades, which include phosphorylation , proteolysis, modulation by the alarmone ppGpp, and partner-switching, to ensure their correct and timely function (Österberg et al. 2011).
1.1.2 B. subtilis σB Partner Switching Cascade
The σB-controlled general stress response of B. subtilis is regulated by an intricate, phosphorylation -dependent partner switching cascade comprising over a dozen currently-known components (Fig. 1.1) (Hecker et al. 2007). In unstressed cells, σB is prevented from interacting productively with RNA polymerase because σB is sequestered in a transcriptionally-inactive complex with its cognate anti-sigma factor, RsbW (where Rsb stands for Regulator of sigma B) (Benson and Haldenwang 1993). Upon the imposition of stress, RsbW releases σB, allowing it to bind to RNA polymerase to initiate transcription of the general stress regulon (Alper et al. 1996; Kim et al. 2004a). This effect is mediated by the anti-anti sigma factor RsbV. In its dephosphorylated state, RsbV has a greater affinity for RsbW than RsbW does for σB. The switch in binding partner of RsbW from σB to RsbV causes the release of σB, allowing σB to interact with RNA polymerase (Yang et al. 1996). Under non-stressful conditions, RsbV is maintained in a phosphorylated state by the kinase function of RsbW, and when phosphorylated, RsbV has a much lower affinity for RsbW than RsbW has for σB and consequently σB is preferentially bound by RsbW (Delumeau et al. 2002).
σ B cascade. The Bacillus subtilis σB cascade is illustrated to show how partner-switching induces the activity of RNA polymerase and acts to regulate gene expression. Pre-stress, the anti-sigma factor RsbW sequesters σB and prevents it from directing RNA polymerase to σB-controlled promoters. In this state, RsbV is phosphorylated (RsbV-P) by the kinase activity of RsbW and hence RsbV is inactivated. Under stress conditions, RsbV-P becomes dephosphorylated by one of two phosphatases and attacks the RsbW:σB complex and liberates σB to direct transcription of its regulon to provide the cell with stress-resistance. RsbV is also the point at which the environmental and energy stress responses converge. Under energetic stress, the phosphatase RsbP is activated by RsbQ and dephosphorylates RsbV-P to allow it to form complexes with RsbW. Environmental stresses are integrated by the stressosome, which sequester the RsbU phosphatase-activator, RsbT, in the absence of stress. Under environmentally stressful conditions, RsbT phosphorylates the STAS domains of the stressosome proteins and disassociates, because of a presumed reduced affinity for the phosphorylated proteins, and RsbT switches its binding partner to the phosphatase RsbU. The RsbT:RsbU complex activates RsbV by its dephosphorylation. The phosphatase RsbX acts to remove phosphoryl groups from the stressosome and to mediate the duration of the stress response by ‘resetting’ the system. Ringed plus signs indicate positive regulatory events affecting σB activity, while ringed minus signs indicate those that are negative regulatory events
RsbV sits at a branch-point in the signalling cascade, where both energy and environmental stresses are integrated (Voelker et al. 1996). The activator/phosphatase pair, RsbQ/P, responds to the energy status of the cell and controls the energy stress branch of the σB pathway. The enzymatic activity of RsbQ is required for the correct response to energy stress (Brody et al. 2001), presumably because the reaction product of the RsbQ α/β hydrolase is needed to stimulate the phosphatase activity of RsbP, towards phosphorylated RsbV (RsbV-P), by binding to its Per-Arndt-Sim (PAS) regulatory domain. When the ATP:ADP ratio falls, the RsbQ/P couple dephosphorylates RsbV, inducing the partner-switching of RsbW away from σB and towards RsbV to liberate σB (Vijay et al. 2000; Kaneko et al. 2005; Nadezhdin et al. 2011).
In the pathway that responds to environmental stresses, such as high salt and ethanol, the RsbV phosphorylation status is controlled by the phosphatase RsbU, which also dephosphorylates RsbV-P (Yang et al. 1996). In this pathway RsbU is activated by the RsbT kinase . When the cell is unstressed, RsbT is bound by structural proteins, including RsbR and RsbS , which self-assemble into a ~1.8 MDa macromolecular complex known as the stressosome. Upon receipt of an environmental stress signal, RsbT phosphorylates the structural proteins, initiating its release from the stressosome, and leading to the subsequent activation of RsbU (Kang et al. 1998; Chen et al. 2003; Kim et al. 2004a). The structural proteins that form the stressosome include RsbS ; RsbR and the paralogues of RsbR, YkoB, YojH and YqaH (Delumeau et al. 2006), which have been collectively re-named RsbRA, RsbRB, RsbRC and RsbRD, respectively (Kim et al. 2004b); and the blue-light sensor, YtvA (Akbar et al. 2001; Losi et al. 2002; Jurk et al. 2013). The relative stoichiometries of the RsbR paralogues in the stressosome, assuming a single entity is formed in the cell, and the exact mechanism by which environmental stress signals are perceived and transduced by the stressosome are not known at this time. For simplicity, most structural studies have concentrated on the RsbR/RsbS /RsbT triumvirate as a surrogate of the likely more complex stressosome assemblies found in the cell.
By use of the σB-dependent promoter of the ctc gene (which encodes a component of the large subunit of the ribosome (Truitt et al. 1988; Schmalisch et al. 2002)) fused to a lacZ reporter, σB activity has been monitored in real time by several groups. From a combination of studies, it is clear that the σB signalling cascade is temporally limited. Upon the imposition of environmental stress, σB activity rises to a maximum after 20–30 min, after which time σB activity begins to decline back to base-line levels. The resetting of the system takes place in two independent steps:
-
1.
RsbW phosphorylates RsbV, resulting in the partner switching of RsbW to resequester and inactivate σB, switching off gene transcription
-
2.
The phosphatase RsbX dephosphorylates RsbR-P and RsbS-P (Chen et al. 2004), which resets the system through the re-sequestration of RsbT by the stressosome (Eymann et al. 2011)
The system is also regulated transcriptionally, due to the presence of a σB promoter upstream of RsbV, which leads to increased amounts of the RsbW anti-sigma factor and RsbX phosphatase, which are located downstream of RsbV in the B. subtilis genome (See 1.7.1) (Dufour et al. 1996).
1.1.3 RsbRST Module Distribution
RsbR, RsbS and RsbT form the core components of the stressosome and the genes encoding these proteins are co-located in the genome at the start of the rsb operon. These three proteins form the RsbRST module, which is distributed widely across bacteria and is found in representatives of the Firmicutes, Actinobacteria, proteobacteria, Bacteroides, cyanobacteria and Deinococcus groups. Whilst the core of the module is well conserved, the N-terminal domain of RsbR is less well maintained between species. Furthermore, the downstream components in the rsb operon vary considerably, including secondary messenger signalling and two-component regulators, as well as alternative sigma factors, indicating that the stressosome complex may have evolved as a signalling hub controlling a diversity of cellular functions (Pané-Farré et al. 2005).
1.1.4 Chapter Outline
In this chapter we will discuss how the structure of the stressosome relates to its function as a signal integration and transduction hub, and the complex signalling behaviours seen in B. subtilis . We will assess the current structural knowledge of the individual components of the stressosome as determined by X-ray crystallography and the structure of the stressosome determined by single particle cryo-EM . We will highlight the modularity of this complex and show that it has been widely adopted among bacterial species as a signalling hub with distinct inputs and outputs. Furthermore, we will discuss the structural basis for its signalling mechanism and the role of different inputs in modulating the output level of the complex. The challenges of structural studies on flexible and heterogeneous complexes will be discussed with reference to the cryo-EM structure. The chapter will close with a discussion of unanswered questions on stressosome signalling.
1.2 Stressosome Components
1.2.1 Stressosome Composition
The simplest stressosome complex comprises the RsbS scaffold protein, which consists of a single STAS (Sulphate Transporter and Anti-Sigma factor antagonist (Sharma et al. 2011)) domain and RsbR, which has a variable N-terminal domain and a conserved C-terminal STAS domain (Chen et al. 2003). The STAS domain scaffold is well-conserved between RsbS and the RsbR paralogues (Fig. 1.2) and the RbsR paralogues can form heterogeneous stressosome complexes both in vivo and in vitro (Delumeau et al. 2006; Jurk et al. 2013). On the imposition of stress, RsbT phosphorylates conserved serine and threonine residues on the STAS domains of RsbS and the RsbR paralogues (Kim et al. 2004a). In the RsbR/RsbS stressosome surrogate, the rate of phosphorylation of RsbS on Ser59 is quicker in vitro than on either RsbR phosphorylation site (Thr171, Thr205) (Chen et al. 2003), strongly suggesting that RsbT interacts predominantly with RsbS in the stressosome. How the kinase is held in an inactive state in the absence of stress signals is unknown. Moreover, the effect and purpose of phosphorylation in the stressosome is also unclear (Kim et al. 2004a; Chen et al. 2004; Liebal et al. 2013; Gaidenko and Price 2014). It is generally agreed, however, that RsbT must dissociate from the stressosome to activate RsbU and the downstream components of the σB cascade in order to respond to the imposition of stress, (Kang et al. 1998). In the absence of RsbT, the RsbX phosphatase is able to dephosphorylate RsbS-P and RsbR-P (Chen et al. 2004). RsbX has a differential activity against the different phosphorylation sites on these proteins, which has been suggested to relate to an adaptive response of the stressosome to sustained, or repeated, stresses (Eymann et al. 2011).
Alignment of STAS domain sequences. Multiple sequence alignment generated for the STAS domains of stressosome proteins from Bacillus subtilis (RsbS , RsbRA-D, YtvA ), Moorella thermoacetica (MtS, MtR), and Vibrio vulnificus (Vvu_RsbS , Vvu_RsbR) to highlight sequence conservation in these proteins. Regions outlined in blue show areas of sequence conservation, residues coloured red indicate partial conservation and those outlined in red are completely conserved across the aligned sequences. The secondary structure elements from the X-ray crystal structure of MtS (PDBID: 2VY9) are shown above the alignment with residues numbered for the MtS sequence. The positions of conserved serine and threonine residues phosphorylated by RsbT kinase are shown with black triangles below the alignments. Note that RsbS and its homologues are only phosphorylated on the conserved serine at position 58 in MtS and 59 in RsbS. VVu_RsbS has a glycine at this position, although there is a serine at position 62 in this protein; there is currently no published experimental evidence that the Vvu_RsbS protein is phosphorylated. Figure 1.2 was prepared using EsPript (Gouet et al. 2003)
1.2.2 Structure of RsbS
RsbS is a single STAS domain protein, and STAS domains are also found in anion transporters, the SpoIIAA anti-anti-sigma factor antagonist in B. subtilis (Kovacs et al. 1998), and as essential components of the stressosome. STAS domains appear to function primarily as a scaffold for the recruitment of other proteins, particularly in partner-switching networks (Aravind and Koonin 2000). The B. subtilis RsbS protein forms stable stressosome complexes with the RsbR paralogues both in vivo and in vitro (Chen et al. 2003; Kim et al. 2004b; Delumeau et al. 2006; Reeves et al. 2010). The RsbS STAS domain has a five-stranded beta sheet core with three alpha helices on one face and a single C-terminal helix on the other face (Fig. 1.3). Phosphorylation takes place on a conserved serine/threonine at the N-terminus of helix 2, the central of the three helices of the STAS domain. Structures of the STAS domains of SpoIIAA (Seavers et al. 2001) and RsbS (Marles-Wright et al. 2008; Quin et al. 2012) in both phosphorylated and non-phosphorylated forms do not reveal any significant conformational changes in protein structure , suggesting that release of RsbT from the stressosome as a function of phosphorylation on RsbS-Ser59, and/or its equivalent residue in RsbR, Thr205 (and/or the non-equivalent Thr171), is likely to occur solely by electrostatic and physical repulsion. Similarly, the anti-anti-sigma factor SpoIIAA is released from its complex with cognate dual-function kinase and anti-sigma factor SpoIIAB by the same mechanism (Seavers et al. 2001; Masuda et al. 2004). However, in the absence of a high resolution structure of a phosphorylated stressosome complex, rather than isolated RsbS and RsbS-P structures, significant conformational changes in STAS domain architecture during stressosome activation cannot be excluded (Kumar et al. 2010).
