Abstract
Ultra-high field MR imaging (e.g., 7T) provides unprecedented spatial resolution and superior signal-to-noise ratio, compared to the routine MR imaging (e.g., 1.5T and 3T). It allows precise depiction of small anatomical structures such as hippocampus and further benefits diagnosis of neurodegenerative diseases. However, the routine MR imaging is still mainly used in research and clinical studies, where accurate hippocampus segmentation is desired. In this paper, we present an automatic method for segmenting hippocampus from the routine MR images by learning 7T-like features from the training 7T MR images. Our main idea is to map features of the routine MR image to be similar to 7T image features, thus increasing their discriminability in hippocampus segmentation. Specifically, we propose a patch-based mapping method to map image patches of the routine MR images to the space of image patches of the 7T MR images. Thus, for each patch in the routine MR image, we can generate a new mapped patch with 7T-like pattern. Then, using those mapped patches, we can use a random forest to train a sequence of classifiers for hippocampus segmentation based on the appearance, texture, and contexture features of those mapped patches. Finally, hippocampi in the test image can be segmented by applying the learned image patch mapping and trained classifiers. Experimental results show that the accuracy of hippocampus segmentation can be significantly improved by using our learned 7T-like image features, in comparison to the direct use of features extracted from the routine MR images.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Hippocampus is an anatomical structure located in medial temporal lobe, and plays important roles in the consolidation of information from short-term memory to long-term memory and also spatial navigation [1]. It becomes of major interest due to its implications in multiple psychiatric disorders and neurodegenerative diseases, e.g., Alzheimer’s disease (AD). Specifically, hippocampal volume and morphological characteristics have been proven as valuable biomarkers for diagnosis and tracking of brain diseases [2]. Therefore, a reliable and accurate way for segmenting hippocampus is necessary to better quantify subtle structural changes in the brain.
Manual delineation by trained raters has been considered as the gold standard in hippocampus segmentation, but the process is tedious and time consuming. Thus, it would be extremely valuable to develop an automated hippocampus segmentation method with high accuracy. Recently, many automatic methods have been developed for hippocampus segmentation, which generally can be divided into two categories. The first category is the atlas-based segmentation methods, which propagate the predefined labels in an atlas image to a subject image (under segmentation) by building their spatial correspondences using deformable image registration. Multi-atlases-based methods [3] can mediate the inter-subject variability and produce better segmentation results through label fusion from multiple atlases. The second category is the learning-based segmentation methods. Based on the image features extracted from both hippocampus and non-hippocampus areas in the training images, machine learning techniques can be used to train a classifier to discriminate the hippocampus from the background. Note that, regardless of the atlas-based methods or learning-based approaches, most existing methods have been developed for the routine MR images, i.e., 1.5T or 3T MR images.
With the advance development of MR imaging techniques, 7T MRI offers unprecedent spatial resolution as well as superior tissue contrast, which allows clear depiction of small anatomical details. Previous studies [4] have demonstrated fine details of in vivo hippocampal structure using 7T MRI, including the boundary of hippocampus as well as the visualization of individual subfields within the hippocampus. Although hippocampus segmentation methods using ultra-high field MRIs have achieved higher accuracy over the routine MR images [5], they are still limited to a small cohort of normal subjects. This is because 7T imaging is now still mainly used for research purpose, not clinical applications, due to practical issues during image acquisition, e.g., lack of a standard protocol and the long imaging time. Therefore, there is still a long way to go before we can use 7T scanner to investigate a large population of diseased brains.
On the other hand, in machine learning area, recent studies have proposed to map raw features to another feature space for increasing their discriminative power. For example, kernel-based methods can make linear models work in the nonlinear settings by mapping the data to the higher dimensions where the mapped data exhibits linear patterns [6]. This domain transfer approach aims to learn a new feature representation for the input features by linear or nonlinear mapping, in order to enlarge the discriminations of mapped features in the new feature space and then eventually improve the classification performance. Accordingly, we are motivated to map features of the routine MR images to the space of features of the 7T images, where the hippocampus and surrounding tissues are better separable.
In this paper, we propose a learning-based method for segmenting hippocampus in the routine MR images, guided by features learned from 7T MR images. In particular, based on the feature mapping between the routine MR and 7T MR images, we can make the image patches of routine MR images similar to 7T image patches, thus allowing better separation of hippocampus from surrounding tissues. Then, we use random forest to train a sequence of classifiers based on the features (i.e., appearance, texture and context features) of those mapped patches. In the application stage, when given a new test image, we can first map each of its image patches to be 7T-like. Then, we can apply a sequence of trained classifiers to the features of those mapped image patches for obtaining a hippocampus probability map. Experimental results show that our proposed method can significantly improve the segmentation accuracy, compared to the conventional approach.
