Abstract
Eukaryotes emerged as a result of profound changes in the global biology of ancestral prokaryotes, being the outcome of large-scale evolutionary innovations at the morphological, metabolic and molecular levels. During these changes, fundamental processes sustaining life also evolved. One of them is translation, which is crucial for gene expression in all forms of life. While both mechanisms and components of the translation machinery are well known, how they evolved still remains poorly understood. In this chapter, we review recent advances in the understanding of how translation might have evolved during the origin and early evolution of eukaryotes from archaeal ancestors. The emerging view suggests that, while the fundamental process of translation remained well conserved across all forms of life, the initiation step underwent a substantial increase in evolution. This was probably due to the emergence of the nuclear membrane and split genes as well as the lack of both Shine-Dalgarno-like sequences in eukaryotic mRNAs and ribosomal protein S1. We discuss the possible evolutionary scenario where, during eukaryogenesis, mRNAs were perhaps translated in an IRES-dependent manner. It was only after IRES-dependent initiation was already established that capped and polyadenylated mRNAs, as well as novel initiation factors, were gradually and sequentially incorporated into protein synthesis, perhaps recruited from diverse cellular processes other than translation. Thus, this cap-independent initiation was later superseded in evolution by the cap-dependent mechanism.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Proteins are one of the elementary molecules of the biosphere, catalyzing the majority of reactions sustaining life, as well as playing structural, transport and regulatory roles in all living organisms. Hence, protein synthesis or “translation” is a fundamental process for all forms of life [1, 2], and translational control plays a crucial role in gene expression during many cellular and developmental processes. Accordingly, the process of translation as well as different regulatory mechanisms should have evolved ever since the beginning of life. Later in evolution, the emergence of eukaryotes represented a profound hallmark in the history of life on our planet, leading to crucial changes at the ecological, morphological, biochemical and molecular levels in living organisms. How translation ever originated and what changes the process of translation underwent during the arousal and radiation of eukaryotes is still the subject of intense debate.
The knowledge of the mechanism and regulation of translation has been established in the last 5 decades by the work of brilliant scientists from many laboratories across different countries. In recent years, the advent of the powerful “omics” era has created a huge data set regarding the molecular composition of cells from hundreds of species from many phyla never studied before, giving rise to an innovative perspective in the study of biological processes. This approach has led to the surprising discovery that a number of components of the translation apparatus have undergone diversification across eukaryotes and that distinct regulatory mechanisms have evolved in different phyla at different times [3–7]. This also has allowed performing phylogenomic analyses across the three domains of life, namely Bacteria, Archaea and Eukarya, to gain insight into how the translation machinery might have evolved during the emergence of eukaryotes. Yet, despite the impressive advances in the field of translation, many questions regarding the emergence and early evolution of translation in eukaryotes still remain open.
In this chapter, we will review recent research shedding light on the evolution of the translation apparatus during the onset of eukaryotes and how it might have evolved right afterwards. Since the elongation and termination steps, or translation, are very well conserved among all kingdoms of life, and the initiation step has undergone substantial modification in eukaryotes as compared to both Archaea and Bacteria, we will focus on the evolution of the initiation step.
2 Translation Initiation in the Prokaryotic World
In prokaryotes, translation happens simultaneously in time and space with transcription, which always synthesizes polycistronic mRNAs. Translation initiation in bacteria consists of the recruitment of the 5′ end of an mRNA by the 30S ribosomal subunit, i.e., the formation of the complex among mRNA, fMet-tRNA fMeti (initiator formylmethionyl-tRNA) and the ribosomal subunit. It is assisted by the translation initiation factors (IFs) IF1, IF2 and IF3. IF2 binds tRNA fMeti and delivers it to the P site of the ribosomal subunit 30S, and its activity is stimulated by IF1; IF3 controls the accuracy of codon-anticodon recognition. In Archaea, initiation is more complex since it possesses at least five archaeal initiation factors (aIFs): aIF1, aIF1A, eIF2, aIF5B and eIF6. aIF1 drives mRNA binding to the ribosome and also confers fidelity of the start codon selection; aIF2 binds tRNA fMeti ; aIF2 along with aIF5B delivers tRNAi to the P site; aIF6 keeps ribosomal subunits dissociated. So far, no role has been found for aIF1A [8–10].
John Shine and Lynn Dalgarno discovered in the early 1970s that mRNA recruitment to the ribosome occurs by a direct base pairing between a purine-rich region located ~7 nucleotides upstream the mRNA start codon, the so-called Shine-Dalgarno (SD) sequence with the consensus AGGAGG, and a complementary sequence at the 3′ end of the 16S rRNA on the 30S small ribosomal subunit (referred to as anti-Shine-Dalgarno sequence, anti-SD) [11, 12]. The critical role of the SD sequence in translation initiation was further experimentally corroborated in a variety of species, both eubacterial and archaeal [10, 13–16]. This, together with the large number of genes possessing SD sequences in many bacteria, led to the general idea that for prokaryotic mRNAs the SD sequence is the essential (although not necessarily the sole) element for ribosome recruitment and for selection of the correct initiation codon. It was then assumed that the SD/anti-SD interaction during initiation is conserved in all prokaryotes [9, 10, 17].
Besides the SD motif, it was later found that ribosomal protein (RP) S1 interacts with the 3′ end of the 16S rRNA, particularly with helices h26 and h45, which contains the anti-SD sequence, as well as with 11 nucleotides of the mRNA 5′-UTR located immediately upstream of the SD sequence. Thus, it is thought that the major function of RPS1 is to bring the mRNA onto the 30S subunit during translation initiation, thereby assisting the interactions between the SD motif in the mRNA and the anti-SD sequence of 16S rRNA. This is consistent with the observation that translation of leaderless mRNAs does not require RPS1, since it does not depend on SD interaction [18–23].
The recent advent of genome- and transcriptome-wide studies of thousands of species led to the discovery in the past few years of a significant number of naturally occurring mRNAs lacking an SD sequence spread across a wide variety of eubacterial and archaeal phyla, being more abundant in Archaea. These include thousands of mRNAs devoid of 5′-UTR (and hence referred to as “leaderless” mRNAs) produced from single genes and from the first genes of operons, as well as mRNAs that possess a 5′-UTR but lack an SD sequence [8, 9, 24–43]. These findings were further confirmed in a more recent study comprising 2,458 bacterial complete genomes [44]. Indeed, several studies have shown that the major pathway to initiate translation initiation in Archaea might involve mostly leaderless mRNAs [29, 32–35].
Thus, in addition to the aforementioned SD/anti-SD-dependent initiation, two other major mechanisms for prokaryotic translation initiation have been described. (1) For leaderless mRNAs the AUG start codon itself was found to serve as the most important signal for ribosome recruitment and translation initiation. Here, the initiator tRNA and IF2 are critical for complex formation between the start codon and the ribosome. It is noteworthy that translation initiation of leaderless mRNAs involves the undissociated ribosome 70S instead of the 30S ribosomal subunit [8, 9, 24, 31, 40, 41, 45–52]. (2) For mRNAs possessing a 5′-UTR but lacking an SD motif, mRNA recruitment into the ribosome can be mediated exclusively by RPS1 [18–23, 30, 33, 53]. These mRNAs exhibiting a pronounced minimum in secondary structure and AUG start codon reside in single-stranded regions of the mRNAs, and ribosome binding to the mRNA is a sequence-independent event but is strictly dependent on the local absence of secondary RNA [54].
Intriguingly, neither archaebacteria nor eukaryotes contain an RPS1 gene, raising the question of how leadered mRNAs devoid of an SD motif are translated in Archaea [8, 17, 18, 30, 51, 55]. Finally, in different species alternate non-SD sequences have been reported to mediate 16S rRNA-mRNA interaction in a variety of prokaryotic mRNAs, including domain #17 of E. coli 16S rRNA [56], the “translation initiation region” sequence of the Mycoplasma genitalium tuf gene [57] and a region of Thermus thermophilus thrS gene mRNA [58]. Moreover, genomic studies of several archaeal species found a strong conservation of a GGTG atypical putative ribosome binding site within 15 nucleotides upstream of the start codon of hundreds of genes [26, 36]. Yet, whether or not these sequences undergo base-pairing with the anti-SD sequence of 16S rRNA is unknown. The biological relevance of these sequences, if any, remains poorly understood.
Overall, the emerging view is that in the prokaryotic world, both SD-dependent and -independent translation mechanisms are present in all major lineages, showing that the 16S rRNA recruitment by prokaryotic mRNAs is a variable process. Since elongation and termination steps of translation are highly conserved in prokaryotes, these findings support the hypothesis that the last universal common ancestor (hereafter termed LUCA) of extant life already possessed an established fundamental translational apparatus, but the mechanisms of initiation further evolved in the bacterial and archaeal lineages and afterwards changed in eukaryotes even more [3, 4, 30, 33, 43, 59–61]. Indeed, evidence suggests that a highly developed translation system was a necessary condition for the emergence of cells on earth [62]. However, the aforementioned variety of prokaryotic mechanisms raises the question of how LUCA might have initiated translation.
