Abstract
For the past decades, many efforts have been made in the fields of protein structure prediction. Among these, the protein backbone reconstruction problem (PBRP) has attracted much attention. The goal of PBRP is to reconstruct the 3D coordinates of all atoms along the protein backbone for given a target protein sequence and its C\(_{\alpha }\) coordinates. In order to improve the prediction accuracy, we attempt to refine the 3D coordinates of all backbone atoms by incorporating the state-of-the-art prediction softwares and support vector regression (SVR). We use the predicted coordinates of two excellent methods, PD2 and BBQ, as our feature candidates. Accordingly, we define more than 100 possible features. By means of the correlation analysis, we can identify several significant features deeply related to the prediction target. Then, a 5-fold cross validation is carried out to perform the experiments, in which the involved datasets range from CASP7 to CASP11. As the experimental results show, our method yields about 8 % improvement in RMSD over PD2, which is the most accurate predictor for the problem.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Proteins are required for growth and development in the human body. A protein, called a polypeptide, is composed of a chain of amino acids. A series of amino acids are linked together by peptide chains to form a protein backbone. An amino acid is the fundamental unit of a protein. There are twenty kinds of standard amino acids and each kind of amino acid can be differentiated by its R group. Each protein has its own specific functions and unique structure which can cooperate with other proteins to achieve some required functionalities. Because protein structure and protein function are closely related, in order to find out these protein functions, most biologists adopt the approaches that predict protein tertiary structures by means of amino acid sequences or other related information.
There are two main approach types of the protein structure prediction. The first one is the experimental method, including X-ray diffraction and nuclear magnetic resonance (NMR) [10, 14]. The other one is the computational method, including homology modeling [5], folding recognition [6], and ab initio [9]. As for these two categories, the former one requires a lot of time and cost while the latter one doesn’t. Therefore, this motivates us to adopt the computational method to predict the 3D protein structure.
The all-atom protein backbone reconstruction problem (PBRP) is to utilize a protein sequence and its 3D coordinates of \(\alpha \)-carbon (C\(_{\alpha }\)) for predicting the 3D coordinates of all atoms (N, C and O atoms) on the protein backbone. There are several related studies which fall into this category, such as SABBAC [11], Wang’s method [17], Chang’s method [2], BBQ [4], Chen’s method [3], Wu’s method [18], PD2 [12] and so on. For a complete survey on the methods (or software) of the protein backbone prediction, one can refer to the thesis written by Yuan [19].
In order to improve the accuracy of protein structure prediction, we propose a new method to refine the 3D coordinates of all backbone atoms by means of support vector regression (SVR) [15, 16]. Our prediction target is the differences of N and O atoms’ coordinates between the predicted results of PD2 and the real 3D coordinates of PDB, and the differences of C atoms’ coordinates between the predicted results of BBQ and the real 3D coordinates of PDB. Our training features are generated from the predicted results of PD2 and BBQ.
The experimental datasets range from CASP7 to CASP11, where CASP stands for the Critical Assessment of Protein Structure Prediction [13]. We perform a 5-fold cross validation experiment for performance evaluation. For each fold of validation, a CASP dataset is extracted for testing and the remaining CASP datasets are involved for training. Coordinates in the training datasets are first predicted by PD2 and BBQ. Then these predicted coordinates are compared with their real coordinates to produce the differences. The differences are the learning objective for the SVR. In the feature selection stage, we analyze the correlations between the objective value and available features, and then select the most representative features. To ease the training process, we partition the amino acids into twenty groups (twenty datasets) and then predict these differences by each individual SVR. Finally, we combine our predicted differences, their corresponding predicted N and O coordinates with PD2, and predicted C coordinates with BBQ to export our predicted coordinates of N, C and O. The performance is evaluated by the RMSD values. The experimental results show that our prediction results yield about \(8\,\%\) improvement over the results predicted by PD2, which is the most accurate predictor for the problem [19].
The rest of this paper is organized as follows. In Sect. 2, we will introduce experimental datasets, root-mean-square deviation (RMSD) and features used in this paper. In Sect. 3, we will describe our proposed method in detail. In Sect. 4, we will present our experimental results. Finally, in Sect. 5, the conclusion will be given.