2 Method
Our goal is to improve the segmentation performance in routine MR images by mapping the features extracted from routine MR images to the space of 7T image features, where the hippocampus and surrounding tissues are better separable. The overview of our proposed framework is shown in Fig. 1. In the training stage, we determine the relationships between patches from the routine MR and 7T MR images by using a patch-based mapping approach. Then, the random forest is used to construct a sequence of classifiers based on multiple types of features, i.e., appearance, texture and context features, extracted from the mapped image patches of routine MR images. In the testing stage, for a new test image under segmentation, its mapped version of 7T-like image is first reconstructed using the learned mapping function. Then, the hippocampus in the test image can be segmented by applying a sequence of trained classifiers to the mapped 7T-like patches of the test image.
2.1 Patch-Based Mapping
Our main contribution is to use a patch-based mapping approach to learn the relationships between the patches from routine MR images and the patches from 7T MR images. First, the linear mapping is built based on the principal component analysis (PCA) models built on the patches from the routine MR images and 7T MR images separately, thus allowing the mapping of their distributions in high dimensional space. In this way, we can obtain the initially mapped patch in 7T space for each patch in the routine MR images. Second, a nonlinear mapping is further employed for each image patch of routine MR images to seek for its correspondence by searching 7T image patches within the closest Euclidean distance from its initially mapped version by the above linear mapping.
Specifically, we define searching windows (as shown by orange boxes in Fig. 2A) at the same location in both the routine MR image and 7T MR image spaces. Then, we extract a number of patches centered at the voxels within the searching windows while allowing the overlap between the extracted patches (as shown by blue and yellow boxes in Fig. 2A). We use the intensity of each voxel as feature, thus each patch can be represented by a column vector x ∊ R p×1, where p is the size of feature dimension. Each patch x is regarded as a feature point in the high dimensional feature space (with p-dimensions). Note that the routine MR images and 7T MR images have their respective feature distributions in their own spaces, as shown by the schematic illustrations in the top of Fig. 2A.
Patch-based linear and nonlinear mappings between image patches from 1.5T and 7T MR images. (A) Linear mapping can be done by PCA representations (top), followed by nonlinear mapping (bottom). (B) Examples of original 1.5T image patches (top), 7T image patches (middle), and mapped 1.5T image patches in the 7T space (bottom).
We use PCA to describe the feature distribution in the spaces of routine MR (e.g., 1.5T) and 7T MR images, respectively:
Where X is the data matrix (x 1, x 2,…, x n ), and n is the number of patches within each searching window. \( \sum \) is a \( n \times p \) rectangular diagonal matrix, containing the singular values of X, and U is a \( p \times n \) matrix, whose columns are orthogonal unit vectors of length p. W is a p × p matrix (t 1, t 2,…, t p ), whose columns, t i , i = 1, ···p, are orthogonal unit vectors of length p. By using PCA, each eigenvector represents the direction of data distribution, while all eigenvectors are independent to each other. The eigenvalue λ(i) represents the variance of data along the direction of the i-th eigenvector. We build the mapping relationships among the patches from the routine MR and 7T MR images by linearly aligning their distributions as shown in Eq. 2 below. The patch in routine MR space is first centered, and then rotated and scaled to match the distribution in the 7T space, thus getting the mapped patch.
where\( \overline{x} = \frac{1}{N}\sum\nolimits_{i = 1}^{N} {x_{i} } \). Here, \( A = W_{1.5T}^{T} \cdot W_{7T} \) is the matrix representing the rotation from the feature space of routine MR patches to the feature space of 7T MR patches, and P is the scale matrix. Note that, P is a diagonal matrix defined in Eq. 3, while the diagonal elements are the ratios of eigenvalues from the two distributions:
In this way, for each patch x 1.5T in the routine MR space, we can get its mapped patch x map in 7T space as shown in Fig. 2B.
For the j-th mapped patch x map (j) from routine MR images (shown in pink box in Fig. 2A), we further compute its Euclidean distances to all 7T patches (shown in yellow boxes in Fig. 2A) within the searching window SW(j) of the j-th location in 7T MR images to find a 7T image patch with the closest Euclidean distance (as shown by red box in Fig. 2A) as its corresponding 7T image patch \( \hat{x}_{7T} (j) \).