2.1 mRNA Recruitment in the Last Universal Common Ancestor of Extant Organisms
As mentioned above, ribosome recruitment by mRNAs is a variable process in prokaryotes. Thus, a crucial question is whether or not all prokaryotes possess an anti-SD sequence at the 3′ end of 16S rRNA on the 30S ribosomal subunit. Complete genome analyses of 277 [30] and 162 [29] prokaryote species (both bacterial and archaeal) and 18 archaeal species [26] surprisingly found that the anti-SD sequence is highly conserved among all species analyzed. Since thousands of prokaryotic mRNAs have been found to lack either an SD motif or a leader sequence, this paradox could be explained by three alternative evolutionary scenarios, i.e., (1) LUCA mRNAs possessed SD sequences at the 5′-UTR of mRNAs, but were lost multiple and independent times in different prokaryotic lineage [29, 30]. For them, RPS1-mediated or leaderless mRNA mechanisms of translation initiation work to a great extent [29, 30]. In this case, the evolutionary pressures that led to the loss of SD sequences, if any, are completely unknown. (2) Only some organisms have possessed SD motifs ever since the beginning of life, opening the possibility that the anti-SD motif present in 16S rRNA from many species might play more, yet-unidentified roles in translation or even in different process, such as ribosomal RNA biogenesis, export or stability. (3) One or more hypothetical sequences, other than the SD motif, might have driven translation initiation in LUCA mRNAs and are currently present in different prokaryotic mRNAs but have not been identified. This idea is supported by the finding that a variety of alternate sequences support base-pairing between the 16S rRNA and mRNA to drive translation initiation [56, 58, 63]. We may conclude that the current knowledge does not shed light on what the mechanism of ribosomal recruitment by LUCA mRNAs might have been.
3 Translation Initiation in Modern Eukaryotes
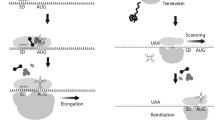
In modern eukaryotes, the vast majority of mRNAs initiate translation by the so-called cap-dependent mechanism, which is mediated by the eukaryotic initiation factors (eIFs) and consists of the recruitment of the mRNA to the 40S ribosomal subunit upon recognition of the cap structure (m7GpppN, where N is the nucleotide located at the very 5′ end of the mRNA) by the cap-binding protein eIF4E.
It begins with the dissociation of the ribosomal subunits 60S and 40S by IF6. Afterward, the free 40S ribosomal subunit, which is stabilized by eIF3, eIF1 and eIF1A, binds to a ternary complex—consisting of eIF2 bound to an initiator Met-tRNA Meti and GTP (eIF2-GTP/Met-tRNA Meti )—to form a 43S pre-initiation complex. eIF5 interacts with eIF2 and eIF3 and is probably also recruited to the 40S ribosomal subunit. On the other hand, and most likely simultaneously, the cap structure of an mRNA is recognized by eIF4E in complex with the scaffold eIF4G. Then, recruitment of the mRNA 5′-UTR by the 43S pre-initiation complex happens, a process that is coordinated by eIF4G via its interaction with eIF4E, the ATPase/RNA helicase eIF4A, the poly (A)-binding protein (PABP) and the 40S ribosomal subunit-associated eIF3. eIF4G-PABP interaction causes a crosstalk between both mRNA ends, hence prompting mRNA circularization, a spatial conformation that stimulates translation and that is known as the closed-loop model. This complex scans in a 5′ ➜ 3′ direction along the 5′-UTR to reach the start codon, usually an AUG. During the scanning (a process that requires ATP), eIF4B stimulates the activity of eIF4A, which unwinds secondary RNA structures in mRNA. eIF1, eIF1A and eIF5 assist in the positioning and fidelity of the 40S ribosomal subunit at the correct start codon so that eIF2 can deliver the anti-codon of the initiator Met-tRNA Meti as the cognate partner for the start codon directly to the peptidyl-site of the 40S ribosomal subunit. Once the ribosomal subunit is placed on the correct start codon a 48S pre-initiation complex is formed. Then eIF5 promotes GTP hydrolysis by eIF2 and the release of the initiation factors. eIF2B and GTP afterward recycle the dissociated eIF2-GDP complex so that it can associate with a new Met-tRNA Meti and take part in a new round of initiation. Finally, the GTPase eIF5B is required for the assembling of the 60S ribosomal subunit to the 48S complex to form an 80S initiation complex. Thereafter, the polypeptide elongation begins [64–66].
In the late 1980s, the groups of Nahum Sonenberg and Eckard Wimmer independently discovered that there is an internal sequence in the 5′-UTR of picornaviral mRNAs located in the proximity of the start codon that allows the 40S ribosomal subunit to land directly on the mRNA in a cap-independent manner and without involvement of eIF4E [67, 68]. This sequence is termed an internal ribosome entry site (IRES). A few years later, the first cellular IRES was discovered in the mRNA of the immunoglobulin heavy-chain binding protein [69], an mRNA that is translated upon poliovirus infection. Since that time a large number of cellular and viral mRNAs have been found translated via different IRESs elements [17, 70–73]. In the following, we will analyze the evolutionary phenomena that might have spurred the emergence of present-day translation in eukaryotes.
4 The Emergence of Eukaryotic Translation
About 1.8 billon years ago, the endosymbiotic association of respiratory, alpha-proteobacterium-like prokaryotes (the ancestors of the mitochondria) with host organisms that possessed an archaeal genetic identity led to the emergence of eukaryotes. The posterior association of this cellular consortium with photosynthetic, cyanobacterium-like endosymbionts led to the evolution of plastids [74–77]. The onset of eukaryotes caused the emergence of novel, much more sophisticated levels of cellular architecture than their prokaryotic ancestors, resulting from the appearance of a plethora of new cell features, including a nucleus and centrioles, as well as endosymbiotic bacteria that evolved toward mitochondria and plastids; peroxisomes, Golgi complex and endoplasmic reticulum; the rearrangement of genetic information in a “fragmented” fashion (i.e., interrupted genes) and packed into multiple linear chromosomes inside the nucleus; cilia; cytoskeleton and motors for vesicle and molecules transportation; sex, mitosis- and meiosis-based cell division; expansion of genome size; expansion of cell size; and in many phyla the emergence of multicellularity resulting out of different developmental programs.
Interestingly, despite the well-established idea that eukaryotes evolved from archaeal ancestors [74, 77–83], phylogenomic analyses have shown different roots for the cellular components of eukaryotes. While they inherited from Archaea the informational machineries, namely replication, transcription and translation, the metabolic and energetic enzymes are mostly of bacterial origin [60, 74, 82]. Consistent with this notion, genome-based phylogenetic analyses as well as structural and biochemical studies have shown that archaeal translation factors [8, 9, 31, 59, 84, 85], ribosomal proteins [18, 86–88], aminoacyl-tRNA synthetases (despite extensive horizontal gene transfer undergone among the three domains of life) [89, 90] and ribosomal RNAs [55, 62, 74, 78–80, 91, 92] have their closest homologs in eukaryotes rather than in bacteria.
The evolutionary emergences of the nucleus and interrupted genes were paramount events of eukaryote genesis. Crucially, they caused the interruption of genetic information of host cells and led to the spatio-temporal separation of transcription and translation. Therefore, upon their emergence eukaryotes needed the prompt evolution of nuclear machineries for intron splicing, for nucleocytoplasmic export and for mRNA protection to ensure that transcripts synthesized in the nucleus reach both the ribosomes and the storage bodies in the cytoplasm. Surveillance systems such as nonsense-mediated mRNA decay (NMD) also evolved to discard aberrant mRNAs [3, 4, 93, 94]. The arousal of eukaryotic cells also led to the evolution of novel features in the translation apparatus, mechanisms and regulation so that gene expression could take place. The major changes summarized are the following.
(1) Eukaryotic ribosomes are much bigger and more complex than their prokaryotic counterparts. Ribosomes evolved toward the eukaryotic 40S and 60S ribosomal subunits from prokaryotic 30S and 50S, respectively. This was due to the addition of several rRNA expansion segments, peptide additions to most ribosomal proteins, as well as the addition of extra eukaryotic-specific ribosomal proteins and the 5.8S rRNA. Thus, while bacterial 70S ribosomes contain ~4500 nucleotides of rRNA, eukaryotic 80S ribosomes contain >5500 nucleotides of rRNA [88, 95–98]. The number of ribosomal proteins increased from 57 (23 in the small ribosomal subunit and 34 in the large subunit) in Bacteria and 68 (28;40) in Archaea to 78 (32;46) in Eukarya [18, 55, 86, 97].
(2) The initiation step of translation underwent a substantial increase in terms of the complexity and number of initiation factors as compared to prokaryotes, i.e., while in Bacteria and Archaea it is assisted by 3 and 6 factors, respectively, eukaryotes need the interplay of at least 14 factors. Thus, novel, eukaryotic-specific initiation factors evolved, namely eIF3 (all subunits), eIF4B, eIF4E, eIF4G, eIF4H and eIF5 [8, 9, 14, 31, 59, 85]. Except for eIF5, all of them recognize the mRNA 5′-UTR for recruitment into the ribosome.