2 Preliminaries
2.1 Datasets and Performance Evaluation
Critical Assessment of Protein Structure Prediction (CASP) [13] is an international competition held every two years since 1994. The main goal of CASP is to evaluate the capabilities of the methods for identifying three-dimensional structure of the protein from its amino acid sequence. In order to assess the performance of a method, CASP examines the predicted 3D structures in many different ways, such as the accuracy of a model, accuracy of a quaternary structure, and so on. Because our research also focuses on the 3D structure prediction, we use CASP datasets to perform our experiments.
Root-mean-square deviation (RMSD) [7, 8] is an evaluation method of molecular modeling which computes the average distance between the predicted values and the ground truths.
where \(X_{i}^{A}\) and \(X_{i}^{B}\) denote the coordinates of the ith atom on the backbone in the proteins A and B, respectively, and l denotes the length of the proteins. Generally, a lower RMSD indicates the higher similarity, which means that the predicted coordinates is close to the real ones. RMSD has been widely used in structural biology. In this paper, we also use RMSD to evaluate the quality of the prediction models.
2.2 Feature Generation and Feature Selection
This subsection describes the features we use to build the SVR models. All features are extracted within a fragment, as illustrated in Fig. 1. That is, once the prediction target, like the C atom, is determined, we define a window around this atom. We say this atom and its surrounding atoms constitute a fragment. The features required by SVR are calculated as follows.
-
Coordinate: The predicted x, y and z coordinates of N, C, O and N\(_{next}\) atoms are obtained from BBQ and PD2, denoted as \(N_x(BBQ)\), \(N_y(BBQ)\), \(C_z(PD2)\), etc. The real coordinates of two C\(_{\alpha }\)s are also involved as the features. Thus, there are totally 30 features.
-
Coordinate difference: The coordinate differences are calculated from the predicted x, y and z coordinates of N, C, O and N\(_{next}\) atoms. Only the difference of each corresponding pair of coordinates is computed, such as \(N_x(PD2)-N_x(BBQ)\), \(N_y(PD2)-N_y(BBQ)\), etc. Thus, 12 features are obtained.
-
Euclidean distance: The Euclidean distance measures the amount of space between the two predicted positions, obtained from BBQ and PD2, of the same atom on the Euclidean space. The formula for calculating Euclidean distance is given as follows.
$$\begin{aligned} d(x_{p_{i}}, x_{b_{i}}, y_{p_{i}}, y_{b_{i}}, z_{p_{i}}, z_{b_{i}}) = \sqrt{\left( x_{p_{i}}- x_{b_{i}} \right) ^{2}+\left( y_{p_{i}}- y_{b_{i}} \right) ^{2}+\left( z_{p_{i}}- z_{b_{i}} \right) ^{2}} \end{aligned}$$(2)In Eq. 2, \(x_{p_{i}}\), \(y_{p_{i}}\), \(z_{p_{i}}\) are the 3D coordinates of the ith atom on the protein backbone predicted by PD2 and \(x_{b_{i}}\), \(y_{b_{i}}\), \(z_{b_{i}}\) are the 3D coordinates of the ith atom on the protein backbone predicted by BBQ.
-
Bond length: Two adjacent atoms form a bond on the protein backbone. There are five bond lengths in a fragment, including N-C\(_{\alpha }\), C\(_{\alpha }\)-C, C=O, C-N\(_{next}\), and N\(_{next}\)-C\(_{\alpha _{next}}\). The bond length is calculated in terms of the Euclidean distance. Because bond lengths associated with BBQ and PD2 are calculated separately, it follows that 10 features are obtained.
-
Bond length difference: Each bond length difference is derived by the two corresponding bond lengths, predicted from the two methods.
-
Bond angle: Since three atoms form an angle, the bond angle can thus be obtained by law of cosines. Consequently, for a fragment with \(L_C= 6\), we can get five different angles, including N-C\(_{\alpha }\)-C, C\(_{\alpha }\)-C=O, C\(_{\alpha }\)-C-N\(_{next}\), O=C-N\(_{next}\), and C-N\(_{next}\)-C\(_{\alpha _{next}}\).
-
Bond angle difference: Each bond angle difference is calculated from the two corresponding predicted bond angles of two methods.