X-ray crystal structure of MtS. The X-ray crystal structure of the phosphorylated form of MtS (PDBID: 3TZB) is shown as a cartoon with secondary structure elements labelled from the N- to C-terminus. Serine 58 is phosphorylated in this structure and the phosphoryl group is depicted as orange and red sticks
1.2.3 Structure of RsbR
The B. subtilis RsbR protein has a C-terminal STAS domain and an N-terminal non-heme globin domain (Murray et al. 2005). While there is no crystal structure of the RsbR STAS domain, it has 30% sequence identity to that of RsbS and will presumably adopt the same fold. Unlike RsbS and other characterised single domain STAS proteins, RsbR is phosphorylated on two conserved threonine residues, Thr171 and Thr205 (Gaidenko et al. 1999; Eymann et al. 2011). Thr205 is equivalent to Ser59 of RsbS and is likely to be located at the N-terminus of helix 2, whereas the unique phosphorylation site of RsbR, Thr171, is positioned at the N-terminus of helix 1. Thr171 tends to be occupied by serine or threonine in the two-domain RsbR paralogues, but glutamate or aspartate in the single domain RsbS paralogues and YtvA , suggesting that the Asp/Glu residues mimic the effect of phosphorylation on RsbR in the latter group of proteins, which apparently increases the activity of RsbT towards RsbS (Chen et al. 2004). Whilst phospho-ablative mutations have been made in RsbR and the effects on phosphorylation patterns, rates and signalling have been measured (Chen et al. 2004; Gaidenko and Price 2014), similar experiments have not been performed to the best of our knowledge on Asp25Thr (or alanine) mutations in RsbS in order to decipher the role of phosphorylation on Thr171 in RsbR.
The structure of the N-terminal domain of RsbR (N-RsbR) displays an all alpha-helical non-heme globin fold (Murray et al. 2005) (Fig. 1.4a, b). A comparison of N-RsbR to other globin-like proteins, such as the HemAT aerotaxis sensor (Hou et al. 2000; Zhang and Phillips 2003) shows that RsbR does not possess a heme binding pocket and also lacks the heme coordinating residues found in the hemoglobins (Murray et al. 2005). In the absence of a heme binding pocket, it is thought that RsbR may interact with an as yet unidentified small molecule ligand in an analogous way to the amino acid-binding globin sensor proteins (Kitanishi et al. 2011), or to the ligand-binding sites in the non-heme globin sensors that regulate the entry of Bacillus anthracis into sporulation (Stranzl et al. 2011).
X-ray crystal structures of RsbR and MtR. The X-ray crystal structures of the B. subtilis RsbR (a) (PDBID: 2BNL) and Moorella thermoacetica MtR (b) (PDBID: 3TZA) are shown as cartoons with secondary structure elements labelled. Both proteins are depicted as the physiologically relevant dimer forms. (c) Secondary structure comparison of RsbR and MtR monomers; RsbR is shown in blue, MtR in orange. While the two structures have the same non-heme globin fold, RsbR has an additional N-terminal α-helix (blue-dashed box) and in the X-ray crystal structure lacks the C-terminal portion of the α-helix that links to the STAS domain (orange-dashed box)
The N-terminal domain of the Moorella thermoacetica RsbR orthologue, MtR (N-MtR), has only 12% sequence identity to that of B. subtilis RsbR , yet an almost identical fold (Fig. 1.4c). The N-MtR structure has an extended alpha helix at the C-terminus of the globin domain that links this with the C-terminal STAS domain (Quin et al. 2012). This extended, terminal helix is equivalent to the ‘J’ helix of the LOV (light-oxygen-voltage) domain of the RsbR paralogue YtvA , which has been suggested to convey blue light-dependent signalling (Jurk et al. 2010).
The N-terminal domains of the RsbR paralogues encoded in the B. subtilis genome share limited sequence identity, although they are each found in stressosomes in vivo and can associate with RsbS to form stressosome-like structures using purified recombinant proteins in vitro (Delumeau et al. 2006). While the structures of their N-terminal domains are yet to be determined, they are likely to form sensory domains given the fact that a number of them belong to the PAS (Per-Arnt-Sim) domain superfamily, which are usually found in signalling systems as sensors (Ponting and Aravind 1997).
1.2.4 Structure of YtvA
While the exact role of the RsbR paralogues is not known, it appears that the detection of blue light by YtvA is modulated by the other RsbR paralogues (van der Steen et al. 2012). Indeed, YtvA is the only RsbR related stressosome component in B. subtilis , and potentially Listeria monocytogenes, for which the activating signal has been confirmed (Möglich and Moffat 2007; Ondrusch and Kreft 2011). This protein has a C-terminal STAS domain and an N-terminal LOV domain that binds a flavin mononucleotide cofactor and undergoes a light-dependent conformational change in the nucleotide binding site (Losi et al. 2002; Möglich and Moffat 2007; Herrou and Crosson 2011) (Fig. 1.5a). This structural rearrangement is transmitted to the ‘J’ helix, which links the sensory domain to the STAS domain, resulting in the movement of this helix away from the axis of the dimer interface (Möglich and Moffat 2007). These structural changes hint at the signal transduction mechanism from the sensory domains of RsbR proteins to the STAS domain, although solution small-angle X-ray scattering experiments on the full length YtvA failed to show any significant structural rearrangements upon illumination with blue light (Jurk et al. 2010).
Structures of YtvA . (a) Crystallographic model of the LOV domain of B. subtilis YtvA in the dark state (PDBID: 2PR5). The model comprises a dimer in the asymmetric unit, which are shown as cartoons with the bound FMN cofactor as sticks coloured by atom. In the light state structure (PDBID: 2PR6) the Jα helices are displaced by up to 2 Å away from the dimer axis. (b) Solution structure of the full length B. subtilis YtbA protein (PDBID: 2MWG). The LOV domain is shown at the top and STAS domain to the bottom in the left panel. The full length structure displays a distinct dimer organisation to the isolated LOV domain with an N-terminal helix, which is not modelled in the crystal structure , forming substantial contacts in the dimer interface
The solution structure of the full length B. subtilis YtvA protein was published recently in the PDB (PDBID: 5MWG) (Fig. 1.5b). While the dimer interface in the crystal structure of the YtvA LOV domain is formed primarily by interactions between the beta-sheets of the two monomers, the solution structure has a distinct dimer arrangement with an additional N-terminal α-helix modelled to form the primary point of contact between the monomers. This particular arrangement is consistent with the dimer arrangement seen in the crystal structures of both the B. subtilis and M. thermoacetica RsbR proteins (Fig. 1.4). These intriguing structural differences may be biologically significant, or represent artefacts of the different constructs and methods used to produce the two structures. The orientation of the LOV domain in relation to the STAS domains in the full length dimer structure hints at a mechanism of signal transduction through the Jα helix to alter the orientation of the LOV domain relative to the STAS domain, either through a movement of the LOV domain itself, or of the STAS domains in the stressosome core.
YtvA is the only RsbR paralogue that is not phosphorylatable, encoding glutamate at both positions equivalent to Thr171 and Thr205 in RsbR. The ready light-to-dark reversibility of YtvA is indicated by a half-life of the photo-excited state of some 40 min (Losi et al. 2003). The effect of mutation of Glu142 and Glu202 in YtvA to serine/threonine, resulting in the potential for phosphorylation of these residues by RsbT, and the potential for phosphoregulation of YtvA and its impact on σB activity, are as-of-yet unconducted experiments that might explain the regulatory role of YtvA in stressosome signalling.
1.2.5 Structure of RsbT
To date there is no X-ray crystal structure of the RsbT protein kinase . However, there is a structure of a Bacillus stearothermophilus homologue, the SpoIIAB anti-sigma factor, with which it shares 28% sequence identity (Campbell et al. 2002). This protein binds to the sporulation sigma factor, σF, in an ATP dependent manner to suppress the activity of the sigma factor. SpoIIAB also binds in an ADP-dependent fashion to the anti-anti-sigma factor protein SpoIIAA, which is a STAS domain protein that disrupts the binding between SpoIIAB and σF to release the sigma factor (Alper et al. 1994). The nucleotide-dependencies of these interactions are probably a reflection of the rather poor stability of recombinant SpoIIAB in the absence of ADP or ATP (Lord et al. 1996). The homologous RsbW can bind to both its cognate sigma factor (σB) and anti-anti-sigma factor in vitro in the absence of additional nucleotide (Delumeau et al. 2002). Both SpoIIAB and RsbT are members of the GHKL superfamily of protein kinase s and the structure of SpoIIAB displays an α/β sandwich with an antiparallel β sheet flanked by alpha helices (Dutta and Inouye 2000). The ATP binding pocket is found in a deep crevice between the helices and the β sheet. The pocket has a highly flexible lid that is thought to communicate the nucleotide binding status of the protein through changes in its structure. Unlike SpoIIAB, which acts as a dimer, RsbT appears to act as a monomer in solution, because it lacks a C-terminal helix found in SpoIIAB that mediates dimerisation (Campbell et al. 2002; Masuda et al. 2004; Delumeau et al. 2006).
1.2.6 Structure of RsbX
RsbX is the protein phosphatase responsible for resetting the stressosome to the resting state after the imposition of stress and its phosphorylation by RsbT. RsbX acts on both RsbS-P and RsbR-P, although it displays differential activity against the different proteins in vitro and in vivo (Chen et al. 2003; Eymann et al. 2011). RsbX is a member of the PP2C family of protein phosphatases (Das et al. 1996) and the structures of both the B. subtilis and M. thermoacetica homologues (MtX) have been determined by X-ray crystallography (Quin et al. 2012; Teh et al. 2015). These proteins share only 25% sequence identity yet superimpose with an RMSD Cα of 1.9 Å over 182 Cα. The two proteins display the same phosphatase fold with an αββα architecture, with the central beta-sandwich flanked by two pairs of alpha helices on the outer faces of the protein (Fig. 1.6). The catalytic centre of the protein is located at one edge of the beta-sandwich and centred around a cluster of acidic residues, which are highly conserved in the PPM phosphatase family (Quin et al. 2012). These residues coordinate two divalent metal ions that are required for the function of the protein. RsbX and MtX show a strong preference in vitro for the presence of Mn2+ in this metal binding site for their activity against both their native and synthetic phosphatase substrates (Quin et al. 2012; Teh et al. 2015). A depression on the surface of these proteins above the active site provides a potentially ideal site for interaction with their targets (Quin et al. 2012; Teh et al. 2015). The dephosphorylation reaction is likely to proceed in a manner analogous to other members of the PP2C family, where a metal-ion bridged water molecule acts as a nucleophile against the phosphate group on the target protein and a second water molecule protonates the dephosphorylated serine/threonine residue (Das et al. 1996; Barford et al. 1998).