(3) mRNAs also underwent profound changes during the transition from prokaryotic to eukaryotic cells. (a) They acquired a novel structure, i.e., monocistronic, capped, polyadenylated and with long UTRs. Moreover, eukaryotic mRNA 5′-UTRs are devoid of an SD motif and, for some lineages, the AUG start codon is surrounded by a context sequence instead. For vertebrate mRNAs, the optimal context is termed the “Kozak motif,” which consists of the consensus sequence G/AXXATGG [99]. Experimental and in silico studies of a few mRNAs from some species suggest that this sequence is not conserved across eukaryotes [100–106]. Recently, a genome-wide in silico analysis of 48 species found that the preferred sequence around the start codon significantly varies across species of all eukaryotic kingdoms [107]. However, no experimental validation of this observation has been performed. (b) They acquired a novel life cycle, being transcribed, capped, polyadenylated, spliced and exported from the nucleus, and further stored, transported, translated and degraded in the cytoplasm. And (c) They acquired a novel functional conformation when engaged in translation, i.e., a circular shape displaying a functional crosstalk between both the 5′- and 3′-ends [4, 5, 93, 108].
(4) New mechanisms for translation regulation evolved in different lineages, including a plethora of eIF4E-interacting proteins (4E-IPs), the TOR pathway, microRNAs, different cytoplasmic granules, eIF2alpha kinases and the control of mRNA circularization by poly(A) tail shortening, among others [4–8, 31, 59].
4.1 A Closer Look at the Untranslated Regions of Eukaryotic mRNAs
Among the key features that evolved in eukaryotic mRNAs are the UTRs, as mRNA stability, transport and translation rates are tightly controlled by cis-acting elements located on them. Indeed, both 5′- and 3′-UTRs are critical targets of different networks of trans-acting factors for finely tuning gene expression at different levels. Notwithstanding, there are remarkable functional differences between both UTRs, as most cis-acting regulatory elements regulating mRNA polyadenylation, degradation, storage, localization and transport of mRNAs are localized at the 3′-UTR. In contrast, 5′-UTR is key for ribosomal landing, scanning and binding of diverse RNA-binding proteins regulating scanning and ATG codon recognition during translation initiation [66, 93, 108–119]. As a consequence of this, while the mean length of 5′-UTR remains remarkably constant in most eukaryotic phyla (70 − 200 nucleotides), 3′-UTR mean length increases as morphological complexity increases [93, 108, 109, 112, 120–125].
According with the crucial roles UTRs play in post-transcriptional regulation of gene expression, leaderless mRNAs are rather seldom in eukaryotes [93, 108, 109, 114, 120–123], and extremely short 5′-UTRs have been reported only among mRNAs from the unicellular protists Giardia lamblia, with 5′-UTRs in the range of 0–14 nucleotides [126], and Entamoeba histolytica, which possesses some 5′-UTRs as short as 5 nucleotides [127]. However, this feature could be due to their parasitic life. Recently, some human mRNAs have also been reported to contain short 5′-UTRs with a median length of 12 nucleotides within a translational element termed TISU (which stands for translation initiator of short 5′-UTR) [128–130]. Yet, the frequency of this element in other species remains to be determined.
5 The Transition from Prokaryotic to Eukaryotic Translation
Several evolutionary forces played crucial roles in the transition from the ancestral, prokaryotic mode of translation toward the establishment of the predominant cap-dependent translation of eukaryotes. It is well established that the last common ancestor of extant eukaryotes had a genome with a high intron density, most likely as a result of an invasion of group II introns from the new mitochondrial endosymbionts into the genes of the host organism [94, 131–134]. The emergences of the nuclear membrane and interrupted genes were probably some of the primordial selection forces to overcome in the first eukaryotes [60, 94], raising the immediate need for developing systems for the protection and nucleocytoplasmic export of mRNAs, for intron splicing and for the removal of aberrant transcripts.
Moreover, because eukaryotic mRNAs lack both SD sequences and RPS1 protein, they cannot efficiently recruit the ribosome directly to the initiation codon. Most probably this was the most important selection pressure that led early eukaryotes to develop a novel mechanism to ensure the correct landing of the ribosome at the 5′-end of mRNAs, i.e., the cap-dependent initiation. These events led to the stepwise increase in sophistication during eukaryogenesis. Hence, although eukaryotes inherited from their archaeal ancestors eIF1, eIF1A, eIF2 (all subunits), eIF2B (but only the alpha, beta and delta and not the gamma or epsilon subunits), eIF4A, eIF5B and eIF6 [9, 59, 84, 85, 135–137], eIF3, eIF4G, eIF4E, eIF4B and PABP evolved exclusively in eukaryotes because of the need to recruit capped and polyadenylated transcripts possessing long 5′-UTRs devoid of SD sequences [3, 4, 6, 9, 14, 59, 84, 85]. Thus, the crucial question arises of how ribosomes from early eukaryotes might have recruited mRNAs to initiate translation in the absence of both eIF4 factors and PABP.
5.1 What Was the Mechanism of mRNA Recruiting in the Early Eukaryotes?
Phylogenomic analyses have recently shown that eukaryotes emerged from the so-called TACK superphylum within the Archaea domain, which comprises the Thaum-, Aig-, Cren- and Korarcaeota groups of archaea [74, 77, 81–83]. This means that the closest relative of the eukaryotic lineage is among the species of the superphylum TACK. Therefore, a close look to the mRNA structure of these lineages might shed light on the type of mRNA (i.e., SD-containing, leaderless or possessing a 5′-UTR devoid of SD motif) the first eukaryotes might have possessed. However, the current knowledge does not allow elucidating what species of the superphylum TACK eukaryotes evolved from, as well as what type of mRNA these particular species use.
Although genome-wide studies of hundreds of species have shown that the major pathway to initiate translation in Archaea might involve mostly leaderless mRNAs [26, 27, 29, 30, 33–37, 42], early eukaryotes might have synthesized transcripts possessing long 5′-UTRs devoid of the SD motif as happens in virtually all present-day eukaryotes. Based on this notion, and given the fact that archaea and eukaryotes lack an RPS1 gene involved in recruitment of bacterial leadered mRNAs devoid of an SD motif, here we propose three possible mechanisms for mRNA recruitment by ribosome in early eukaryotes.
(1) mRNAs used a variety of non-SD sequences that interacted with different internal regions of the rRNAs on the 30S ribosomal subunit. This idea is supported by evidence proving sequence complementarity and interaction between hundreds of mRNAs and different segments of the 18S and 28S rRNA from different eukaryote species with a potential role in translation regulation [138–141]. Indeed, RPS1 is also missing in some bacterial lineages, which led G.E. Fox to suggest that RPS1 was added to bacterial ribosomes only after the Archaea-Bacteria divergence happened [55].
(2) Alternate ribosomal proteins might have been responsible for mRNA recruitment. Eukaryotes evolved a whole set of novel, eukaryote-specific ribosomal proteins [18, 86–88], making conceivable that some of them might have evolved because of enhanced mRNA recruitment. For example, ribosomal proteins such as RPS5 and RPS15, which along with eIF2α contact mRNA positions −3 and +4 of the AUG context sequence [142], might have been involved in driving mRNA recruitment in early eukaryotes.
(3) Existing initiation factors promoted mRNA recruitment. RPS1 contains six copies of an RNA-binding fragment that is known as the S1 domain. Many proteins possess one or more S1 domains, including the translation initiation factor IF1 and its eukaryotic equivalent eIF1A, as well as the eukaryotic eIF2α [18, 55]. Since the S1 motif is found in all three domains of life and factors IF-1/eIF1A are universally distributed, Fox [55] has suggested that IF-1/eIF1A might be the original source of the S1 motif, possibly derived from the initiation machinery. Thus, in the absence of the RSP1 and SD motif, eIF1A or eIF2α might have been involved in mRNA recruitment in early eukaryotes.
6 The Natural History of the Cap Structure, eIF4s and PABP Sheds Light on the Evolution of the Cap-Dependent Translation
Because present-day cap-dependent translation is a highly sophisticated process, it cannot have appeared fully formed, but arose by stepwise addition of components and regulatory steps. So, what were the possible mechanisms underlying evolution of translation initiation in early eukaryotes? As with all evolutionary studies, we can infer the ancient nature of any current biological process by studying its present-day components and looking at their “ancestral” features. Here we think that the contemporary features of the cap structure, eIF4G, eIF4E eIF3 and PABP, all of them of eukaryotic origin, as well as the more ancient eIF4A, shed light on the evolutionary history of eukaryotic translation initiation. Analysis of these molecules argues for a stepwise addition of factors into the initiation step of translation by a mechanism of molecular tinkering [143], i.e., by recruiting more ancient components from other, already present cellular processes to perform a novel function into translation initiation.
Francoise Jacob first proposed the concept of “molecular tinkering” 40 years ago to explain one of the most creative forces of evolution, i.e., transforming a feature that evolved to perform a specific function to give it new functions [143]. This concept was afterwards applied by Gould and Vrba in 1982 to the evolution of morphological features that now enhance fitness but were not built by natural selection for their current role. For them, a morphological feature or structure, previously shaped by natural selection for a specific function (an adaptation), but later utilized for a new use is called a “exaptation” [144]. In the following, we analyze current features of different molecules to infer their evolutionary history and, finally, reconstruct the whole evolutionary history of the translation initiation in eukaryotes. The evidence supports the notion that some of the eukaryotic initiation factors are indeed molecular exaptations.