For each patch in the routine MR image, we get a new mapped patch with 7T-like appearance. Then, we reconstruct a new 7T-like image for each training routine MR image by weighted averaging of these mapped patches.
2.2 Learning-Based Segmentation
In this section, we describe the details on how to train classifier (random forest) with features extracted from 7T-like images and their corresponding labels (i.e., hippocampus or non-hippocampus) as defined in the original 1.5T MR training images.
Feature Extraction. Three types of features, i.e., image appearance, texture, and context features, are extracted from the 7T-like training images.
Image appearance features include intensity, spatial location, and Haar-like features at different scales [7]. For each voxel, we further extract texture features, e.g., the first-order difference filters, the second-order difference filters, 3D Hyperplane filters, 3D Sobel filters, Laplacian filters, and Range difference filters.
By using image appearance and texture features to train classifier, the initial probability map for hippocampus label can be obtained by the trained random forest, where a higher value denotes the higher probability of the respective voxel belonging to hippocampus. Then, we can use this probability map to extract context features [8]. By combining context features with the previous appearance and texture features, we can train next classifier, which can also use for producing a new probability map. By iteratively extracting contexture features and training new classifier, we can finally obtain a sequence of trained classifiers [9], which can be used to iteratively segment hippocampus for test image in the testing stage.
Training of Random Forest. From each image in a training dataset, consisting of a number of mapped 7T-like images, we randomly select half of hippocampus voxels from the training images as positive samples, and also the same size of voxels from the exterior region around the hippocampal boundary as negative samples. Then, the features, i.e., image appearance, texture, context, are extracted from image patches centered at those selected voxels. Finally, we use random forest [10] to train the classifier. Note that random forest is an ensemble learning method, which can construct a set of decision trees at training time and then perform classification at the application time by combining the outcomes of all decision trees.
Testing. For a new test image to be segmented, we first reconstruct its 7T-like image based on the mapping steps described in Sect. 2.1. Then, various features, i.e., image appearance, texture and context, are extracted for each voxel in the mapped 7T-like version of the test image. Afterwards, the trained classifiers are sequentially applied to the competed features for generating a sequence of probability maps. Finally, the binary segmentation result of hippocampus is obtained by applying the level-set algorithm on the final obtained probability map.
3 Experiments
3.1 Data
We incorporated two datasets in this study, (1) a 1.5T MR dataset for segmentation and (2) a 7T MR dataset for helping learn 7T-like features for the 1.5T MR images.
1.5T MR dataset: 20 normal subjects were selected from IXI dataset (http://brain-development.org), including 10 females and 10 males with the age of 31.55 ± 9.70 years. MRI scans were obtained on the 1.5T GE Signa Echo speed scanner. T1 weighted 3D volume was acquired with 124 coronal slices at the resolution of 0.9375 × 0.9375 × 1.5 mm3.
7T MR dataset: 7T MR images also include 20 normal subjects, consisting of 6 males and 14 females with the age of 28.92 ± 16.51 years. The images were acquired using a 3D fast low-angle shot (Spoiled FLASH) sequence with 60 slices at the resolution of 0.35 × 0.35 × 0.35 mm3. The image plane was set parallel to the longest axis running through the hippocampus.
All images were pre-processed using the following steps: (1) skull stripping, (2) inhomogeneity correction, (3) intensity normalization for making image contrast and luminance consistent across all subjects, and (4) rotation to make the coronal slice orthogonal to the long axis of hippocampus. For 7T MRI data, all images were affine aligned to a template. For 1.5T MRI data, all images were upsampled with the same resolution as 7T MR images by using trilinear interpolation and then affine registered to the 7T template. Lastly, all images were cropped using a small rectangle box for covering left and right hippocampi separately, in order to reduce the computational burden.
3.2 Experimental Setting
A leave-one-out cross-validation strategy (i.e., one subject used as testing data, and other 19 subjects used as training set) was adopted and repeated 20 times. The parameters in our method were set as follows: patch size 7 × 7 × 7, and searching window 21 × 21 × 21. 500 Haar-like features and 376 texture features were used. Specifically, the number of trees was set as 20 in the random forest, with the depth of each tree as 20. We iterated the program 4 times, where context features were updated in each iteration. In all experiments, we used same parameter settings.