X-ray crystal structure of RsbX and Mt X. (a) Crystallographic models of both the B. subtilis RsbX (PDBID: 3ZT9) (green) and Moorella thermoacetica MtX (PDBID: 3W40) (blue) protein phosphatases have been published. The models are depicted as cartoons with secondary structure features labelled from the N- to the C-terminus. Manganese ions required for the catalytic activity of the protein bound in the active site of the protein are depicted as purple spheres, with water molecules shown as red spheres. (b) Metal binding site of RbsX and MtX showing residues coordinating the bound manganese ions. The two manganese ions (purple spheres) are coordinated by the carboxylic acid groups of aspartic acid residues and the backbone carbonyl groups of two glycine residues (shown as stick representations). A number of ordered solvent residues are also present in the coordination shell of the manganese ions (red spheres)
1.3 Stressosome Complexes
The RsbR paralogues and RsbS form a stable 1.5 MDa stressosome complex in B. subtilis that sequesters RsbT in the absence of stress. These complexes can be isolated directly from B. subtilis cells and are present throughout the life of individual bacterial cells (Kim et al. 2004b; Gaidenko and Price 2014). Minimal stressosome complexes can be reconstituted in vitro by mixing RsbS with the RsbR paralogues to simplify the structural biology, although RsbR has a tendency to self-associate into stressosome-like particles in the absence of RsbS . In the absence of RsbS, stressosome complexes are unable to sequester RsbT, and deletion of the rsbS gene in B. subtilis leads to constitutive activation of RsbT and a resulting small-colony phenotype. This is presumably caused by deleterious effects of the products of the σB regulon, or a negative effect of competition for cellular resources with σA regulated housekeeping genes (Kang et al. 1996).
The minimal stressosome complexes of individual RsbR paralogues and RsbS are competent to bind RsbT (Delumeau et al. 2006) and can be phosphorylated by the kinase on conserved residues in their C-terminal STAS domains (Reeves et al. 2010). RsbR paralogues in stressosomes can also be exchanged in vitro, implying that these complexes have some dynamic properties in solution (Delumeau et al. 2006). However, immunofluorescence experiments using antibodies specific for the N-terminal domain of RsbR show that RsbR – and hence the stressosome - forms punctate foci that persist in cells throughout the stress response and its recovery (Marles-Wright et al. 2008). These results suggest that in wildtype B. subtilis the stressosome always contains RsbR and that the RsbR paralogues may exchange into stressosomes through the life of the cell. It is not known if distinct populations of stressosomes with different RsbR paralogues exist at the same time within the cell. B. subtilis stressosomes purified by anti-N-RsbR affinity purification contain at least three paralogues of RsbR (Delumeau et al. 2006); thus, single, double, and triple knockouts are effectively complemented by the remaining paralogues in normally growing B. subtilis (Kim et al. 2004b). A quadruple knockout of the RsbR paralogues leads to constitutive σB activation in much the same manner as the RsbS knockout, as no competent stressosome complexes are able to form in the absence of the RsbR paralogues, leading to the presence of free RsbT in the cell, up-regulated RsbU activity and σB liberated to interact with RNA polymerase.
1.4 Production of Recombinant B. subtilis Stressosomes for Structural Analysis
1.4.1 RsbR/RsbS Binary Complex
Recombinant B. subtilis stressosome complexes for structural analysis were produced by plasmid-based co-expression of RsbR and RsbS from a bi-cistronic operon in Escherichia coli and purified to homogeneity in a multi-step protocol (Marles-Wright et al. 2008). As with any high molecular weight protein complex destined for structural analysis, the purification protocol was extensively refined to ensure the final sample was as homogeneous as possible. The optimised protocol included anion-exchange, size-exclusion gel-filtration, hydrophobic interaction chromatography, and a second gel-filtration step. Using this protocol, stressosome complexes were separated from ribosomes, and other high-molecular weight proteins and macromolecular complexes from the E. coli host strain. Despite the relatively high levels of protein expression and the multi-step purification protocol, it was still possible to identify contamination by the cubic core of the pyruvate dehydrogenase complex in some micrographs (Marles-Wright and Lewis, unpublished observations ).
1.4.2 RsbR/S/T Ternary Complex
Production of the RsbR/S/T ternary complex was achieved by expression of RsbT as an N-terminal GST-fusion to enhance protein production levels and solubility. The RsbT protein was purified by glutathione affinity chromatography in the presence of 1 mM ADP to ensure the nucleotide-binding site of the kinase was occupied, and subsequent on-column cleavage of the GST-tag was achieved using the HRV-3C protease. Purified ADP-loaded RsbT was mixed in excess with purified RsbR/S minimal stressosomes and subjected to an additional round of size-exclusion gel-filtration chromatography to remove unbound RsbT (Marles-Wright et al. 2008).
1.5 Cryo-EM Structure of the Bacillus subtilis Stressosome
Initial negative stain electron microscopy studies of recombinant B. subtilis stressosomes identified a 20 nm ring-shaped structure (Fig. 1.7) (Chen et al. 2003). Due to the absence of distinct views other than the characteristic ring, the complex was initially thought to form a doughnut-like oligomer that exhibited a preferential orientation on the carbon-film of the EM grid (Delumeau et al. 2006). Collection of data on unstained samples by cryo-EM and single-particle analysis allowed the calculation of 3D–reconstructions for a stressosome core structure comprising the full length RsbS protein and an N-terminally truncated RsbR; the full length RsbR/RsbS stressome; and a ternary complex between RsbR/RsbS and the kinase RsbT. We will discuss each of these in turn below.
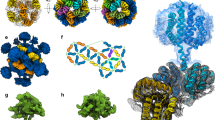
Negatively-stained transmission electron micrograph of the B. subtilis stressosome. Purified recombinant B. subtilis stressosome complexes were stained with 1% uranyl acetate and imaged by transmission electron microscopy. (a) RsbR/RsbS stressosome complexes have a ring-shaped appearance in projection, with rough edges. They are approximately 25–30 nm in size when imaged by TEM. (b) RsbR/RsbS/RsbT ternary complexes have a similar appearance to the RsbR/RsbS complexes with more pronounced features at their edges. The ternary complex has a similar diameter to the binary complex. These images were taken with the assistance of Professor J. Robin Harris
1.5.1 RsbR/RsbS Core Structure
The single-particle cryo-EM reconstruction of the RsbR(146–274):RsbS core STAS-domain complex was determined to a resolution of 6.5 Å by imposing icosahedral symmetry restraints on the calculation of the molecular envelope as indicated by analysis of the initial Eigenimages from the first round of reference-free class-averaging. The reconstructed density was a hollow shell with an outer radius of 90 Å and an inner radius of 45 Å. The final reconstruction displayed clear tubes and flat sheets of electron density consistent with the alpha helices and beta sheet seen in the crystal structure of MtS (Fig. 1.8). The position of the STAS domains in the EM-derived molecular envelope places the N-terminus of the STAS domain at the external surface. This distribution is consistent with their covalent attachment to the C-terminus of the N-terminal domains of RsbR, to place these domains on the outside of the stressosome complex so as to most easily interact with other proteins, ligands and stimuli. Indeed, it has already been suggested that the groove in the dimer interface of N-RsbR could be utilised for binding upstream signalling partners, based on its structural similarity to HemAT and the location of the unique ‘Z’ helix of HemAT in the dimer interface groove; the binding of KaiC peptides in the circadian clock complex KaiA/KaiC; and the interaction of Vitreoscilla haemoglobin with a partner dioxygenase (Murray et al. 2005).
Single particle reconstruction of the RsbR (146–274) /RsbS stressosome core complex. The final experimental EM-derived icosahedral reconstruction of the RsbR146–274/RsbS stressosome core complex is shown as a blue surface contoured at 3σ. Views down the icosahedral two (a), three (b) and fivefold (c) axes are shown with an icosahedral net for reference. The stressosome core has radius of 90 Å
The arrangement of the STAS domains across the icosahedral dimer interface of the core stressosome reconstruction was distinct to the dimeric arrangement seen within the MtS crystal structure ; the primary interactions in the stressosome core are between the first β-strand and the C-terminal helix of the STAS domain, whereas the crystallographic dimer of MtS is formed between α3 and β5 across the two chains in the dimer, and probably does not represent a stable assembly as determined by the PISA server (Krissinel and Henrick 2007) .
1.5.2 RsbR/RsbS Structure
The cryo-EM reconstruction of the full-length RsbR/RsbS minimal stressosome was determined to a nominal resolution of 8.0 Å. Inspection of experimental Eigenimages generated from the initial particle set showed that the complex possessed a mixed-symmetry, with an icosahedral core and projections from this core that obeyed a lower, D 2 symmetry (Fig. 1.9). While unusual, this type of symmetry mismatch has been found in in the capsids of bacteriophage, where nucleic-acid portal translocases inhabit unique vertices (Dube et al. 1993), and also in multi-component proteasome complexes (Beuron et al. 1998). Analysis of this symmetry mismatch and manual model building with a child’s magnetic toy led to a single plausible model for the RsbR/RsbS stressosome containing 40 copies of RsbR and 20 copies of RsbS (Fig. 1.10). In this model, the core of the stressosome is comprised solely of 60 STAS domains, but with an additional 20 projections that obey D 2 symmetry to yield 20 peripheral ‘turrets’ made up of dimers of N-RsbR (Fig. 1.11). This model was validated experimentally by the production of Eigenimages from back-projections of the final stressosome model with D 2 symmetry imposed (Fig. 1.9c). The final reconstruction had a core with a radius of 90 Å and total radius with the projections of 150 Å. The density for the peripheral projections was less well defined than the core density, implying a level of heterogeneity in the position of the N-termini of RsbR in the complex relative to the core.
Experimental Eigenimages of the RsbR/RsbS stressosome complex. (a) Eigenimages showing symmetry elements associated RsbR/RsbS stressosome showing the mixed symmetry that appears due to the N-terminal projections of RsbR protein. (b) Eigenimages from the full length RsbR/RsbS reconstruction, which appear to show a clear tenfold symmetry (c) Eigenimages from re-projections of a model RsbR/RsbS with imposed D 2 symmetry, which also exhibit a clear tenfold symmetry, suggesting the tenfold feature is a consequence of the centring and not a structural feature
Stressosome model building. To aid with the determination of the subunit arrangement within the stressosome reconstructions a model of the stressosome core was built using a magnetic modelling set (a). In this model the magnetic bars represent dimers of RsbR (blue) and RsbS (red), and the steel balls serve as anchor points between the bars, but represent holes at the fivefold axes of the stressosome. The model was built using a set of rules for protein:protein interactions determined from solution studies of the individual components, namely: (1) RsbS is a dimer in solution, but doesn’t interact to form higher-order structures. (2) RsbR is a dimer in solution and can interact with itself to form higher-order structures; however, higher-order interactions with RsbS are stronger than self-interactions. (3) The final model would take the form of an icosahedron. These rules lead the model that was ultimately shown to represent the cryo-EM data. A 3D–printed model of the RsbR(146–274):RsbS stressosome core is shown in (b), where all STAS domains are coloured blue
Single particle reconstruction of the RsbR/RsbS stressosome complex. The final experimental EM-derived D2 symmetry reconstruction of the RsbR/RsbS stressosome complex is shown as a surface contoured at 2σ. The RsbR and RsbS core STAS domains are coloured blue, while the N-terminal RsbR signalling domains are coloured red. Views down the three unique twofold symmetry axes are shown in panels a–c, with an icosahedral net for reference. The total radius of the stressosome is 150 Å
1.5.3 RsbR/RsbS /RsbT Ternary Complex
The ternary complex of RsbR/RsbS and RsbT was determined to 8.3 Å resolution and displayed the same symmetry mismatch as the binary RsbR/RsbS complex. This reconstruction had identical core and total dimensions to the RsbR/RsbS complex, while the presence of additional density features was apparent above the regions of the core not occupied by the N-RsbR turrets (Fig. 1.12). These additional density features were attributed to the presence of the RsbT protein. In this reconstruction the turrets appeared less well distinguished than in the RsbR/RsbS complex implying a greater degree of positional heterogeneity than in the binary complex.