6.1 Origin of the Cap Structure of mRNAs
The m7GpppN cap structure of eukaryotic mRNAs plays a crucial role in mRNA biogenesis and stability. It is essential for efficient splicing, mRNA export and translation. Interestingly, all nuclear processes of mRNA biogenesis (namely transcription, capping, polyadenylation, splicing nuclear export and stability) are tightly intertwined [145–152]. During transcription, which is performed by RNA polymerase II (Pol II), the cap addition is the first modification that occurs to all eukaryotic pre-mRNAs. It is co-transcriptionally added after 20–30 nucleotides have been polymerized in virtue of the interaction of the capping enzymes with the carboxyl-terminal domain (CTD) of the largest subunit of Pol II. Once transcription reaches the transcript’s end, a signal triggers polyadenylation of the pre-mRNA by the poly(A) polymerase and, right after the transcript is released, a process that is dependent on the presence of Pol II CTD. The synthesized transcript is then recognized by the nuclear cap-binding protein CBP20 in complex with CBP80 (forming the so-called nuclear CBC) for both intron splicing and nucleo-cytoplasm export to happen. Upon phosphorylation, Pol II CTD enhances the overall rate of splicing.
The extensive coupling of all process for mRNA biogenesis [145–149, 151, 152], the finding that the cap structure is recognized by many proteins belonging to different processes of RNA metabolism [149, 153] and the discovery of a strong dependence of most of mRNA degradation pathways on the cap structure (namely AU-rich element decay, bulk 5′–3′ decay, NMD, miRNA-mediated decay, and deadenylation-mediated mRNA decay) [147–149] support the hypothesis that the cap structure has been involved in different aspects of RNA metabolism ever since eukaryotes originated. It also supports the idea that among the very first components and processes that appeared in eukaryotes were the Pol II CTD, the cap structure and the CBC to provide a “platform” to assemble the splicing, nuclear export, mRNA protection and NMD machineries [3, 4]. Thus, both the cap and poly(A) tail of mRNAs might have played no role in translation during eukaryogenesis, being incorporated into the translation process later in evolution only after eIF4E, eIF4G and PABP had evolved.
6.2 Origin of Eukaryotic Initiation Factors 3, 4G and 4E
The scaffold eIF3 is the largest and functionally most complex of initiation factors, with a composition across eukaryotes from 6 to 13 different subunits. Among its activities, eIF3 binds to and coordinates the interaction between eIF4G and the 40S ribosomal subunit, thereby enhancing most of the reactions of the translation initiation pathway [154–157]. Structural and sequence studies have shown that eIF3, the ‘lid’ subcomplex of the 26S proteasome (involved in protein degradation) and the COP9 signalosome or CSN complex (involved in the ubiquitin-proteasome pathway, DNA-damage response and cell cycle control) share a similar architecture composed of multiple subunits possessing the PCI domain (Proteasome, CSN, eIF3) [156–161]. Since PCI proteins are crucial scaffolds for the assembly of multiprotein complexes, these observations support the hypothesis that an ancestral core of eIF3 evolved from a versatile PCI-containing multimeric complex involved in different cellular processes other than translation. The finding that some eIF3 subunits also play roles not related to translation, such as the cell cycle, apoptosis, protein turnover, mRNA deadenylation or decay, 20S pre-rRNA processing or NMD [158–161], supports this hypothesis. Thus, an ancestral, multisubunit eIF3 was perhaps a scaffold that gradually incorporated additional subunits from other cellular machineries and was incorporated into translation initiation later in evolution because it improved the efficiency and regulation of the mRNA recruitment [4].
eIF4G is a scaffold, modular protein that possess binding sites for different proteins involved in translation initiation, such as PABP, eIF4E, eIF3 and eIF4A. The C-terminal third of all eIF4Gs contains of one, two or three consecutive α-helical domains called HEAT (Huntington, Elongation factor 3, A subunit of protein phosphatase 2A, and Target of rapamycin) [162, 163]. Homologs of the HEAT domain named HEAT-1 [162] also exist in Upf2/NMD2, a component of the NMD system, and in CBP80, indicating that they may have evolved from a common ancestral protein [85, 162, 164, 165]. For instance, the consecutive HEAT-1, HEAT-2 and HEAT-3 domains of eIF4G are present in CBP80 as well, meaning that both proteins descended from an ancestor protein that already contained the three consecutive HEAT domains [162].
HEAT-containing proteins participate in a wide variety of cellular processes that are dependent on assembling large multiprotein complexes [166, 167]. HEAT domains are part of central adapters driving processes such as mRNA processing, translation and degradation [85]. Since the complexes eIF4F, NMD and nuclear CBC each include a HEAT-1-containing protein (eIF4G, Upf2/NMD and CBP80, respectively) [164], it has been suggested that early in eukaryotic evolution a versatile ancestral protein containing the HEAT-1 domain served as an adapter in different RNA processes that subsequently diverged and evolved toward distinct binding specificities [85, 162, 164, 165]. Therefore, this protein may have first appeared in the nucleus as a proto-CBP80 to provide, together with the cap, a “platform” for splicing factors and for mRNA protection during nuclear export. Later in evolution, it might have diverged in the cytoplasm into the Upf2/NMD2 when NMD evolved, and also into a proto-eIF4G, a scaffold that facilitated a more efficient initiation of translation by bringing the mRNAs into the close proximity of the ribosomes. Therefore, and similar to eIF3, these features suggest that eIF4G might have appeared in early eukaryotes for functions different from in translation and that it was incorporated into this process later in evolution because it also conferred a better efficiency of mRNA recruitment [3, 4, 85, 162, 164, 165]. Cap-dependent initiation of translation could only then have evolved after sites to bind eIF4E, PABP, eIF4A and eIF3 appeared in the proto-eIF4G [3, 4].
eIF4E has long been known to play its main role in translation initiation through cap recognition [168] and is also of eukaryotic origin [85]. Interestingly, eIF4E is found being part of different cytoplasmic granules where it is involved in mRNA decay or storage [169, 170]. In addition, a fraction of this protein localizes inside the nucleus in several eukaryotes where it mediates the export of certain mRNAs to the cytoplasm [171–173]. These findings suggest that eIF4E is versatile enough to utilize the features required for cap-binding activity in different cellular processes [4, 171].
Although most probably eIF4E emerged as a translation factor, it has been discussed that other evolutionary scenarios are also possible [4, 171]. For instance, it may first have appeared in early eukaryotes either as a mediator of nuclear export, thus enhancing mRNA stability, or as a mediator of cytoplasmic storage of mRNAs, but playing no role in translation [4, 171]. An example of this possible scenario is provided by one of the eIF4E isoforms from Giardia lamblia, as it binds only to nuclear noncoding small RNAs and plays no role in translation [174]. The findings that the cap and eIF4E confer stability to mRNAs by protecting them from 5′ exonucleases and decapping enzymes [175] suggest that the appearance of both the cap and eIF4E could have been a big evolutionary leap by protecting mRNAs from degradation. Since 5′ exoribonucleases emerged after eukaryogenesis [176], and the enzymes for the for capping of mRNAs, namely 5′ triphosphatase, guanylyltransferase and guanine-N7-methyl-transferase are of eukaryotic origin [177, 178], it was suggested that the 5′ exoribonucleases evolved in early eukaryotes following the emergence of mRNA capping for cell protection from RNA viruses or viroids [177]. The appearance of eIF4E could have followed this evolution by further increasing mRNA stability, since in the absence of any means of interacting directly with the ribosome itself, it could not be involved in translation. eIF4E should have been incorporated into the translation process only after a scaffold protein emerged (namely eIF4G), able to coordinate eIF4E activity. Because the absence of eIF4E precludes the existence of the cap-dependent translation, the emergence of the ancestral eIF4E implies that its own mRNA was most likely translated in a cap-independent, IRES-dependent manner [3, 4, 171].
6.3 Origin of PABP and the Evolution of mRNA Circularization
PABPs are scaffold proteins of eukaryotic origin that evolved into two main families, nuclear and cytoplasmic. They interact with many proteins and participate in different events of mRNA biogenesis both inside the nucleus and in the cytoplasm. In the nucleus, PABPs play essential roles in mRNA polyadenylation and stability, and they may be involved in the mRNA shuttle to the cytoplasm. In the cytoplasm, PABPs either protect mRNAs from decay or trigger transcript decay by promoting mRNA interactions with deadenylase complex proteins. By interacting with eIF4G, PABP also promotes circularization of the mRNA, a conformation that is critical for translation initiation since it provides an effective means for the protein synthesis apparatus to selectively translate only intact mRNAs, i.e., those that harbor both a cap and a poly(A) tail. In addition, translation termination happens at a ‘correct’ stop codon, as opposed to a premature termination codon, only if the ribosome is close enough to the poly(A) tail. The signal indicating this proximity is the interaction of the terminating ribosome with PABP. In the absence of this signal, upframeshift protein (UPF) 1 binds eukaryotic releasing factors (eRF) 1 and 3 in the terminating ribosome, triggering NMD. Finally, PABPs also play a role in mRNA transport and localization [179–184].
PABPs interact with poly(A) tails via their RNA-recognition motifs (RRMs). These are present in one to four repeats plus a carboxy-terminal domain (CTD) that interacts with factors regulating translation initiation and termination, polyadenylation and deadenylation [182]. The RRM is the most prevalent eukaryotic RNA-binding domain and is involved in all aspects of RNA metabolism. This is an ancient and versatile RNA-binding domain present in all eukaryotes and many bacteria [185]. RRM-containing proteins, including PABPs, evolved from successive duplications of a single RRM-carrying gene with the addition of auxiliary motifs during their diversification in eukaryotes [185].