-
Torsion angle: The torsion angle is computed by four consecutive atoms on the main chain. In addition to \({\phi }\)(C\(_{prev}\)-N-C\(_{\alpha }\)-C), \({\psi }\)(N-C\(_{\alpha }\)-C-N) and \({\omega }\)(C\(_{\alpha } \)-C-N\(_{next}\)-C\(_{\alpha _{next}}\)), we also choose the other features in our fragment, including N-C\(_{\alpha }\)-C=O, O=C-N\(_{next}\)-C\(_{\alpha _{next}}\), C\(_{\alpha }\)-C=O-N\(_{next}\), C\(_{\alpha }\)-C-N\(_{next}\)=O (planes of C\(_{\alpha }\)-C-N\(_{next}\) and C -N\(_{next}\)=O), N\(_{next}\)-C-C\(_{\alpha }\)=O and C-N\(_{next}\)-C\(_{\alpha _{next}}\)-C\(_{next}\). Since PD2 and BBQ are used for the computation of torsion angles, 18 features are obtained.
-
Torsion angle difference: This feature is obtained from the torsion angles. We compute the differences from the torsion angles obtained by PD2 and BBQ.
So far, we have defined nine kinds of features. Table 1 shows the feature names and their sizes.
The above feature extraction method is performed around the C atom in a fragment-by-fragment manner, as shown in Fig. 1. We assign the C atom as the fragment center because this arrangement is most suitable for predicting both C and O atoms. Nevertheless, one may wonder whether this is also suitable when N atom is served as the prediction target. Therefore, we assign the N atom as the fragment center and perform another experiment. The experimental results (not shown in this paper) exhibit that the prediction accuracies of N atoms with the C-center window and those with the N-center window only show little difference. Thus, we still use the C-center window for predicting N atoms here.
Since the performance of models depends heavily on the selected features, we have to consider which one is relevant to the coordinate prediction. In order to identify important features, we calculate the Pearson’s correlation coefficient between the objective value and each feature value. For a given feature, its correlation coefficient with the objective variable is given in Eq. 3.
where n denotes the number of data elements, \(x_{i}\) denotes the ith element of data instances (\(x_{1}\), \(x_{2}\), ..., \(x_{n}\)), \(y_{i}\) denotes the ith instance of the objective values (\(y_{1}\), \(y_2\), ...,\(y_{n}\)), \(\bar{x}\) and \(\bar{y}\) represent the means of x and y, respectively.
3 The Coordinate Difference Prediction Method
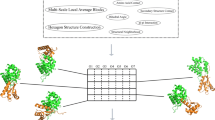
In order to improve the predicted results, we adopt SVR to predict the x-difference, y-difference and z-difference of each of N, C and O atoms on the backbone of a target protein. Then, these differences are combined with the predicted results of BBQ and PD2 to yield our predicted coordinates. Our coordinate difference prediction procedure is described as follows.
-
Algorithm: The Coordinate Difference Prediction Method.
-
Input: 1. One training set T, containing the predicted coordinates obtained from PD2 and BBQ and the real coordinates in PDB.
2. One target protein, containing the predicted coordinates of PD2 and BBQ, along with real C\(_{\alpha }\) coordinates.
-
Output: The predicted coordinates of N, C and O atoms along the target protein backbone.
-
Step 1 (Extract features): Partition the residues of all proteins in T into 20 groups, corresponding to 20 types of standard amino acids. Calculate the 103 feature values associated with each kind of residue, defined in the previous section.
-
Step 2 (Perform correlation analysis): For each of the nine objective values (\(O_x\)-difference, \(O_y\)-difference, \(O_z\)-difference, \(N_x\)-difference, etc.) in T, calculate the Pearson’s correlation coefficient between each feature value and the objective value. Since the p-value represents the confidence level associated with its correlation coefficient, we thus can adopt a thresholding method to identify significant features.
-
Step 3 (Predict the difference by SVR): For each kind of objective values and amino acid groups, we use the selected features to train an SVR model. Thus, 180 models (20 kinds of residues, 3 kinds of atoms, 3D coordinates) are obtained. Then, these models are invoked to perform prediction of the target protein based on the residue and atom types.
-
Step 4 (Combine the predicted difference with PD2/BBQ): Combine the predicted differences with their corresponding predicted positions obtained by BBQ and PD2 to generate the final coordinates.
-
Step 5 (Merge all residues together): Bring the predicted coordinates of all residues together to reconstruct the 3D positions of all atoms (N, C and O atoms) on the target protein backbone.
The flow chart is shown in Fig. 2.