Single particle reconstruction of the RsbR/RsbS /RsbT ternary stressosome complex. The final experimental EM-derived D2 symmetry reconstruction of the RsbR/RsbS : RsbT ternary stressosome complex is shown as a surface contoured at 1.5σ. The RsbR and RsbS core STAS domains are coloured blue, the N-terminal RsbR signalling domains are coloured red, the additional density present when this reconstruction is compared to the RsbR/RsbS reconstruction is attributed to the presence of RsbT and is shown in green. Views down the three unique twofold symmetry axes are shown in panels a–c, with an icosahedral net for reference. The total radius of the stressosome is 150 Å as before
1.5.4 Pseudo-atomic Model of the B. subtilis Stressosome
With cryo-EM reconstructions available for the core stressosome, RsbR/RsbS binary complex, and RsbR/RsbS/RsbT ternary complex it was possible to dock the crystal structures of the individual stressosome components into the reconstructed density maps to produce a pseudo-atomic model of the stressosome complexes.
Because the turret-like peripheral projections from the RsbR/RsbS and RsbR/RsbS/RsbT reconstructions were absent from the RsbR(146–274)/RsbS complex, these regions were attributed to the N-terminal domains of RsbR. The initial model was based on the structure of the B. subtilis N-RsbR protein (Murray et al. 2005; Marles-Wright et al. 2008). The crystallographic dimer fitted the density well; however, the model lacked the linker between the N-terminal globin domain and the C-terminal STAS domain; therefore, the density visible between the turrets and core stressosome was not occupied by this model. The subsequent structure of the M. thermoacetica RsbR homologue, MtR, had fifteen additional C-terminal residues that formed four turns of an α-helix that extended from the bottom of the globin domain to the N-terminus of the STAS domain (Quin et al. 2012). Docking the MtR model into the stressosome reconstructions accounted completely for the turret density and the neck region between the peripheral turrets and the inner core (Fig. 1.13).
Docking of MtR to the stressosome reconstruction envelope. Orthogonal views of the secondary structure fit of the X-ray crystal structure of MtR in the RsbR/RsbS stressosome reconstruction; the density map is contoured at 2σ. The model of MtR is shown as a secondary structure cartoon coloured orange and red
A monomer of the MtS structure was used as a model for the STAS domain and was fitted to the clear secondary structure elements seen in the stressosome core reconstruction (Fig. 1.14). With the position of RsbR determined by the position of the turrets in the RsbR/RsbS reconstructions, it was possible to assign the positions of the RsbR and RsbS STAS domains with reference to the turrets. The three reconstructions were aligned to the same coordinate frame and STAS domains below the turrets were assigned as RsbR and those with no turret as RsbS .
Docking of MtS model to the stressosome core reconstruction envelope. Orthogonal views of the secondary structure fit of the X-ray crystal structure of MtS (PDBID: 3TZA) in the icosahedral stressosome core (RsbR146–274/RsbS ) reconstruction. The density map is contoured at 2.5σ to emphasise α-helices. The model of MtS is shown as a secondary structure cartoon and colour ramped from blue at the N-terminus, to red at the C-terminus
The RsbR/RsbS /RsbT ternary complex reconstruction showed clear additional density above the positions assigned as RsbS in the RsbR/RsbS reconstruction; this density was therefore attributed to the presence of the RsbT protein. The co-crystal structure of the SpoIIAA/SpoIIAB complex was used to guide fitting of a SpoIIAB-based homology model of the RsbT kinase in the RsbR/RsbS /RsbT reconstruction (Fig. 1.15) (Masuda et al. 2004). This placed RsbT above RsbS and in proximity to RsbR to form a phosphorylation -competent complex in which RsbS was closest to the ATP-binding site in RsbT, in accordance with the faster rate of phosphorylation of RsbS than RsbR in minimal stressosomes in vitro (Chen et al. 2003).
Docking of RsbS and RsbT models to the ternary stressosome complex reconstruction envelope. Secondary structure fit of a homology model of RsbS produced from MtS (PDBID: 2VY9) and a homology model of of RsbT produced from the Bacillus stearothermophilus SpoIIAB structure (PDBID: 1TH8) in the RsbR/RsbS/RsbT ternary stressosome complex reconstruction. The density map is contoured at 2σ; the model of RsbT is shown as a secondary structure cartoon coloured green, while RsbS is shown in red. In this model RsbT is poised above the RsbS protein in a pre-phosphorylation state
Using the three reconstructions it was possible to produce a complete pseudo-atomic model of the stressosome to gain insight into the architecture of the complex and how its structure influences its signalling mechanism (Fig. 1.16). These insights are discussed in Sect. 1.6 below, with reference to the validating experimental evidence
Pseudo-atomic model of the RsbR/RsbS /RsbT stressosome ternary complex. (a) A pseudo-atomic model of the RsbR/RsbS/RsbT stressosome was produced using the fitted coordinates of homology models of each component of the complex. The N-terminus of RsbR was modelled using the MtR crystal structure (PDBID: 3TZA) and is shown in orange; the STAS domains of RsbR and RsbS were modelled using the structure of MtS (PDBID: 2VY9) and are shown in blue and red respectively; and RsbT was modelled using the structure of SpoIIAB (PDBID: 1TH8) and is shown in green. (b) Cross section of the RsbR/RsbS /RsbT pseudo-atomic model showing the empty central cavity and dimensions of the complex. The cavity has a diameter of 90 Å, while the external extent of the core of the stressosome is 180 Å. The RsbR signalling domains extend from the complex to give an overall diameter of 300 Å. (c) Cartoon model of the orientation of the two domains of RsbR/MtR from the pseudo-atomic stressosome model, showing the arrangement of the STAS domain dimer in the core. Signalling domain at top and STAS domain at the bottom. (d) Ribon models of a structural alignment of the STAS domain dimer from YtvA (blue) and RsbR in the stressome core. The two models show essentially the same monomer orientation in the dimer, with a small 10° shift in the relative position of the monomers around the twofold axis
1.5.5 Other Stressosomes
Since the discovery of the stressosome as an ordered macromolecular complex (Chen et al. 2003) a number of studies have presented negatively stained electron micrographs of stressosome complexes from B. subtilis and M. thermoacetica (Delumeau et al. 2006; Marles-Wright et al. 2008; Quin et al. 2012). The gross appearance of these complexes in two-dimensions are essentially the same, with a 90 Å core radius and turrets giving a total radius of 150 Å. To date the cryo-EM reconstructions of the B. subtilis stressosome are the only published examples for this class of protein complex (Marles-Wright et al. 2008). Further examples of reconstructions from other species will determine whether the structural paradigm seen for the B. subtilis stressosome holds in different bacteria and will help shed further light onto the structure and mechanism of this widely distributed bacterial signalling complex.
1.6 Structure/Function Relationships in Stressosomes
The complex structure of the B. subtilis stressosome underlies its central role in integrating the diverse environmental signals that activate the σB pathway. The knowledge gained from the cryo-EM reconstructions and pseudo-atomic models permitted a number of insights into the relationship between the structure and function of the stressosome.
1.6.1 Activation
Inspection of the initial stressosome reconstructions highlights the potential mobility of the sensory turrets, given the fact that these regions appear less well defined than the core of the complex. These reconstructions were produced before the introduction of the gold-standard FSC resolution estimation methods, or the introduction of more sensitive detector technologies and modern algorithms that are driving the current resolution revolution in electron microscopy . Nonetheless, the structures may suggest a model of signal perception by the N-terminal domains and subsequent signal transduction through movement of the linking helixes that is transmitted to the C-terminal STAS domains, with subsequent activation of the RsbT kinase . The pseudo-atomic model produced from the stressosome reconstructions illustrates the relationships between the individual proteins that make up the complex (Fig. 1.16a, b). There is a large cavity at the centre of the stressosome; however, it is not thought that any proteins reside within this cavity and its presence is likely to be a secondary consequence of the architecture of the complex, where the STAS domains of the core are spaced to allow the recruitment of the RsbT kinase and to allow space for the RsbR N-terminal domains. Having a central cavity may allow structural rearrangements in the complex upon receipt of activating signals that would not be possible with a solid core.
The exact nature of the primary activating signals for the B. subtilis stressosome is not known. While each of the RsbR paralogues have a distinct N-terminal domain, only the activating signal for the YtvA blue-light receptor has been identified, though the details of how this signal modulates stressosome activation is still an area of active debate (Jurk et al. 2010; van der Steen and Hellingwerf 2015). Illumination of crystals of the LOV domain of YtvA with blue light leads to a shift in the Jα-helix region of the protein that links the N-terminal LOV domain to the C-terminal STAS domain (Möglich and Moffat 2007). This movement was suggested to propagate signal perception to changes in the relative orientation of the STAS domain (Möglich and Moffat 2007) and thus induce gross structural changes in the stressosome core to allow signal transduction to activate RsbT. Solution scattering studies of full length YtvA show minimal structural rearrangements upon illumination with blue-light, leading to the observation that this particular switch ‘does not flip’ (Jurk et al. 2010). Further work by this group, using NMR, showed that the surface accessibility of some residues in the STAS domain change in response to blue light and that these structural changes propagated by light absorption are not substantial in magnitude and thus may not be observed in low resolution SAXS studies. The subtle structural changes in YtvA may rather affect the network of protein-protein interactions with the partner proteins in the stressosome (Jurk et al. 2011). It is clear from this work on YtvA that the magnitude of changes in individual stressosome components upon activation is likely to be small on the scale of individual proteins, but it tells little about the changes in the structure, or thermodynamics of the whole system.
It is notable that the modelled organisation of the full length RsbR/MtR proteins in the stressosome is consistent with the dimer organisation of the full length structure of YtvA in solution (Figs. 1.5b and 1.16c). Furthermore, the STAS domain dimers in the stressosome core are consistent with the STAS domains seen in the YtvA structure, with only minor differences in their relative orientation (Fig. 1.16d). The differences seen in the position of the N-terminal portion of the STAS domain in relation to the Jα helix indicates a level of conformational flexibility in this region of the stressosome core that further reinforces the hypothesis that any signals perceived by the N-terminal RsbR/YtvA paralogues could be transmitted to the stressosome core to permit STAS domain phosphorylation by RsbT and its release from the complex.