Hernández has proposed that the poly(A) tail and an PABP first arose in early eukaryotes as part of the primary adaptive responses to the emergence of nuclear membrane and split genes, but initially they might have had no role in translation [4, 6]. Afterwards, mutations in PABP that allowed binding to eIF4G, thereby promoting mRNA circularization, underwent a strong positive selection because they (1) increased mRNA stability, (2) ensured a more efficient recruitment of the 40S ribosomal subunit by the mRNA and (3) mRNA circularity represents a checkpoint that determines to initiate translation only in intact mRNAs [4, 6].
6.4 Origin of eIF4A and the Evolution of the Scanning Process
Sequence comparison and biochemical analyses show that eIF4A is the most ancient of eIF4 factors. Orthologs are found in Archaea [9, 14, 59, 85, 186], Bacteria [187–189] and Eukaryotes [171, 186, 190–193], indicating that eIF4A evolved before eukaryotes appeared. eIF4A belongs to the extensive DEAD-box family of RNA proteins, which is a wide and versatile family of ATP-dependent RNA helicases that exists across all phyla of Bacteria, Archaea and Eukaryotes and that is involved in many aspects of RNA metabolism, including translation, nonsense-mediated mRNA decay (NMD), splicing, RNA transport and ribosomal biogenesis [190–192]. This indicates that RNA unwinding by RNA helicases already existed before the eukaryotes appeared and that eIF4A evolved from RNA helicases already present in the archaeal ancestor of eukaryotes [3, 4, 6].
Eukaryotes possess mRNAs with long 5′-UTRs with energetically stable secondary structures that would prevent scanning and hence translation. Therefore, the translation machinery requires RNA helicases to unwind these structures. In contrast to bacterial ribosomes, which possess intrinsic mRNA helicase activity [194], in eukaryotes RNA unwinding is mainly performed by eIF4A. Remarkably, other RNA helicases belonging to the asp-glu-ala-asp (DEAD)-box or DEAD/asp-glu-x-his (DExH)-box families also stimulate or repress translation by performing RNA unwinding during different steps of translation initiation, including the scanning step. This is the case of helicases DDX3/Ded1, Dhh1/RCK, VASA/DDX4, RHA/DHX9 and DHX29. Interestingly, these helicases also play various roles in different processes of RNA metabolism other than translation, such as RNA export and pre-mRNA splicing and transport [190, 192, 193]. Since the RNA helicases are a family of proteins that participate in many processes of RNA metabolism in both the nucleus and the cytoplasm [191], it has been proposed that the early eukaryotic RNA helicases were versatile proteins with broad substrate specificities involved in different RNA processes, and this probably included translation initiation. Later in evolution, they diversified into more specific enzymes, some of them specializing in translation [3, 4, 171]. The finding that eIF4A-III, a highly related eIF4A-cognate, participates in NMD, RNA splicing and mRNA localization, but not in translation [192], supports this hypothesis.
Thus, an evolutionary scenario is possible where a proto-eIF4A with broad substrate specificity might have existed, performing its function in diverse aspects of the RNA metabolism, from which it was afterwards incorporated into translation. Crucially, the evolution of eIF4G and the incorporation of diverse RNA helicases, including a proto-eIF4A, into translation initiation allowed both the incorporation of eIF4E and the establishment of the scanning process in the translation mechanism. These events enabled the translation machinery to efficiently translate mRNAs with more complex 5′-UTRs, resulting in the current widespread cap-dependent translation initiation mechanism.
7 A Timeline for the Emergence of the Cap-Dependent Translation Initiation
We can summarize the evidence discussed above and outline a brief timeline hypothesis on the origin and early evolution of the cap-dependent translation initiation in early eukaryotes. Overall, the evidence discussed in this chapter supports the notion that molecular tinkering [143] has played a crucial role underlying the establishment of the cap-dependent initiation of translation, i.e., by gradually recruiting into translation more ancient, already existing molecules involved in different cellular processes. This notion is supported by the current existence of a diversity of viruses performing translation with a wide variety of requirements of the translation factors that, indeed, might represent intermediary steps of this evolutionary process [3, 4, 6].
Hernández (4) has proposed that upon eukaryote emergence, perhaps there was a transition period before the arousal of the cap-dependent translation when monocistronic mRNAs with long 5′-UTRs and devoid of SD sequences recruited the 40S ribosomal subunit in a cap-independent manner and in the absence of eIF3, PABP and eIF4 factors, becoming thus the first examples of an IRES. In other words, early eukaryotes inherited a functional translational apparatus from archaeal ancestors that recruited mRNAs in a cap-independent, IRES-dependent manner. The cap structure and the poly(A) tail of mRNA, as well as a PABP and perhaps eIF4E, already existed, but they played no role in translation. They might have appeared for functions in RNA metabolism that emerged among the primary adaptive responses to the emergence of the nuclear membrane (i.e., the need for nucleocytoplasmic mRNA export and protection) and the appearance of interrupted genes, but initially had no role in translation [3, 4]. In this scenario, present-day IRES are rather relicts of the past [3]. Discistroviridae IRESs represent an example of the minimal level of complexity in terms of dependence on proteins to initiate translation (185). They show that some mRNAs could drive recognition of the AUG start site by the ribosome in the total absence of other factors, including tRNA. For other mRNAs, at least eIF2 and eIF5B of archaeal origin, were involved in binding of the Met-tRNA Meti to the initiator codon and the assembly of 80S complexes, respectively, as the mechanism used by the some picornaviruses to initiate translation, such as the porcine teschovirus type 1 (186). In this virus, the 40S ribosomal subunit can actually be recruited directly to its mRNA by an IRES with only the further requirement of the eIF2- GTP-Met-tRNA Meti ternary complex for 48S pre-initiation complex formation.
The incorporation into translation of novel scaffold molecules with coordinator abilities, such as an ancestral HEAT-containing domain protein (a proto-eIF4G), perhaps picked up from other cellular processes such as NMD or mRNA nuclear export, further improved the efficiency and regulation of the ribosomal subunit recruitment by the mRNA. Evidence for this possible evolutionary stage is provided by the translation driven by the encephalomyocarditis virus and other picornavirus IRESs, which requires nearly all the canonical initiation factors and the middle part of eIF4G, but neither eIF4E nor the cap structure is required [195, 196].
Later on in evolution, a minimal core of eIF3 (i.e., a proto-eIF3) could have been derived from other, more ancient cellular processes such as the ubiquitin-proteasome and protein degradation pathways and incorporated into translation. Translation initiation thus became more dependent on new factors like eIF3, which by bridging eIF4G and the 40S ribosomal subunit enhanced the efficiency and accuracy of mRNA recruitment. This hypothetical evolutionary stage is similar to what happens in the translation of messages from different viruses, including hepatitis C virus, pestiviruses and Rhopalosiphum padi virus, where direct binding of the 40S ribosomal subunit to the mRNA is driven by the IRES [197, 198]. HCV and pestivirus mRNAs have the additional requirement of eIF3 and eIF2-GTP-Met-tRNA Meti ternary complex to form the 48S-initiation complex. In Rhopalosiphum padi virus mRNA, the binding of the 40S ribosomal subunit absolutely requires eIF3, but it occurs in the absence of the eIF4 group of factors [197, 198].
In all evolutionary stages, existing proto-eIF4A helicases, perhaps performing activity in different RNA metabolism activities, could help RNA unwinding. The incorporation of a proto-eIF4A along with eIF4E improved both the efficiency and the regulatory possibilities of mRNA recruitment even more, leading ultimately to the cap-dependent mechanism to initiate translation.
8 Concluding Remarks
One of the enigmas of modern biology is how eukaryotic translation emerged. We have discussed evidence supporting the notion that tinkering [143] might have played a crucial role in the origin and evolution of the cap-binding mechanism in eukaryotes [4, 6]. According to Jacob, “…natural selection does not work as an engineer works. It works like a tinkerer—a tinkerer who does not know exactly what he is going to produce but uses whatever he finds around him whether it be pieces of string, fragments of wood, or old cardboards” [143]. “…Evolution would slowly modify his work, unceasingly retouching it, cutting here, lengthening there, seizing the opportunities to adapt it progressively to its new use…It works on what already exists, either transforming a system to give it new functions of combining several systems to produce a more elaborate one” [143].
We have discussed that early eukaryotes inherited a core of translation machinery and that, in the absence of SD sequences in mRNAs and RPS1 in ribosomes, the first eukaryotic mRNAs were translated in a cap-independent, IRES-driven manner that was then superseded in evolution by the cap-dependent mechanism, rather than vice versa. Thus, the contemporary cellular IRESs might be relics of the past. This hypothesis is supported by the observations that (1) IRES-dependent, but not cap-dependent translation can take place in the absence of not only a cap, but also many initiation factors and (2) eIF4E and eIF4G, molecules absolutely required for cap-dependent translation, are among the most recently evolved translation factors.