4 Experimental Results
For evaluating the performance of our method, we adopt CASP7, CASP8, CASP9, CASP10 and CASP11 as the experimental datasets, which contain 65, 52, 63, 39 and 55 proteins, respectively. We use only the information of chain A of proteins to carry out the experiments. If there is no chain A, the next chain is used. All features are scaled into the range of \([a, b] = [-1, 1]\) by Eq. 4.
where \(x_{i}\) denotes the value of a certain feature of the ith training data element, \(x_{max}\) is the maximum value in the feature, \(x_{min}\) is the minimum value, a and b are the lower and upper bounds of the range, respectively.
In the 5-fold cross validation, each CASP is selected as the testing dataset for one time. Once a CASP is determined for testing, the remaining CASPs serve as the training datasets. For example, if we select CASP7 as the testing dataset, then the rest ones, from CASP8 to CASP11, are used as the training dataset. The testing procedure is performed for each CASP dataset.
As mentioned in the previous section, in our algorithm, Step 1 generates the feature candidates and Step 2 calculates correlation coefficients of these features and the objective values. From the correlation analysis, we find that some of the features are indeed significant.
Figure 3 shows the correlations of the 103 features and the \(O_x\)-differences of all 20 amino acids obtained from CASP8 to CASP11. We count the number of significances, which means that p-value (the complement of confidence level) is less than 0.05. The value of 0.05 is a widely adopted standard cut-off. It denotes that the test shows strong evidence against the null hypothesis that no correlation between the objective value and the feature value. Note that maximum count of significances is 20, because there are 20 types of amino acids. Here, if one of the following criteria is satisfied, a feature is considered as a significant one, and it is selected for training.
-
1.
The feature has correlation value greater than or equal to 0.15 and the number of its significance counts is greater than or equal to 15.
-
2.
The feature has correlation value greater than or equal to 0.4 and the number of its significance counts is greater than or equal to 10.
Furthermore, Fig. 4 illustrates the count of significances of the 103 features with respect to \(O_x\)-differences obtained from CASP8 to CASP11. According to our criteria, we can extract three significant features (indices 34, 37 and 40), which are the predicted differences \(C_x(PD2)- C_x(BBQ)\), \(O_x(PD2) - O_x(BBQ)\) and \(N_{next,x}(PD2) - N_{next,x}(BBQ)\), respectively. That is to say, these features highly correlate with our target \(O_x\)-differences. Similar results are revealed for y-coordinates and z-coordinates. All significant features over all CASPs are listed in Table 2.
Next, we use these selected features to train the SVR models. Here we adopt LibSVM [1] to perform our experiments. We use the RBF (Radial Basis Function) kernel and set the three SVR parameters (cost, tube and hyperparameter) to the default values.
The performance of our experimental results is evaluated by RMSD. Table 3 shows the RMSDs of individual N, C and O atoms of BBQ, PD2 and our 5-fold cross validation method. The improvement of O prediction over PD2 is about 7.7 %. And the improvement of C prediction over PD2 is about 13.2 %, which combines the gain from the BBQ over PD2 (about 6.4 %) and the gain from ours over BBQ (about 7.8 %). Table 4 shows the overall RMSDs of BBQ, PD2 and our method. Our method yields 8.03 % improvement over PD2.
5 Conclusion
In the past decades, lots of efforts have been devoted to the study of the protein backbone reconstruction problem. Until now, the methods, such as BBQ, PD2 and so on, have already been successfully developed and applied to the problem. Since these methods have their individual strengths and weakness, the prediction accuracy can thus be improved by taking advantage of their strengths.
In this paper, we propose an algorithm to refine the 3D coordinates of all atoms on a protein backbone with SVR. The objective values of our prediction is the differences between the predicted coordinates and the real ones. We first define a set of feature candidates extracted from the predicted coordinates of BBQ and PD2. It is well-known that the key factor to affect the prediction performance is the feature relevance. Thus, we perform the correlation analysis to identify significant features. The experimental datasets range from CASP7 to CASP11. As the experimental results show, the three most significant features for predicting the \(O_x\)-differences and \(C_x\)-differences are the differences of the predicted x-coordinates of PD2 and BBQ in C, O and N\(_{next}\) atoms. Similar results are exhibited for y-coordinates and z-coordinates. In summary, our method yields about 8 % improvement in RMSD over PD2, which is the best previous predictor in this problem up to now.