It is also possible that the stressosome core provides a rigid scaffold that allows movement only in the N-terminal domains of the RsbR paralogues and YtvA . In this model, signalling by the stressosome would be a result of the differential positioning of the N-terminal domains in the activated/resting state and their influence on the probability of activating the kinase function of RsbT. The effect of mutations in RsbR seen in the work by Gaidenko and colleagues may be a consequence of constraints on the movement of the N-terminal domain of RsbR about the STAS domain scaffold in the stressosome core with resultant changes in the activation of RsbT, rather than direct interactions with RsbT itself (Gaidenko et al. 2011). The lack of significant conformational changes seen in YtvA low resolution solution studies (Jurk et al. 2010) could be explained in this model by a requirement for the core stressosome STAS scaffold to provide a rigid body to push against when activated.
In the context of a native stressosome with twenty peripheral turrets, small changes induced by external signals could be propagated and amplified allosterically to activate stressosome phosphorylation by RsbT and its subsequent release. Indeed, the σB response to environmental stimuli in live B. subtilis cells obeys a sigmoidal curve, indicating that co-operativity plays an important role in the signal transduction cascade that most likely stems from the stressosome (Marles-Wright et al. 2008).
Single cell fluorescence experiments and mathematical models of stressosome activation show that the response to different environmental stresses obeys the same kinetics and magnitude of response, indicating a key role for the stressosome in mediating and normalising the environmental stress response and highlighting the role of RsbR in the activation of RsbT (Locke et al. 2011; Liebal et al. 2013; Young et al. 2013). The hypothesis that structural changes in the sensory domains of the RsbR paralogues affect RsbT binding to the stressosome through direct interactions with the kinase , or through signalling via the STAS domains, has been tested both in vitro (Murray et al. 2005) and in vivo through the mutagenesis of both the N-terminal domain of RsbR and its linker helix (Gaidenko et al. 2011; Gaidenko et al. 2012). Mutagenesis of residues that line a surface groove in the dimer interface of the RsbR sensory domain abrogated the binding of RsbT to reconstituted stressosomes in vitro. However, the introduction of these mutants into a strain of B. subtilis lacking the other RsbR paralogues, failed to show corresponding changes to σB activation in vivo (Gaidenko et al. 2011). Strains bearing other mutations in the dimer interface showed increased basal activity of σB, of particular note was the effect seen in the glutamic acid 136 to lysine mutation, located in the linking helix that connects the two domains of RsbR. While a number of mutants displayed an enhanced basal σB activity, there was little effect on signalling outputs upon the imposition of stress. Mutagenesis of the linker helix in RsbR has shown that changes to residues in the helix-interface enhance basal stressosome output, whilst those on the surface of the linker helix diminish it (Gaidenko et al. 2012). The enhanced basal stressosome activity seen does not appear to diminish its capacity for responding to specific stresses, this may be a consequence of the presence of a pool of RsbT that is still bound to stressosomes that is not completely exhausted by the high basal activity, or an as yet unidentified stressosome signalling mechanism that is independent of the N-terminal RsbR domains and/or phosphorylation (Gaidenko and Price 2014). Taken together, these observations suggest that the perception of stresses by the stressosome and modulation of the output signal is more complex than can be explained by a mechanism where there is a direct link between stress perception by the N-terminal RsbR domains and transduction to the C-terminal STAS domains, with subsequent activation of the RsbT kinase .
The identification of putative heme-containing RsbR paralogues in Vibrio vulnificus, Chromobacterium violaceum and Ruegeria species (Pané-Farré et al. 2005) provides a potential means by which to directly probe stressosome activation, given the known haem-containing signalling systems and their responsiveness to diatomic gases (Zhang and Phillips 2003). Future structural analyses of stressosome complexes will hopefully cast light on the changes that take place upon signal perception and give insight into how this is propagated to activate RsbT.
1.6.2 Stressosome Signalling Response
While the exact nature of the activating signals for the stressosome-mediated branch of the σB pathway are not known, and the mechanism of signal transduction is still a matter of some debate, the nature and dynamics of this response have been the subject of a number of single-cell systems biology studies (Locke et al. 2011; Young et al. 2013). Experiments on population-level activation of the σB pathway showed that the energy stress branch of the B. subtilis general stress response obeys a simple hyperbolic activation model, while the stressosome mediated environmental stress branch displays a cooperative Hill response (Marles-Wright et al. 2008). The cooperative nature of the environmental stress response was attributed to the stressosome, which carries multiple copies of RsbT, and would be ideally placed to rapidly release a bolus of the kinase in response to stresses perceived by the RsbR sensory domains.
A number of elegant single-cell studies were performed by the Elowitz group to investigate the dynamics of the σB response in B. subtilis (Locke et al. 2011; Young et al. 2013). In their first study they show that the energy stress response displays stochastic frequency-modulated pulsing behaviour on a single-cell level (Locke et al. 2011). This behaviour is proposed to allow the bacteria to respond to unpredictable environments, while stochastic differences across populations present a bet-hedging strategy to mitigate the high cost of activating the σB regulon. In contrast to the frequency-modulated behaviour exhibited by the energy stress pathway, subsequent studies showed that the stressosome-mediated environmental stress response displayed amplitude-modulated pulsing (Young et al. 2013). The biological consequences of this response to stress are a rapid activation of the σB regulon in high-stress conditions and a change in the magnitude of response as levels of stress increase. The structure of the stressosome can be used to provide a molecular explanation for this response: through the release of a bolus of RsbT in response to environmental stresses perceived by the stressosome, the σB pathway can be activated rapidly to provide a timely response to high-stress conditions. The tuneable magnitude of the stressosome response is thought to be a consequence of the release of variable amounts of RsbT from the complex as a function of the magnitude of the perceived stress. The presence of multiple RsbR paralogues in B. subtilis and other species provide for the possibility for the modulation of the stressosome in response to different stresses and cellular contexts, where different paralogues may be present in the stressosome in different growth conditions (Pané-Farré et al. 2005).
1.6.3 Phosphorylation Dynamics of Stressosome
Activation of the stressosome is accompanied by the phosphorylation of RsbS and RsbR paralogues on conserved serine and threonine residues by the RsbT protein kinase (Kim et al. 2004a). The stressosome appears to be primed through phosphorylation of Thr171 in RsbR by RsbT in unstressed cells; in strains where this residue is mutated, the stressosome-mediated response is much diminished (Kim et al. 2004a). Phosphorylation of RsbS at Ser59, and at the equivalent position in RsbR, Thr205, follows the imposition of stress and the subsequent release of RsbT. The activity of the RsbX phosphatase against both RsbS-P and RsbR-P is required to reset the stressosome to a pre-stress state for the recruitment of RsbT back to the resting state (Chen et al. 2004). In high-stress conditions, RsbR is more likely to be phosphorylated on both Thr171 and Thr205 and this correlates with a diminished stress response (Eymann et al. 2011).
Mathematical models of the dephosphorylation of RsbS-P and RsbR-P by RsbX are not entirely consistent with all the published experimental data (Liebal et al. 2013). However, the mathematical models do highlight the dynamic behaviour of the RsbX phosphatase against stressosome components. The rapid dephosphorylation of RsbS-P is in marked contrast to the slow dephosphorylation of RsbR-P, and the models for these reactions explain the experimental observation that in high-stress conditions RsbR is not fully dephosphorylated (Eymann et al. 2011). The presence of two regulatory loops for stressosome signalling, the first mediated by the RsbX phosphatase to reset the signal, and the slow kinetics of dephosphorylation of Thr205-P on RsbR-P, allow for the rapid resetting of the stressosome in the first case, and modulation of the response to high stress in the latter case. These insights into stressosome phosphorylation dynamics correlate with the behaviours modelled by Elowitz and colleagues as described above in Sect. 1.6.2.
1.7 Evolution of the Stressosome Signalling Pathway
1.7.1 Structure and Conservation of the Stressosome Gene Cluster
In B. subtilis , the stressosome genes rsbR, rsbS and rsbT form a gene cluster that is co-transcribed with the σB structural gene and four additional genes encoding key regulators of σB activity as part of an eight-gene operon (Fig. 1.17). Transcription of this operon is initiated from two different promoters: a σA-dependent promoter upstream of of rsbR leads to synthesis of an mRNA comprising the entire operon (rsbR-rsbS-rsbT-rsbU-rsbV-rsbW-sigB-rsbX), whilst a σB-dependent promoter located between the rsbU and rsbV genes increases transcription of the downstream half of the operon (rsbV-rsbW-sigB-rsbX) following stress (Wise and Price 1995). Identical operons are present in many low-GC Gram-positive bacteria including several Bacillus species (e.g. B. licheniformis, B. halodurans, B. pumilus, B. coagulas and Oceanobacillus iheyensis) and close relatives like the Listeria (e.g. L. monocytogenes and L. innocua). However, stressosome-related genes are absent from the genomes of members of the Bacillus cereus group, although these organisms encode a σB protein, its principal regulators RsbV, RsbW and a PP2C-type phosphatase, RsbY (Fig. 1.17). Like RsbU, RsbY acts as a positive regulator of σB activity in B. cereus (Schaik et al. 2005). Genome comparison and experimental data suggest that in the absence of a stressosome, the phosphatase activity of RsbY is regulated by a hybrid sensor histidine kinase , termed RsbK, encoded in close proximity of the sigB operon. According to the current model RsbK controls the phosphorylation status of the N-terminal REC domain (CheY-homologous receiver domain) of RsbY and thereby the activity of the C-terminally located PP2C domain (de Been et al. 2010, 2011). In addition, the activity of the hybrid kinase RsbY is modulated by methylation via RsbM, which is encoded down-stream of RsbY (Chen et al. 2012).
σ B -operon structure and domain organization of the encoded proteins. In B. subtilis , genes encoding the stressosome proteins RsbR, RsbS, and RsbT are organized with the σB structural gene and additional regulators of σB activity in an eight-gene operon. While the core of the σB-operon (light grey), is conserved in close relatives of B. subtilis including B. cereus and S. aureus, different solutions – including the RsbRSTX module (shaded black) or a hybrid kinase (black stripes) – merging at the control of a PP2C-type phosphatase (dark grey) have evolved to control the phosphorylation dependent interaction within the σB partner-switching module. Domain organization of the encoded proteins is shown below open reading frames (arrows). Transcription start sites and the initiating sigma factor are indicated. Abbreviations of domains detected with SMART (http://smart.embl-heidelberg.de) are: RsbR_N (RsbR N-terminus), STAS (sulphate transporter and anti-sigma factor antagonist), GHKL: (Gyrase, Hsp90, Histidine Kinase , MutL), RsbU_N (RsbU N-terminus), PP2C (Protein phosphatase 2C), Sigma (sigma factor), Ferritin (Ferritin family), REC (cheY-homologous receiver domain), MeTrc (Methyltransferase , chemotaxis proteins), TM (transmembrane domain), CHASE (extracellular sensory domain), HAMP (Histidine kinase s, Adenylyl cyclases, Methyl binding proteins, Phosphatases domain), GAF (domain present in phytochromes and cGMP-specific phosphodiesterases), HisKA (His Kinase A (phosphoacceptor) domain)
A similar situation is found in the genus Staphylococcus, which does not encode components of the stressosome, but has retained a truncated version of the B. subtilis sigB operon with the structure rsbU-rsbV-rsbW-sigB (Fig. 1.17). In contrast to B. cereus RsbY, the staphylococcal RsbU protein shows high similarity over the entire length of its sequence to the B. subtilis RsbU protein. In this context it is noteworthy that residues in the N-terminus of B. subtilis RsbU that are important for the interaction with RsbT are not conserved in Staphylococcus aureus RsbU (Hardwick et al. 2007). Furthermore, expression of S. aureus RsbU in B. subtilis leads to unrestricted σB activation and thus it remains an open question as to how RsbU, and hence σB activity, is controlled in the staphylococci (Pané-Farré et al. 2009).