Afterwards, the evolution of the translation machinery followed a gradual addition of scaffold proteins, namely eIF3, eIF4G, PABP as well of eIF4A and eIF4E, which highly improved the efficiency and regulation of mRNA binding to the 40S ribosomal subunit [3, 4]. Indeed, eIF3, eIF4G, eIF4A, PABP, the cap structure and the polyadenylation of mRNAs and perhaps eIF4E might be molecular exaptations. The rudiments of these molecules might have first arisen during eukaryogenesis with no role in translation before the cap-dependent initiation of translation appeared, performing activities other than translation, perhaps involved in mRNA nuclear export, splicing and stability, and were gradually added into the initiation of translation by a process of molecular tinkering later in evolution [143]. The diversity of viruses infecting present-day cells with a variety of needs of translation factors and cap that might represent the different evolutionary steps discussed here supports this hypothesis.
Finally, there are still many open questions on the evolution of translation in early eukaryotes. For example, we still lack satisfying explanations for the evolutionary origin of monocistronic transcripts, for the mechanism of mRNA recruiting in the early eukaryotes, for the origin of most ribosomal proteins and RNA extensions of rRNAs, and for the archaeal lineage that originated the early eukaryotes.
References
Mathews MB, Sonenberg N, Hershey JWB. Origins and principles of translational control. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. p. 1–31.
Mathews MB, Sonenberg N, Hershey JWB. Origins and principles of translational control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. p. 1–40.
Hernández G. Was the initiation of translation in early eukaryotes IRES-driven? Trends Biochem Sci. 2008;33:58–64.
Hernández G. On the origin of the cap-dependent initiation of translation in eukaryotes. Trends Biochem Sci. 2009;34:166–75.
Hernández G. On the emergence and evolution of the eukaryotic translation apparatus. In: Biyani M, editor. Cell-free protein synthesis. Rijekra, Croatia: InTech; 2012. p. 31–50.
Hernández G, Altmann M, Lasko P. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem Sci. 2010;35:63–73.
Hernández G, Proud CG, Preiss T, Parsyan A. On the diversification of the translation apparatus across eukaryotes. Comp Funct Genom. 2012;2012:256848.
Benelli D, Londei P. Translation initiation in Archaea: conserved and domain-specific features. Biochem Soc Trans. 2011;19:89–93.
Londei P. Evolution of translational initiation: news insights from the archaea. FEMS Microbiol Rev. 2005;29:185–200.
Laursen BS, Sørensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–23.
Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementary to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–6.
Shine J, Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975;254:34–8.
Jacob WF, Santer M, Dahlberg AE. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci USA. 1987;84:4757–61.
Dennis PP. Ancient ciphers: translation in archaea. Cell. 1997;89:1007–10.
Band L, Henner DJ. Bacillus subtilis requires a “stringent” Shine-Dalgarno region for gene expression. DNA. 1984;3:17–21.
Steitz JA, Jakes K. How ribosomes select initiator regions in mRNA: base-pair formation between the 3’ terminus of 16 s rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:4734–8.
Jackson RJ. A comparative view of initiation site selection mechanisms. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational control of gene expression. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory press; 2000. p. 127–83.
Wilson DN, Nierhaus KH. Ribosomal proteins in the spotlight. Crit Rev Biochem Mol Biol. 2005;40:243–67.
Moll I, Grill S, Gründling A, Bläsi U. Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol Microbiol. 2002;44:1387–96.
Delvillani F, Papiani G, Dehò G, Briani F. S1 ribosomal protein and the interplay between translation and mRNA decay. Nucleic Acid Res. 2011;39:7702–15.
Sengupta J, Agrawal RK, Frank J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc Natl Acad Sci USA. 2001;98:11991–6.
Kaberdin VR, Bläsi U. Translation initiation and the fate of bacterial mRNAs. FEMS Microbiol Rev. 2006;30:967–79.
Komarova AV, Tchufistova LS, Supina EV, Boni IV. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–47.
Shell SS, Wang J, Lapierre P, Mir M, Chase MR, Pyle MM, Gawande R, Ahmad R, Sarracino DA, Ioerger TR, Fortune SM, Derbyshire KM, Wade JT, Gray T. Leaderless transcripts and small proteins are common features of the mycobacterial translational landscape. PLoS Genet. 2015;11:e1005641.
Ma J, Campbell A, Karlin S. Correlation between Shine-Dalgarno sequence and gene features such as predicted expression levels and operon structure. J Bacteriol. 2002;184:5733–45.
Torarinsson E, Klenk HP, Garret RA. Divergent transcriptional and translational signals in Archaea. Environ Microbiol. 2005;7:45–54.
Sensen CW. Organizational characteristics and information content of an archaeal genome: 156 kb of sequence from Sulfolobus solfataricus P2. Mol Microbiol. 1996;22:175–91.
Weiner J, Herrmann R, Browning GF. Transcription in Mycoplasma pneumoniae. Nucleic Acid Res. 2000;28:4488–96.
Chang B, Halgamuge S, Tang SL. Analysis of SD sequences in completed microbial genomes: non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–9.
Nakagawa S, Niimura Y, Miura KI, Gojobori T. Dynamic evolution of translation initiation mechanisms in prokaryotes. Proc Natl Acad Sci USA. 2010;107:6382–7.
Benelli D, Londei P. Begin at the beginning: evolution of translational initiation. Res Microbiol. 2009;160:493–501.
Brenneis M, Hering O, Lange C, Soppa J. Experimental characterization of Cis-acting elements important for translation and transcription in halophilic archaea. PLoS Genet. 2007;3:e229.
Zheng X, Hu GQ, She ZS, Zhu H. Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genom. 2011;12:361–70.
Slupska MM, King AG, Fitz-Gibbon S, Besemer J, Borodovsky M, Miller JH. Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J Mol Biol. 2001;309:347–60.
Tolstrup N, Sensen CW, Garrett RA, Clausen IG. Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles. 2000;4:175–9.
Jäger D, Förstner KU, Sharma CM, Santangelo TJ, Reeve J. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genom. 2014;15:684.
Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep. 2013;5:1121–31.
Kramer P, Gäbel K, Pfeiffer F, Soppa J. Haloferax volcanii, a prokaryotic species that does not use the Shine Dalgarno mechanism for translation initiation at 5’-UTRs. PLoS ONE. 2014;9:e94979.
Sullivan MJ, Curson AR, Shearer N, Todd JD, Green RT, Johnston AW. Unusual regulation of a leaderless operon involved in the catabolism of dimethylsulfoniopropionate in Rhodobacter sphaeroides. PLoS ONE. 2011;6:e15972.
Krishnan KM, Van Etten WJ, Janssen GR. Proximity of the start codon to a leaderless mRNA’s 5’ terminus is a strong positive determinant of ribosome binding and expression in Escherichia coli. J Bacteriol. 2010;192:6482–5.
Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell. 2011;147:147–57.
La Teana A, Benelli D, Londei P, Bläsi U. Translation initiation in the crenarchaeon Sulfolobus solfataricus: eukaryotic features but bacterial route. Biochem Soc Trans. 2013;41:350–5.
Hering O, Brenneis M, Beer J, Suess B, Soppa J. A novel mechanism for translation initiation operates in haloarchaea. Mol Microbiol. 2009;71:1451–63.
Omotajo D, Tate T, Cho H, Choudhary M. Distribution and diversity of ribosome binding sites in prokaryotic genomes. BMC Genom. 2015;16:e604.
Balakin AG, Skripkin EA, Shatsky IN, Bogdanov AA. Unusual ribosome binding properties of mRNA encoding bacteriophage lambda repressor. Nucleic Acid Res. 1992;20:563–71.
Brock JE, Pourshahian S, Giliberti J, Limbach PA, Janssen G. Ribosomes bind leaderless mRNA in Escherichia coli through recognition of their 5′-terminal AUG. RNA. 2008;14:2159–69.
Van Etten WJ, Janssen GR. An AUG initiation codon, not codon-anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli. Mol Microbiol. 1998;27:987–1001.
Grill S, Gualerzi CO, Londei P, Bläsi U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: evolutionary implications for translation. EMBO J. 2000;19:4101–10.
O’Donnell SM, Janssen GR. The initiation codon affects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5’ untranslated leader. J Bacteriol. 2001;183:1277–83.
O’Donnell SM, Janssen GR. Leaderless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli. J Bacteriol. 2002;184:6730–3.
Moll I, Grill S, Gualerzi CO, Bläsi U. Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol Microbiol. 2002;43:239–46.
Moll I, Hirokawa G, Kiel MC, Kaji A, Bläsi U. Translation initiation with 70S ribosomes: an alternative pathway for leaderless mRNAs. Nucleic Acid Res. 2004;32:3354–63.
Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–8.
Scharff LB, Childs L, Walther D, Bock R. Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding site. PLoS Genet. 2011;7:e1002155.
Fox GE. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol. 2012;2:e003483.
Golshani A, Krogan NJ, Xu J, Pacal M, Yang XC, Ivanov I, Providenti MA, Ganoza MC, Ivanov IG. Abou Haidar MG. Escherichia coli mRNAs with strong Shine/Dalgarno sequences also contain 5’ end sequences complementary to domain #17 on the 16S ribosomal RNA. Biochem Biophys Res Commun. 2004;316:978–83.
Loechel S, Inamine JM, Hu PC. A novel translation initiation region from Mycoplasma genitalium that functions in Escherichia coli. Nucleic Acids Res. 1991;19:6905–11.