References
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst. Technol. 2(3), 1–27 (2011)
Chang, H.Y., Yang, C.B., Ann, H.Y.: Refinement on O atom positions for protein backbone prediction. In: Proceedings of the 2nd WSEAS International Conference on Biomedical Electronics and Biomedical Informatics (BEBI 2009), Moscow, Russia, pp. 99–104 (2009)
Chen, K.Y., Yang, C.B., Huang, K.S.: Prediction of protein backbone structure by preference classification with SVM. In: Proceedings of the 9th International Conference on Information Systems and Technology Management, Sao Paulo, Brazil, pp. 1193–1206 (2012)
Gront, D., Kmiecik, S., Kolinski, A.: Backbone building from quadrilaterals: a fast and accurate algorithm for protein backbone reconstruction from alpha carbon coordinates. J. Comput. Chem. 28, 1593–1597 (2007)
Holm, L., Sander, C.: Database algorithm for generating protein backbone and side-chain coordinates from a C alpha trace application to model building and detection of coordinate errors. J. Mol. Biol. 21(1), 183–194 (1991)
Eisenberg, D., Bowie, J.U., Luthy, R.: A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170 (1991)
Kabsch, W.: A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sect. A 32, 922–923 (1976)
Kabsch, W.: A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sect. A 34, 827–828 (1978)
Kazmierkiewicz, R., Liwo, A., Scheraga, H.A.: Energy-based reconstruction of a protein backbone from its \(\alpha \)-carbon trace by a Monte-Carlo method. J. Comput. Chem. 23, 715–723 (2002)
Krasnogor, N., Hart, W.E., Smith, J., Pelta, D.A.: Protein structure prediction with evolutionary algorithms. In: Proceedings of the Genetic and Evolutionary Compution Conference, Orlando, USA, pp. 1596–1601 (1999)
Maupetit, J., Gautier, R., Tuffery, P.: SABBAC: online structural alphabet-based protein backbone reconstruction from alpha-carbon trace. Nucleic Acids Res. 34, W147–W151 (2006)
Moore, B.L., Kelley, L.A., Barber, J., Murray, J., MacDonald, J.T.: High-quality protein backbone reconstruction from alpha carbons using Gaussian mixture models. J. Comput. Chem. 34, 1881–1889 (2013)
Moult, J., Fidelis, K., Kryshtafovych, A., Rost, B., Tramontano, A.: Critical assessment of methods of protein structure prediction (CASP) x Round IX. Proteins 79, 1–5 (2011)
Ruczinski, I., Kooperberg, C., Bonneau, R., Baker, D.: Distribution of beta sheets in proteins with application to structure prediction. Proteins: Struct. Funct. Genet. 48, 85–97 (2008)
Smola, A.J., Scholkopf, B.: A tutorial on support vector regression. Stat. Comput. 14, 199–222 (2004)
Vapnik, V., Golowich, S.E., Smola, A.: Support vector method for function approximation, regression estimation, and signal processing. In: Advances in Neural Information Processing Systems 9, pp. 281–287. MIT Press (1996)
Wang, J.H., Yang, C.B., Tseng, C.T.: Reconstruction of protein backbone with the \(\alpha \)-carbon coordinates. J. Inf. Sci. Eng. 26(3), 1107–1119 (2010)
Wu, H.F., Yang, C.B., Hor, C.Y., Peng, Y.H., Tseng, K.T.: Protein backbone reconstruction with tool preference classification for standard and nonstandard proteins. In: Proceedings of the 12th Conference on Information Technology and Applications in Outlying Islands, Kinmen, Taiwan, pp. 175–182 (2013)
Yuan, H.C.: A survey of computational methods for protein structure prediction. Master’s thesis, National Sun Yat-sen University, Kaohsiung, Taiwan, July 2015
Acknowledgments
This research work was partially supported by the Ministry of Science and Technology of Taiwan under contract MOST 104-2221-E-110-018-MY3.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this paper
Cite this paper
Huang, DY., Hor, CY., Yang, CB. (2016). Coordinate Refinement on All Atoms of the Protein Backbone with Support Vector Regression. In: Perner, P. (eds) Advances in Data Mining. Applications and Theoretical Aspects. ICDM 2016. Lecture Notes in Computer Science(), vol 9728. Springer, Cham. https://doi.org/10.1007/978-3-319-41561-1_16
Download citation
DOI: https://doi.org/10.1007/978-3-319-41561-1_16
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-41560-4
Online ISBN: 978-3-319-41561-1
eBook Packages: Computer ScienceComputer Science (R0)