Mirroring the situation observed in the B. cereus group and the staphylococci, many species that do not encode σB still encode the stressosome gene cluster (see Sect. 1.1.3). Contrary to B. subtilis and the Listeria, however, the gene encoding the negative feedback phosphatase RsbX is always located immediately down-stream of the stressosome genes, thus forming a highly conserved RsbRSTX module (Fig. 1.18). This module, according to the B. subtilis model, would provide the minimal set of proteins required to form a functional stressosome complex (RsbR, RsbS , RsbT) and a negative regulator (RsbX) to reset activated stressosomes. Indeed, the RsbRSTX module is almost always associated with genes encoding various proteins with signal transduction related functions including transcription factors , hybrid sensor kinase s or proteins involved in the turnover of secondary messengers (Pané-Farré et al. 2005). Since these down-stream modules are usually separated by only a few base pairs from the stressosome module it is likely that they are functionally linked to the stressosome, thereby providing distinct regulatory outputs.
Architecture of different output modules associated with the RsbRSTX stressosome module. Examples shown represent species from which experimental data on stressosome function or expression is available. Locus-tags and, if available, protein names are indicated. Abbreviations of domains detected with SMART (http://smart.embl-heidelberg.de) are: RsbR_N (RsbR N-terminus), STAS (sulphate transporter and anti-sigma factor antagonist), GHKL: (Gyrase, Hsp90, Histidine Kinase , MutL), RsbU_N (RsbU N-terminus), PP2C (Protein phosphatase 2C), Sigma (sigma factor), REC (cheY-homologous receiver domain), Sensor globin (heme coupled globin sensor), GGDEF (diguanylate cyclase), GAF (domain present in phytochromes and cGMP-specific phosphodiesterases), HisKA (His Kinase A (phosphoacceptor) domain), PAS (Per-Arnt-Sim protein), PAC (Motif C-terminal to PAS motifs), HTP (Histidine Phosphotransfer domain), HDc (Metal dependent phosphohydrolases with conserved ‘HD’ motif)
1.7.2 Evolution of the Stressosome
Almost nothing is known at present about the origin and evolution of the stressosome. The observation that the stressosome gene cluster can be found in a huge variety of distantly related bacteria and even some archaea (Methanosarcina horonobensis, Halorhabdus utahensis and Candidatus Methanoregula boonei) suggests that the stressosome genes were distributed across the microbial world by horizontal gene transfer. Indeed, the strong association of stressosome genes in a highly conserved RsbRSTX module may favour transfer between species and would thus provide an excellent building block that could be easily combined with various output modules to control different cellular processes. In agreement with this hypothesis, a study by Marri and colleagues suggested that the stressosome genes entered the genome of Mycobacterium avium subsp. paratuberculosis by horizontal gene transfer (Marri et al. 2006). The intriguing possibility that the stressosome genes are transferred between species as a unit raises the interesting question: how is this sensing complex integrated into the regulatory circuits of the recipient organism? In B. subtilis , signals perceived by the stressosome are conveyed to σB by the physical interaction of the serine threonine kinase RsbT with the N-terminal four-helical bundle of RsbU that acts as a recruitment domain for RsbT (Delumeau et al. 2004). This interaction does not require RsbT kinase activity (Kang et al. 1998). It is tempting to speculate that the B. subtilis sigB-operon could have arisen by an insertion of a group of genes including rsbUVW and sigB between the rsbT gene and the gene encoding an RsbX-like phosphatase. Domains showing sequence similarity to the N-terminal domain of B. subtilis RsbU can be identified in a number of down-stream proteins associated with the stressosome module in phylogenetically diverse species (Fig. 1.19); in such cases the answer as to how the stressosome communicates with the associated down-stream modules is evident. Consistent with that notion, a surface area identified to be crucial for the interaction of RsbU with RsbT in B. subtilis (Hardwick et al. 2007) shows a high degree of amino acid conservation in a comparison of RsbRSTX associated RsbU N-terminus-like domains (Fig. 1.19). Intriguingly, an isolated RsbU N-terminal domain located down-stream of an RsbRSTX gene cluster and up-stream of a GAF domain protein can be found in the actinobacterium Modestobacter marinus (Ponting and Aravind 1997). This observation could indicate the formation of a new stressosome signalling cascade, or alternatively, document the decay of the same.
The RsbU N-terminal domain. Sequences of proteins with an RsbU N-terminal domain were retrieved via the Pfam web-page (http://pfam.xfam.org). (a) Sequence alignment of RsbU N-terminal domains present in proteins encoded down-stream of RsbRSTX modules. For clarity, only the B. subtilis RsbU N-terminus was included as representative for species with an rsbUVWσB rsbX down-stream gene cluster. Protein residues within the RsbU N-terminus identified as important for RsbT:RsbU interaction in B. subtilis (Hardwick et al. 2007) are labelled with an asterisk. (b) Domain architectures of proteins associated with RsbRSTX modules displaying an RsbU N-terminal domain. (c) Structure of the B. subtilis RsbU N-terminus showing the amino acid conservation across RsbU N-termini presented in the above alignment. Abbreviations are as follows: Bsub (Bacillus subtilis), Cwoe (Conexibacter woesei), Dgib (Desulfotomaculum gibsoniae), Malb (Methylomicrobium album), Mkan (Mycobacterium kansasii), Mmar (Modestobacter marinus), Mmar (Mycobacterium marinum), Mpar (Mycobacterium paratuberculosis), Mroh (Mycobacterium rhodesiae), Msme (Mycobacterium smegmatis), Mthe (Moorella thermoacetica), Myco (genus Mycobacterium), Nocar (genus Nocardia), Ncyr (Nocardia cyriacigeorgica), Nfar (Nocardia farcinica), Nnov (Nocardia nova), Rueg (Ruegeria sp.), Smob (Symbiobacter mobilis)
In the majority of species, however, no RsbU N-terminus-like domain can be identified in the down-stream encoded regulators. If these proteins indeed receive regulatory input from the nearby-encoded stressosome, how is signal transfer realised? A possible answer may lay in the evolutionary origin of the RsbT protein, a member of the widely distributed GHKL protein family. GHKL domains are an integral part of the sensor kinase s of bacterial two component signal systems, in which the GHKL domain phosphorylates a conserved histidine residue within an N-terminally associated phospho-accepting histidine/dimerisation domain (HisKA). Assuming a conserved mode of communication between the stressosome and the putative downstream module via the RsbT orthologue, it is interesting to note that HisKA domains represent the least common denominator in the majority of down-stream modules devoid of RsbU N-terminus-like domains.
Further complexity is added to the stressosome by the observation that the RsbX-like phosphatase of many species often has an additional N-terminal GHKL domain related to RsbT. The presence of an additional domain with potential kinase activity in these RsbRSTX modules underscores the importance of phosphorylation events during stressosome signalling. However, a recent study by Gaidenko and Price showed that stressosome dependent signalling is also supported in B. subtilis in the absence of a phosphoryltable RsbS protein, leading to the suggestion that RsbS phosphorylation is a recent evolutionary addition that overlays a primordial signalling mechanism to increase the sensitivity of the stressosome by increasing the rate of RsbT dissociation (Gaidenko and Price 2014). Presumably, there are some genetic or growth differences in the strains used in this study in comparison to previous studies that pointed towards the inviability of rbsS null and point mutants (Kang et al. 1996).
Whilst orthologues of RsbT and the STAS domains of RsbS and RsbR orthologues are highly conserved within the RsbRST module across species, significant variation occurs in the N-termini of RsbR orthologues. The STAS domain of RsbR orthologues has been found to be linked to non-heme globin as well as heme globin sensory domains and members of the PAS domain family, which occur in tandem within a single RsbR protein (e.g. in the genus Aeromonas (Pané-Farré et al. 2005; Sharma et al. 2011)). In some species, in addition to the promoter-proximal RsbR protein, further genes encoding RsbR paralogues can be identified. The occurrence of RsbR paralogues is particularly prominent in the Gram-positive bacteria. Usually, the N-termini of the RsbR paralogues appear to be related to the N-terminus of the RsbR protein encoded within the RsbRSTX modules, but a diversity of N-terminal domains may be seen across the RsbR paralogues of one organism (see Sect. 1.2.3).
The observation that RsbR proteins are equipped with different N-termini and some species even encode multiple RsbR paralogues provides further evidence for the notion that the RsbR N-terminus plays a crucial role in stressosome signal perception. However, experiments testing this hypothesis in B. subtilis do not unequivocally support this conclusion for the family of RsbR co-antagonists displaying non-heme globin N-terminal domains (Gaidenko et al. 2011, 2012) and the role of this domain in signalling remains unclear.
Finally, proteins similar to RsbR but not encoded within a stressosome gene cluster can be identified in several species of green non-sulphur bacteria. For instance, the photosynthetic bacterium Chloroflexus aurantiacus encodes 17 full-length RsbR paralogues some of which display multiple N-terminal PAS domains. Given the early evolutionary origin of green non-sulphur bacteria, it will be interesting to determine if these proteins also interact to form complex protein assemblies like the stressosome of B. subtilis , which could point to an evolutionary origin of the stressosome and would imply that stressosome-like protein assemblies may be a much more widely distributed paradigm than currently anticipated.
In summary, the available structural and sequence data indicate that stressosome dependent signalling may show substantial differences between species regarding signal perception, integration and transmission to the various output modules. Comparative genomics of stressosome proteins and down-stream regulators in combination with targeted experiments will be required to understand shared and specific mechanisms of signal transduction involving the stressosome.
1.7.3 Stressosome Function in Other Organisms
While the B. subtilis stressosome and the σB pathway is well characterised, there is little data available on stressosome function in other species. The stressosome module can be identified in microorganisms that thrive in virtually every ecological niche and there seems to be no preference for a specific habitat or lifestyle. This includes bacteria living in soil, deep-sea and estuarine marine environments, as well as plant and animal symbionts and pathogens (Pané-Farré et al. 2005) (Fig. 1.20).
Distribution of the RST stressosome module. To estimate the occurrence of RSTX modules within the microbial world, taxonomic information for the RsbR protein family (K17763) was retrieved from the KEGG webpage (http://www.kegg.jp). Phyla in which the genes for RsbR proteins are organized in a rsbRSTX gene cluster are highlighted in grey. Numbers in blue circles indicate the total number of species with RsbRSTX modules within one phylum. Numbers in parentheses indicate the number of species that in addition to the promoter-proximal RsbR in the RsbRSTX cluster also encode at least one RsbR paraloge. No RsbRSTX module was identified in the green nonsulfur bacteria, but multiple RsbR-like proteins were detected in six members (white circle) of this phylum
In the Gram-positive pathogen L. monocytogenes the stressosome controls σB-activity in response to both environmental and nutritional stress (Chaturongakul and Boor 2004; Chaturongakul and Boor 2006). Furthermore, by replacing the B. subtilis rsbR gene with its L. monocytogenes counterpart, the response to environmental and nutritional stress of the latter was transferred into the B. subtilis host. However, the presence of RsbR paralogues in B. subtilis abrogated the nutritional stress sensing properties of the L. monocytogenes RsbR protein (Martinez et al. 2010). These results provide evidence that strengthens the hypothesis that the RsbR proteins directly sense stress signals within the cell.