Jenner L, Romby P, Rees B, Schulze-Briese C, Springer M, Ehresmann C, Ehresmann B, Moras D, Yusupova G, Yusupov M. Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding. Science. 2005;308:120–3.
Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci USA. 1998;95:224–8.
Koonin EV. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol. 2010;11:209.
Koumandou VL, Wickstead B, Ginger ML, van der Giezen M, Dacks JB, Field MC. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48:373–96.
Woese CR. On the evolution of cells. Proc Natl Acad Sci USA. 2002;99:8742–7.
Wu CJ, Janssen GR. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5’ untranslated leader. Mol Microbiol. 1996;22:339–55.
Sonenberg N, Hinnebusch A. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45.
Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27.
Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev. 2011;75:434–67.
Pelletier J. Sonenberg N Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–5.
Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5’ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–43.
Macejak DG, Sarnow P. Internal initiation of translation mediated the 5’UTR leader of a cellular mRNA. Nature. 1991;353:90–4.
Martínez-Salas E, Piñeiro D, Fernández N. Alternative mechanisms to initiate translation in eukaryotic mRNAs. Comp Funct Genom. 2012;2012:391546.
Elroy-Stein O, Merrick WC. Translation initiation via cellular internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. p. 155–72.
Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–7.
Dounda JA, Sarnow P. Translation initiation by viral internal ribosome entry sites. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. p. 129–53.
Guy L, Saw JH, Ettema TJG. The archaeal legacy of eukaryotes: a phylogenomic perspective. In: Keeling PJ, Koonin EV, editors. The origin and evolution of eukaryotes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2014. p. 97–128.
Poole AM, Gribaldo S. Eukaryotic origins: how and when was the mitochondrion acquired? In: Keeling PJ, Koonin EV, editors. The origin and evolution of eukaryotes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2014. p. 129–40.
McFadden GI. Origin and evolution of plastids and photosynthesis in eukaryotes. In: Keeling PJ, Koonin EV, editors. The origin and evolution of eukaryotes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2014. p. 263–70.
Williams TA, Foster PG, Cox CJ, Embley M. An archaeal origin of eukaryotes supports only two primary domains of life. Nature. 2013;504:231–6.
Woese CR, Fox GE. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–90.
Woese CR, Fox GE. The concept of cellular evolution. J Mol Evol. 1977;10:1–6.
Woese CR. Interpreting the universal phylogenetic tree. Proc Natl Acad Sci USA. 2000;97:8392–6.
Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci USA. 2008;105:20356–61.
Williams TA, Foster PG, Nye TM, Cox CJ, Embley TM. A congruent phylogenomic signal places eukaryotes within the Archaea. Proc Biol Sci. 2012;279:4870–9.
Guy L, Ettema TJG. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–7.
Pedulla N, Palermo R, Hasenöhr D, Bläsi U, Cammarano P, Londei P. The archaeal eIF2 homologue: functional properties of an ancient translation initiation factor. Nucleic Acid Res. 2005;33:1804–12.
Aravind L, Koonin EV. Eukaryotic-specific domains in translation initiation factors: implications for translation regulation and evolution of the translation system. Genome Res. 2000;10:1172–84.
Lecompte O, Ripp R, Thierry JC, Moras D, Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acid Res. 2002;30:5382–90.
Wilson DN, Cate JHD. The structure and function of the eukaryotic ribosome. In: Hershey JWB, Sonenberg N, Mathews MB, editors. Protein synthesis and translational control. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2012. p. 11–27.
Dresios J, Panopoulos P, Synetos D. Eukaryotic ribosomal proteins lacking a eubacterial counterpart: important players in ribosomal function. Mol Microbiol. 2006;59:1651–63.
Wolf YI, Aravind L, Grishin N, Koonin EV. Evolution of aminoacyl-tRNA synthetases-analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710.
O’Donoghue P, Luthey-Schulten Z. On the Evolution of Structure in Aminoacyl-tRNA Synthetases. Microbiol Mol Biol Rev. 2003;67:550–3.
Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, Hud NV, Williams LD. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci USA. 2014;111:10251–6.
Roberts E, Sethi A, Montoya J, Woese CR, Luthey-Schulten Z. Molecular signatures of ribosomal evolution. Proc Natl Acad Sci USA. 2008;105:13953–8.
Lynch M. The origins of genome architecture. Sunderland, Massachusetts: Sinauer Associates, Inc., Publishers; 2007.
Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–5.
Taylor DJ, Frank J, Kinzy TG. Structure and function of the eukaryotic ribosome and elongation factors. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. p. 59–85.
Yokoyama T, Suzuki T. Ribosomal RNAs are tolerant toward genetic insetions: evolutionary origin of the expansion segments. Nucleic Acid Res. 2008;36:3539–51.
Hartman H, Favaretto P, Smith TF. The archaeal origins of the eukaryotic translational system. Archaea. 2006;2:1–9.
Klinge S, Voigst-Hoffmann F, Leibundgut M, Ban N. Atomic structures of the eukaryotic ribosome. Trends Biochem. 2012;37:189–98.
Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37.
Lukaszewicz M. M. F, Jerouville B, Stas A, Boutry M. In vivo evaluation of the context sequence of the translation initiation codon in plants. Plant Sci. 2000;154:89–98.
Lutcke HA, Chow KC, Mickel FS, Moss KA, Kern HF, Scheele GA. Selection of AUG initiation codons differs in plants and animals. EMBO J. 1987;6:43–8.
Cigan M, Donahue TF. Sequence and structural features associated with translational initiator regions in yeast - a review. Gene. 1987;59:1–18.
Donahue TF, Cigan M. Sequence and structural requirements for efficient translation in yeast. Meth Enzymol. 1990;185:366–71.
Cavener DR. Eukaryotic start and stop translation sites. Nucleic Acid Res. 1991;19:3185–92.
Kim Y, Lee G, Jeon E, Sohn EJ, Lee Y, Kang H, Lee Dw, Kim DH, Hwang I. The immediate upstream region of the 5´-UTR from the AUG start codon has a pronounced effect on the translational efficiency in Arabidopsis thaliana. Nucleic Acid Res. 2014;14:485–98.
Kawaguchi R, Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acid Res. 2005;33:955–65.
Nakagawa S, Niimura Y, Gojobori T, Tanaka H, Miura K. Diversity of preferred nucleotide sequences around the translation initiation codon in eukaryote genomes. Nucleic Acids Res. 2008;36:861–71.
Lynch M, Scofield DG, Hong X. The evolution of transcription-initiation sites. Mol Biol Evol. 2005;22:1137–46.
Mignone F, Gissi C, Liuni S, Pesole L. Untranslated regions of mRNAs. Genome Biol. 2002;3:1–10.
Mazumder B, Seshadri V, Fox PL. Translational control by the 3’-UTR: the ends specify the means. Trends in Biochem Sci. 2003;28:91–8.
Hentze MW, Gebauer F, Preiss T. cis-Regulatory sequences and trans-actin factors in translational control. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational control in biology and medicine. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2007. p. 269–96.
Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81.
Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–55.
Araujo PR, Yoon K, Ko D, Smith AD, Qiao M, Suresh U, Burns SC, Penalva LOF. Before it gets started: regulating translation at the 5´-UTR. Comp Funct Genom. 2012;12:1–8.
Alekhina AM, Vassilenko KS. Translation initiation in eukaryotes: versatility of the scanning model. Biochemistry (Moscow). 2012;77:1465–77.
van der Velden A, Thomas AAM. The role of the 5´untranslated region of an mRNA is translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106.
Faye MD, Holcik M. The role of IRES trans-acting factors in carcinogenesis. Biochim Biophys Acta. 2015;1849:887–97.
Komar AA, Hatzoglou M. Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle. 2011;10:229–40.
King HA, Cobbold LC, Willis AE. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans. 2010;38:1581–6.
Davuluri RV, Suzuki Y, Sugano S, Zhang MQ. CART Classification of Human 5´UTR Sequences. Genome Res. 2000;10:1807–16.
Resch AM, Ogurstov AY, Rogozin IB, Shabalina SA, Koonin EV. Evolution of alternative and constitutive regions of mammalian 5’UTRs. BMC Genom. 2009;10:162.
Moshonov S, Elfakes R, Golan-Mashiach M, Sinvani H, Dikstein R. Links between core promoter and basic gene features influence gene expression. BMC Genom. 2008;9:92.
Grillo G, Turi A, Licciulli F, Mignone F, Liuni S, Banfi S, Gennarino VA, Horner DS, Pavesi G, Picardi E, Pesole G. UTRdb and UTRsite (RELEASE 2010): a collection of sequences and regulatory motifs of the untranslated regions of eukaryotic mRNAs. Nucleic Acid Res. 2010;38:D75–80.
Vogel C, Abreu RS, Ko D, Le SY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400.
de Sousa R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol BioSyst. 2009;5:1512–26.
Adam RD. The Giardia lamblia genome. Int J Parasitol. 2000;30:475–84.
Bruchhaus I, Leippe M, Lioutas C, Tannich E. Unusual gene organization in the protozoan parasite Entamoeba histolytica. DNA Cell Biol. 1993;12:925–33.
Elfakes R, Dikstein R. A translation initiation element specific to mRNAs with very short 5UTR that Also regulates transcription. PLoS ONE. 2008;3:e3094.
Elfakes R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acid Res. 2011;39:7598–609.
Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4G1 cooperation in start codon selection. Cell Metab. 2015;21:479–92.
Zhaxybayeva O, Gogarten P. Spliceosomal introns: new insights into their evolution. Curr Biol. 2003;13:R764–6.
Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35.
Rogozin IB, Carmel L, Csuros M, Koonin EV. Origin and evolution of spliceosomal introns. Biol Direct. 2012;16:11.
Irimia M. Roy SW (2014) Origin of spliceosomal introns and alternative splicing. In: Keeling PJ, Koonin EV, editors. The origin and evolution of eukaryotes. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2014. p. 295–316.
Maone E, Di Stefano M, Berardi A, Benelli D, Marzi S, La Teana A, Londei P. Functional analysis of the translation factor aIF2/5B in the thermophilic archaeon Sulfolobus solfataricus. Mol Microbiol. 2007;65:700–13.
Lee JH, Choi SK, Roll-Mecak A, Burley SK, Dever TE. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc Natl Acad Sci USA. 1999;96:4342–7.
Kyrpides NC, Woese CR. Archaeal translation initiation revisited: the initiation factor 2 and eukaryotic initiation factor 2B a- b-d subunit families. Proc Natl Acad Sci USA. 1998;95:3726–30.
Pánek J, Klolár M, Vohradsky J, Valásek LS. An evolutionary conserved pattern of 18S rRNA sequence complementary to mRNA 5´UTRs and its implications for eukaryotic gene translation regulation. Nucleic Acid Res. 2013;41:7625–34.
Matveeva OV, Shabalina S. Intermolecular mRNA-rRNA hybridization and the distribution of potential interaction regions in murine 18S rRNA. Nucleic Acid Res. 1993;21:1007–11.
Mauro VP, Edelman GM. rRNA-like sequences occur in diverse primary transcripts: implications for the control of gene expression. Proc Natl Acad Sci USA. 1997;94:422–7.
Tranque P, Hu MC, Edelman GM, Mauro VP. rRNA complementarity within mRNAs: a possible basis for mRNA-ribosome interactions and translational control. Proc Natl Acad Sci USA. 1998;95:12238–43.
Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CUT, Pestova T. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–36.
Jacob F. Evolution and Tinkering. Science. 1977;196:1161–6.
Gould SJ, Vrba ES. Exaptation - a missing term in the science of form. Paleobiology. 1982;8:4–15.
Nojima T, Hirose T, Kimura H, Hagiwara M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem. 2007;282:15645–51.
Vinciguerra P, Stutz F. mRNA export: an assembly line from genes to nuclear pores. Curr Opin Cell Biol. 2004;16:285–92.
Cougot N, van Dijk E, Babajko S, Séraphin B. Cap-tabolism. Trends Biochem Sci. 2004;29:436–44.
Ling SH, Qamra R, Song H. Structural and functional insights into eukaryotic mRNA decapping. Wiley Interdiscip Rev RNA. 2011;2:193–208.
Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA. 2010;2:277–98.
Ghosh A, Lima CD. Enzymology of RNA cap synthesis. Wiley Interdiscip Rev RNA. 2010;1:152–72.
Martinez-Rucobo FW, Kohler R, van de Waterbeemd M, Heck AJ, Hemann M, Herzog F, Stark H, Cramer P. Molecular basis of transcription-coupled pre-mRNA capping. Mol Cell. 2015;58:1079–89.
Reed R. Coupling transcription, splicing and mRNA export. Curr Opin Cell Biol. 2003;15:326–31.
Rhoads RE. eIF4E: new family members, new binding partners, new roles. J Biol Chem. 2009;284:16711–5.
Rezende AM, Assis LA, Nunes EC, Lima TD, Marchini FK, Freire ER, Reis CR, de Melo Neto OP. The translation initiation complex eIF3 in trypanosomatids and other pathogenic excavates-identification of conserved and divergent features based on orthologue analysis. BMC Genom. 2014;15:1175.
Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–60.
des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature. 2015;525:491–5.
Querol-Audi J, Sun C, Vogan JM, Smith MD, Gu Y, Cate JH, Nogales E. Architecture of human translation initiation factor 3. Structure. 2013;21:920–8.
Pereira RV, de Gomes MS, Jannotti-Passos LK, Borges WC, Guerra-Sá R. Characterisation of the COP9 signalosome in Schistosoma mansoni parasites. Parasitol Res. 2013;112:2245–53.
Schwechheimer C. The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta. 2004;1695:45–54.
Scheel H, Hofmann K. Prediction of a common structural scaffold for proteasome lid, COP9-signalosome and eIF3 complexes. BMC Bioinformatics. 2005;6:71.
Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600.
Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry. 2005;44:12265–72.
Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CUT, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell. 2001;7:193–203.
Pontig CP. Novel eIF4G domain homologues linking mRNA translation with nonsense-mediated mRNA decay. Trends Biochem Sci. 2000;25:423–6.
Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20:8944–57.
Kobe B, Gleichmann T, Horne J, Jennings IG, Scotney PD, Teh T. Turn up the HEAT. Structure. 1999;7:R91–7.
Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18.
Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–83.
Layana C, Ferrero P, Rivera-Pomar R. Cytoplasmic ribonucleoprotein foci in eukaryotes: hotspots of bio(chemical) diversity. Comp Funct Genom. 2012;2012:504292.
Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Sign. 2011;23:324–34.
Hernández G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev. 2005;122:865–76.
Strudwick S, Borden KLB. The emerging roles of translation factor eIF4E in the nucleus. Differentiation. 2002;70:10–22.
Osborne MJ, Borden KLB. The eukaryotic translation initiation factor eIF4E in the nucleus: taking the road less traveled. Immunol Rev. 2015;263:210–23.
Li L, Wang CC. Identification in the ancient protist Giardia lamblia of two eukaryotic translation initiation factor 4E homologues with distinctive functions. Eukaryot Cell. 2005;4:948–59.
Ramirez CV, Vilela C, Berthelot K, McCarthy JE. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J Mol Biol. 2002;318:951–62.
Deutscher MP, Li Z. Exoribonucleases and their multiple roles in RNA metabolism. Prog Nucleic Acid Res Mol Biol. 2001;66:67–105.
Shuman S. The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb Symp Quant Biol. 2001;66:301–12.
Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acid Res. 2002;30:1427–64.
Muhlemann O. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–49.
Eliseeva IA, Lyabin DN, Ovchinnikov LP. Poly(A)-binding proteins: structure, domain organization, and activity regulation. Biochemistry (Moscow). 2013;78:1377–91.
Derry MC, Yanagiya A, Martineau Y, Sonenberg N. Regulation of poly(A)-binding protein through PABP-interacting proteins. Cold Spring Harb Quant Biol. 2006;71:537–43.
Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim Biophys Acta. 2004;1678:67–84.
Vazquez-Pianzola P, Suter B. Conservation of the RNA transport machineries and their coupling to translation control across eukaryotes. Comp Funct Genom. 2012;2012:287852.
Vazquez-Pianzola P, Urlaub H, Suter B. Pabp binds to the osk 3’UTR and specifically contributes to osk mRNA stability and oocyte accumulation. Dev Biol. 2011;357:404–18.
Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA- binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–31.
Lu WT, Wilczynska A, Smith E, Bushell M. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans. 2014;42:166–72.
Toone WM, Rudd KE, Friesen JD. deaD, a new Escherichia coli gene encoding a presumed ATP-dependent RNA helicase, can suppress a mutation in rpsB, the gene encoding ribosomal protein S2. J Bacteriol. 1991;173:3291–302.
Nishi K, Morel-Deville F, Hershey JW, Leighton T, Schnier J. An eIF-4A-like protein is a suppressor of an Escherichia coli mutant defective in 50S ribosomal subunit assembly. Nature. 1988;336:496–8.
Lu J, Aoki H, Ganoza MC. Molecular characterization of a prokaryotic translation factor homologous to the eukaryotic initiation factor eIF4A. Int J Biochem Cell Biol. 1999;31:215–29.
Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nature Rev Mol Cell Biol. 2011;12:505–16.
Lüking A, Stahl U, Schmidt U. The protein family of RNA helicases. Crit Rev Biochem Mol Biol. 1998;33:259–96.
Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. mRNA helicases: the tacticians of translational control. Nat Rev Mol Cell Biol. 2011;12:235–45.
Marintchev A. Roles of helicases in translation initiation: a mechanistic view. Biochim Biophys Acta. 2015;1829:799–809.
Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58.
Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol. 1996;16:6870–8.
Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–69.
Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83.
Terenin IM, Dmitriev SE, Andreev DE, Royall E, Belsham GJ, Roberts LO, Shatsky IN. A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry. Mol Cell Biol. 2005;25:7879–88.
Acknowledgments
This work was supported by the National Institute of Cancer (INCan), Mexico, and by the National Council of Science and Technology (CONACyT, grant no. 168154 to G.H.), Mexico.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Hernández, G., Osnaya, V.G., García, A., Velasco, M.X. (2016). On the Origin and Early Evolution of Translation in Eukaryotes. In: Hernández, G., Jagus, R. (eds) Evolution of the Protein Synthesis Machinery and Its Regulation. Springer, Cham. https://doi.org/10.1007/978-3-319-39468-8_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-39468-8_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-39467-1
Online ISBN: 978-3-319-39468-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)