There is also evidence that the stressosome has been adapted to control secondary messenger levels, rather than the activity of a transcription factor . The M. thermoacetica orthologue RsbT, termed MtT, inhibits the activity of a downstream-encoded GG(D/E)EF-type diguanylate cyclase (Schirmer and Jenal 2009), MtG (Quin et al. 2012). Activation of the stressosome and hence inhibition of the MtG diguanylate cyclase activity is thus assumed to control the cellular level of the second messenger cyclic di-GMP (c-di-GMP) in M. thermoacetica. C-di-GMP is one of the most common and important bacterial secondary messengers and has been shown to be involved in the regulation of many process often involving lifestyle changes that occur during development , virulence gene expression, biofilm formation, the cell cycle, or the switch between a sessile and a motile state (Römling et al. 2013; Ryan 2013). However, the in vivo role of the M. thermoacetica stressosome has not yet been investigated.
For the actinobacterium and opportunistic human pathogen Mycobacterium marinum transcription of stressosome genes was observed to be increased during cold stress, suggesting a role for the stressosome in cold adaptation in this species (Pettersson et al. 2013). For the γ-proteobacterium Xanthomonas campestris, a plant pathogen infecting cabbage, phosphorylation of the RsbR orthologue has been reported during transition from exponential growth to stationary phase (Musa et al. 2013). Since phosphorylation of RsbR correlates with stress activation, stressosome function may be required during stationary phase adaptation in X. campestris. The M. marinum downstream protein associated with the RsbRSTX cluster has a domain similar to the N-terminus of RsbU, and which is linked to a C-terminal PAS-PP2C-GAF-GHKL multi-domain polypeptide and may be involved in sigma-factor regulation via the PP2C domain. A functional prediction is, at present, not possible for the two potential X. campestris downstream proteins, which display a combination of REC, GHKL and HisKA domains, but do not possess an RsbU N-terminus-like domain (Fig. 1.18).
A stressosome gene cluster has also been identified in a number of Vibrio species including V. vulnificus and V. mimicus, two species that cause seafood associated gastroenteritis; V. coralliilyticus, a coral pathogen; and V. orientalis, a luminescent bacterium. Interestingly, whole genome sequencing has shown that the stressosome gene cluster is particularly well conserved in clinical V. vulnificus isolates and less frequently found in the genome of environmental strains (Morrison et al. 2012; Williams et al. 2014), suggesting that the stressosome may be advantageous during the establishment of an infection. The presence of an N-terminal heme-binding sensor globin domain in the V. vulnificus RsbR orthologue strongly suggests a function in the sensing of oxygen or other diatomic gases for the V. vulnificus stressosome. The stressosome genes are associated in V. vulnificus with two open reading frames encoding a hybrid histidine kinase and an HDc domain that likely functions in the degradation of c-di-GMP (Fig. 1.17). Hence, as in M. thermoacetica, control of c-di-GMP levels may represent the regulatory output of the V. vulnificus stressosome.
1.8 Conclusions
The stressosome has been the subject of extensive research into its role in the general stress response of the model Gram-positive bacterium B. subtilis since its discovery and isolation over 20 years ago. Our structural studies of the stressosome complex showed how it is able to sequester a pool of the RsbT activator kinase that can be released upon the receipt of stress signals by the sensory domains of RsbR and related proteins. The mechanism of activation and signal transduction by the stressosome remains to be determined and is a subject of great interest and debate within the field. With the explosion in the number of sequenced bacterial genomes seen in the last 10 years, the distribution of stressosome signalling systems across different phyla points to their utility. Furthermore, the diversity of downstream signalling modules seen in different species hints at a variety of different physiological roles for the stressosome. Future work will hopefully reveal the mechanism of stressosome signalling and shed light onto the nature of stressosome signalling in diverse bacterial species.
References
Aertsen A, Michiels CW (2004) Stress and how bacteria cope with death and survival. Crit Rev Microbiol 30:263–273. doi:10.1080/10408410490884757
Akbar S, Gaidenko TA, Kang CM et al (2001) New family of regulators in the environmental signaling pathway which activates the general stress transcription factor σB of Bacillus subtilis. J Bacteriol 183:1329–1338. doi:10.1128/JB.183.4.1329-1338.2001
Alper S, Duncan L, Losick R (1994) An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195–205
Alper S, Dufour A, Garsin DA et al (1996) Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol 260:165–177. doi:10.1006/jmbi.1996.0390
Aravind L, Koonin EV (2000) The STAS domain – a link between anion transporters and antisigma-factor antagonists. Curr Biol 10:R53–R55
Barford D, Das AK, Egloff MP (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct 27:133–164. doi:10.1146/annurev.biophys.27.1.133
Benson AK, Haldenwang WG (1993) Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci U S A 90:2330–2334
Beuron F, Maurizi MR, Belnap DM et al (1998) At sixes and sevens: characterization of the symmetry mismatch of the ClpAP chaperone-assisted protease. J Struct Biol 123:248–259. doi:10.1006/jsbi.1998.4039
Bhate MP, Molnar KS, Goulian M, DeGrado WF (2015) Signal transduction in histidine kinases: insights from new structures. Structure 23:981–994. doi:10.1016/j.str.2015.04.002
Brody MS, Vijay K, Price CW (2001) Catalytic function of an alpha/beta hydrolase is required for energy stress activation of the σB transcription factor in Bacillus subtilis. J Bacteriol 183:6422–6428. doi:10.1128/JB.183.21.6422-6428.2001
Campbell EA, Masuda S, Sun JL et al (2002) Crystal structure of the Bacillus stearothermophilus anti-sigma factor SpoIIAB with the sporulation sigma factor sigmaF. Cell 108:795–807. doi:10.1016/S0092-8674(02)00662-1
Capra EJ, Laub MT (2012) Evolution of two-component signal transduction systems. Annu Rev Microbiol 66:325–347. doi:10.1146/annurev-micro-092611-150039
Chaturongakul S, Boor KJ (2004) RsbT and RsbV contribute to σB-dependent survival under environmental, energy, and intracellular stress conditions in Listeria monocytogenes. Appl Environ Microbiol 70:5349–5356. doi:10.1128/AEM.70.9.5349-5356.2004
Chaturongakul S, Boor KJ (2006) σB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol 72:5197–5203. doi:10.1128/AEM.03058-05
Chen C-CCC, Lewis RJRJ, Harris R et al (2003) A supramolecular complex in the environmental stress signalling pathway of Bacillus subtilis. Mol Microbiol 49:1657–1669. doi:10.1046/j.1365-2958.2003.03663.x
Chen C-C, Yudkin MD, Delumeau O (2004) Phosphorylation and RsbX-dependent dephosphorylation of RsbR in the RsbR-RsbS complex of Bacillus subtilis. J Bacteriol 186:6830–6836. doi:10.1128/JB.186.20.6830-6836.2004
Chen LC, Chen JC, Shu JC et al (2012) Interplay of RsbM and RsbK controls the σB activity of Bacillus cereus. Environ Microbiol 14:2788–2799. doi:10.1111/j.1462-2920.2012.02788.x
Das AK, Helps NR, Cohen PT, Barford D (1996) Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution. EMBO J 15:6798–6809
de Been M, Tempelaars MH, van Schaik W et al (2010) A novel hybrid kinase is essential for regulating the σB-mediated stress response of Bacillus cereus. Environ Microbiol 12:730–745. doi:10.1111/j.1462-2920.2009.02116.x
de Been M, Francke C, Siezen RJ, Abee T (2011) Novel σB regulation modules of Gram-positive bacteria involve the use of complex hybrid histidine kinases. Microbiology 157:3–12. doi:10.1099/mic.0.045740-0
Delumeau O, Lewis RJ, Yudkin MD (2002) Protein-protein interactions that regulate the energy stress activation of σB in Bacillus subtilis. J Bacteriol 184:5583–5589. doi:10.1128/JB.184.20.5583-5589.2002
Delumeau O, Dutta S, Brigulla M et al (2004) Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J Biol Chem 279:40927–40937. doi:10.1074/jbc.M405464200
Delumeau O, Chen C-C, Murray JW et al (2006) High-molecular-weight complexes of RsbR and paralogues in the environmental signaling pathway of Bacillus subtilis. J Bacteriol 188:7885–7892. doi:10.1128/JB.00892-06
Dube P, Tavares P, Lurz R, van Heel M (1993) The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J 12:1303–1309
Dufour A, Voelker U, Voelker A, Haldenwang WG (1996) Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J Bacteriol 178:3701–3709
Dutta R, Inouye M (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25:24–28
Eymann C, Schulz S, Gronau K et al (2011) In vivo phosphorylation patterns of key stressosome proteins define a second feedback loop that limits activation of Bacillus subtilis σ(B). Mol Microbiol 80:798–810. doi:10.1111/j.1365-2958.2011.07609.x
Gaidenko TA, Price CW (2014) Genetic evidence for a phosphorylation-independent signal transduction mechanism within the Bacillus subtilis stressosome. PLoS One 9:e90741. doi:10.1371/journal.pone.0090741
Gaidenko TA, Yang X, Lee YM, Price CW (1999) Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway of Bacillus subtilis. J Mol Biol 288:29–39. doi:10.1006/jmbi.1999.2665
Gaidenko TA, Bie X, Baldwin EP, Price CW (2011) Substitutions in the presumed sensing domain of the Bacillus subtilis stressosome affect its basal output but not response to environmental signals. J Bacteriol 193:3588–3597. doi:10.1128/JB.00060-11
Gaidenko TA, Bie X, Baldwin EP, Price CW (2012) Two surfaces of a conserved interdomain linker differentially affect output from the RST sensing module of the Bacillus subtilis stressosome. J Bacteriol 194:3913–3921. doi:10.1128/JB.00583-12
Gao R, Stock AM (2009) Biological insights from structures of two-component proteins. Annu Rev Microbiol 63:133–154. doi:10.1146/annurev.micro.091208.073214
Gouet P, Robert X, Courcelle E (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. doi:10.1093/nar/gkg556
Haldenwang WG, Losick R (1979) A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature 282:256–260
Hardwick SW, Pané-Farré J, Delumeau O et al (2007) Structural and functional characterization of partner switching regulating the environmental stress response in Bacillus subtilis. J Biol Chem 282:11562–11572. doi:10.1074/jbc.M609733200
Hecker M, Völker U (2001) General stress response of Bacillus subtilis and other bacteria. Adv Microb Physiol 44:35–91. doi:10.1016/S0065-2911(01)44011-2
Hecker M, Pané-Farré J, Völker U (2007) SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu Rev Microbiol 61:215–236. doi:10.1146/annurev.micro.61.080706.093445
Herrou J, Crosson S (2011) Function, structure and mechanism of bacterial photosensory LOV proteins. Nat Rev Microbiol 9:713–723. doi:10.1038/nrmicro2622
Hou S, Larsen RW, Boudko D et al (2000) Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403:540–544. doi:10.1038/35000570
Jurk M, Dorn M, Kikhney A et al (2010) The switch that does not flip: the blue-light receptor YtvA from Bacillus subtilis adopts an elongated dimer conformation independent of the activation state as revealed by a combined AUC and SAXS study. J Mol Biol 403:78–87. doi:10.1016/j.jmb.2010.08.036
Jurk M, Dorn M, Schmieder P (2011) Blue flickers of hope: secondary structure, dynamics, and putative dimerization interface of the blue-light receptor YtvA from Bacillus subtilis. Biochemistry 50:8163–8171. doi:10.1021/bi200782j
Jurk M, Schramm P, Schmieder P (2013) The blue-light receptor YtvA from Bacillus subtilis is permanently incorporated into the stressosome independent of the illumination state. Biochem Biophys Res Commun 432:499–503. doi:10.1016/j.bbrc.2013.02.025
Kaneko T, Tanaka N, Kumasaka T (2005) Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis. Protein Sci 14:558–565. doi:10.1110/ps.041170005
Kang CM, Brody MS, Akbar S et al (1996) Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J Bacteriol 178:3846–3853
Kang CM, Vijay K, Price CW (1998) Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol Microbiol 30:189–196
Kim T-J, Gaidenko TA, Price CW (2004a) In vivo phosphorylation of partner switching regulators correlates with stress transmission in the environmental signaling pathway of Bacillus subtilis. J Bacteriol 186:6124–6132. doi:10.1128/JB.186.18.6124-6132.2004
Kim T-J, Gaidenko TA, Price CW (2004b) A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J Mol Biol 341:135–150. doi:10.1016/j.jmb.2004.05.043
Kitanishi K, Kobayashi K, Uchida T et al (2011) Identification and functional and spectral characterization of a globin-coupled histidine kinase from Anaeromyxobacter sp. Fw109-5. J Biol Chem 286:35522–35534. doi:10.1074/jbc.M111.274811
Kovacs H, Comfort D, Lord M et al (1998) Solution structure of SpoIIAA, a phosphorylatable component of the system that regulates transcription factor sigmaF of Bacillus subtilis. Proc Natl Acad Sci U S A 95:5067–5071
Krell T, Lacal J, Busch A et al (2010) Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64:539–559. doi:10.1146/annurev.micro.112408.134054
Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi:10.1016/j.jmb.2007.05.022
Kumar A, Lomize A, Jin KK et al (2010) Open and closed conformations of two SpoIIAA-like proteins (YP_749275.1 and YP_001095227.1) provide insights into membrane association and ligand binding. Acta Crystallogr Sect F Struct Biol Cryst Commun 66:1245–1253. doi:10.1107/S1744309109042481
Liebal UW, Millat T, Marles-wright J et al (2013) Simulations of stressosome activation emphasize allosteric interactions between RsbR and RsbT. BMC Syst Biol 7:3. doi:10.1186/1752-0509-7-3
Locke JCW, Young JW, Fontes M et al (2011) Stochastic pulse regulation in bacterial stress response. Science 334:366–369. doi:10.1126/science.1208144
Lord M, Magnin T, Yudkin MD (1996) Protein conformational change and nucleotide binding involved in regulation of sigmaF in Bacillus subtilis. J Bacteriol 178:6730–6735
Losi A, Polverini E, Quest B, Gärtner W (2002) First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J 82:2627–2634. doi:10.1016/S0006-3495(02)75604-X
Losi A, Quest B, Gärtner W (2003) Listening to the blue: the time-resolved thermodynamics of the bacterial blue-light receptor YtvA and its isolated LOV domain. Photochem Photobiol Sci 2:759–766
Lowe EC, Baslé A, Czjzek M et al (2012) A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci U S A 109:1–6. doi:10.1073/pnas.1200479109
Marles-Wright J, Grant T, Delumeau O et al (2008) Molecular architecture of the “stressosome,” a signal integration and transduction hub. Science 322:92–96. doi:10.1126/science.1159572
Marri PR, Bannantine JP, Paustian ML, Golding GB (2006) Lateral gene transfer in Mycobacterium avium subspecies paratuberculosis. Can J Microbiol 52:560–569. doi:10.1139/w06-001
Martinez L, Reeves A, Haldenwang W (2010) Stressosomes formed in Bacillus subtilis from the RsbR protein of Listeria monocytogenes allow σB activation following exposure to either physical or nutritional stress. J Bacteriol 192:6279–6286. doi:10.1128/JB.00467-10
Masuda S, Murakami KS, Wang S et al (2004) Crystal structures of the ADP and ATP bound forms of the Bacillus anti-sigma factor SpoIIAB in complex with the anti-anti-sigma SpoIIAA. J Mol Biol 340:941–956. doi:10.1016/j.jmb.2004.05.040
Mitchell JG, Kogure K (2006) Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol Ecol 55:3–16
Möglich A, Moffat K (2007) Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol 373:112–126. doi:10.1016/j.jmb.2007.07.039
Moran CP, Lang N, Banner CD et al (1981) Promoter for a developmentally regulated gene in Bacillus subtilis. Cell 25:783–791
Morrison SS, Williams T, Cain A et al (2012) Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553. doi:10.1371/journal.pone.0037553
Murray JW, Delumeau O, Lewis RJ (2005) Structure of a nonheme globin in environmental stress signaling. Proc Natl Acad Sci U S A 102:17320–17325. doi:10.1073/pnas.0506599102
Musa YR, Bäsell K, Schatschneider S, Vorhölter FJ, Becher D, Niehaus K (2013) Dynamic protein phosphorylation during the growth of Xanthomonas campestris pv. campestris B100 revealed by a gel-based proteomics approach. J Biotechnol 167(2):111–122. doi:10.1016/j.jbiotec.2013.06.009. Epub 2013 Jun 20
Nadezhdin EV, Brody MS, Price CW (2011) An α/β hydrolase and associated Per-ARNT-Sim domain comprise a bipartite sensing module coupled with diverse output domains. PLoS One 6:e25418. doi:10.1371/journal.pone.0025418
Ondrusch N, Kreft J (2011) Blue and red light modulates SigB-dependent gene transcription, swimming motility and invasiveness in Listeria monocytogenes. PLoS One 6:e16151. doi:10.1371/journal.pone.0016151
Österberg S, del Peso-Santos T, Shingler V (2011) Regulation of alternative sigma factor use. Annu Rev Microbiol 65:37–55. doi:10.1146/annurev.micro.112408.134219
Paget MS (2015) Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5:1245–1265. doi:10.3390/biom5031245
Pané-Farré J, Lewis RJ, Stülke J et al (2005) The RsbRST stress module in bacteria: a signalling system that may interact with different output modules. J Mol Microbiol Biotechnol 9:65–76. doi:10.1159/000088837
Pané-Farré J, Jonas B, Hardwick SW et al (2009) Role of RsbU in controlling SigB activity in Staphylococcus aureus following alkaline stress. J Bacteriol 191:2561–2573. doi:10.1128/JB.01514-08
Petersohn A, Brigulla M, Haas S et al (2001) Global analysis of the general stress response of Bacillus subtilis. J Bacteriol 183:5617–5631. doi:10.1128/JB.183.19.5617-5631.2001
Pettersson BMF, Nitharwal RG, Das S et al (2013) Identification and expression of stressosomal proteins in Mycobacterium marinum under various growth and stress conditions. FEMS Microbiol Lett 342:98–105. doi:10.1111/1574-6968.12118
Ponting CP, Aravind L (1997) PAS: a multifunctional domain family comes to light. Curr Biol 7:R674–R677
Price CW, Fawcett P, Cérémonie H et al (2001) Genome-wide analysis of the general stress response in Bacillus subtilis. Mol Microbiol 41:757–774
Quin MB, Berrisford JM, Newman JA et al (2012) The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling. Structure 20:350–363. doi:10.1016/j.str.2012.01.003
Reeves A, Martinez L, Haldenwang W (2010) Expression of, and in vivo stressosome formation by, single members of the RsbR protein family in Bacillus subtilis. Microbiology 156:990–998. doi:10.1099/mic.0.036095-0
Römling U, Galperin MY, Gomelsky M (2013) Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi:10.1128/MMBR.00043-12
Ryan RP (2013) Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology 159:1286–1297. doi:10.1099/mic.0.068189-0
Schaik Van W, Tempelaars MHMH, Zwietering MHMH et al (2005) Analysis of the role of RsbV, RsbW, and RsbY in regulating σB activity in Bacillus cereus. J Bacteriol 187:5846–5851. doi:10.1128/JB.187.16.5846
Schirmer T, Jenal U (2009) Structural and mechanistic determinants of c-di-GMP signalling. Nat Rev Microbiol 7:724–735. doi:10.1038/nrmicro2203
Schmalisch M, Langbein I, Stülke J (2002) The general stress protein Ctc of Bacillus subtilis is a ribosomal protein. J Mol Microbiol Biotechnol 4:495–501
Seavers PR, Lewis RJ, Brannigan JA et al (2001) Structure of the Bacillus cell fate determinant SpoIIAA in phosphorylated and unphosphorylated forms. Structure 9:605–614
Sharma AK, Rigby AC, Alper SL (2011) STAS domain structure and function. Cell Physiol Biochem 28:407–422. doi:10.1159/000335104
Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69:183–215. doi:10.1146/annurev.biochem.69.1.183
Stranzl GR, Santelli E, Bankston LA et al (2011) Structural insights into inhibition of Bacillus anthracis sporulation by a novel class of non-heme globin sensor domains. J Biol Chem 286:8448–8458. doi:10.1074/jbc.M110.207126
Teh AH, Makino M, Hoshino T et al (2015) Structure of the RsbX phosphatase involved in the general stress response of Bacillus subtilis. Acta Crystallogr D Biol Crystallogr 71:1392–1399. doi:10.1107/S1399004715007166
Truitt CL, Weaver EA, Haldenwang WG (1988) Effects on growth and sporulation of inactivation of a Bacillus subtilis gene (ctc) transcribed in vitro by minor vegetative cell RNA polymerases (E-sigma 37, E-sigma 32). Mol Gen Genet 212:166–171
van der Steen JB, Hellingwerf KJ (2015) Activation of the general stress response of Bacillus subtilis by visible light. Photochem Photobiol 91:1032–1045. doi:10.1111/php.12499
van der Steen JB, Avila-Pérez M, Knippert D et al (2012) Differentiation of function among the RsbR paralogs in the general stress response of Bacillus subtilis with regard to light perception. J Bacteriol 194:1708–1716. doi:10.1128/JB.06705-11
Vijay K, Brody MS, Fredlund E, Price CW (2000) A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol Microbiol 35:180–188
Voelker U, Voelker A, Haldenwang WG (1996) Reactivation of the Bacillus subtilis anti-sigma B antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol 178:5456–5463
Wang E, Bauer MC, Rogstam A et al (2008) Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol Microbiol 69:466–478. doi:10.1111/j.1365-2958.2008.06295.x
Williams TC, Blackman ER, Morrison SS et al (2014) Transcriptome sequencing reveals the virulence and environmental genetic programs of Vibrio vulnificus exposed to host and estuarine conditions. PLoS One 9:e114376. doi:10.1371/journal.pone.0114376
Wise AA, Price CW (1995) Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J Bacteriol 177:123–133
Yang X, Kang CM, Brody MS, Price CW (1996) Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev 10:2265–2275
Young JW, Locke JCW, Elowitz MB (2013) Rate of environmental change determines stress response specificity. Proc Natl Acad Sci U S A 110:4140–4145. doi:10.1073/pnas.1213060110
Zhang W, Phillips GN (2003) Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure 11:1097–1110
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Pané-Farré, J., Quin, M.B., Lewis, R.J., Marles-Wright, J. (2017). Structure and Function of the Stressosome Signalling Hub. In: Harris, J., Marles-Wright, J. (eds) Macromolecular Protein Complexes. Subcellular Biochemistry, vol 83. Springer, Cham. https://doi.org/10.1007/978-3-319-46503-6_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-46503-6_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-46501-2
Online ISBN: 978-3-319-46503-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)