We compared the hippocampus segmentation results using IXI 1.5T data by our proposed method (i.e., with patch-based 7T-like feature mapping) and by the baseline method (i.e., without any mapping procedure). To quantitatively evaluate the performance of our proposed method, Dice similarity coefficient Dice(S1, S2) = 2V(S1 ∩ S2)/(V(S1) + V(S2)) was used between the manual label S1 and automatic segmentation S2, where V(S) is the volume of a segmentation S. Note that, a higher Dice ratio indicates better segmentation in the range of [0, 1].
3.3 Experimental Results
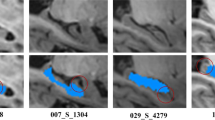
Visual Inspection. We show the segmentation results by our proposed method and the baseline method in Fig. 3. It can be observed that our segmented hippocampus (Fig. 3B) has more similar shape with manual segmentation (more obviously in the zoom-in views) than the result by the baseline method (Fig. 3C).
Quantitative Results. The comparison of averaged Dice overlap ratio of hippocampus segmentation results by our proposed method and the baseline method w.r.t. ground truths is provided in Table 1, separately for left and right hippocampi. Our proposed method by employing the features from the 7T-like images achieves much higher segmentation accuracy than the baseline method directly using the features from the original 1.5T images. Our method also achieves much higher segmentation accuracy in the comparison with the two existing algorithms [11, 12]. Note that, the results by the two algorithms are directly referred as reported in the papers, where individual results on left and right hippocampi are not available.
4 Conclusion
We presented a new automatic hippocampus segmentation method for the routine MR images by learning features from high-field 7T MR images. Our method is built on the patch-based mapping approach, which aimed to find the most similar patch from high-field MR images for each patch in the routine MR image by statistical mapping. Using the features acquired from the mapped 7T-like images, we achieved significant improvement for hippocampus segmentation. It is worth indicating that our approach can also be used to improve segmentation accuracy for other anatomical structures in the low-resolution images by learning features from high-resolution images.
References
Amaral, D., Lavenex, P.: Hippocampal neuroanatomy. The Hippocampus Book 1, 37--114 (2007)
Akhondi-Asl, A., Jafari-Khouzani, K., Elisevich, K., Soltanian-Zadeh, H.: Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods. NeuroImage 54, S218–S226 (2011)
Chupin, M., et al.: Fully automatic segmentation of the hippocampus and the amygdala from MRI using hybrid prior knowledge. In: Ayache, N., Ourselin, S., Maeder, A. (eds.) MICCAI 2007, Part I. LNCS, vol. 4791, pp. 875–882. Springer, Heidelberg (2007)
Cho, Z.H., Han, J.Y., Hwang, S.I., Kim, D.S., Kim, K.N., Kim, N.B., Kim, S.J., Chi, J.G., Park, C.W., Kim, Y.B.: Quantitative analysis of the hippocampus using images obtained from 7.0 T MRI. NeuroImage 49, 2134–2140 (2010)
Kim, M., Wu, G., Li, W., Wang, L., Son, Y.D., Cho, Z.H., Shen, D.: Automatic hippocampus segmentation of 7.0 Tesla MR images by combining multiple atlases and auto-context models. NeuroImage 83, 335–345 (2013)
Muller, K.R., Mika, S., Ratsch, G., Tsuda, K., Scholkopf, B.: An introduction to kernel-based learning algorithms. IEEET Neural Netw. 12, 181–201 (2001)
Viola, P., Jones, M.J.: Robust real-time face detection. Int. J. Comput. Vision 57, 137–154 (2004)
Tu, Z.W., Bai, X.A.: Auto-context and its application to high-level vision tasks and 3D brain image segmentation. IEEET Pattern Anal. 32, 1744–1757 (2010)
Wang, L., Gao, Y.Z., Shi, F., Li, G., Gilmore, J.H., Lin, W.L., Shen, D.G.: LINKS: Learning-based multi-source IntegratioN frameworK for segmentation of infant brain images. NeuroImage 108, 160–172 (2015)
Breiman, L.: Random forests. Mach. Learn. 45, 5–32 (2001)
Chupin, M., Mukuna-Bantumbakulu, A.R., Hasboun, D., Bardinet, E., Baillet, S., Kinkingnehun, S., Lemieux, L., Dubois, B., Garnero, L.: Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: method and validation on controls and patients with Alzheimer’s disease. NeuroImage 34, 996–1019 (2007)
Wu, G.R., Kim, M.J., Sanroma, G., Wang, Q., Munsell, B.C., Shen, D.G.: Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. NeuroImage 106, 34–46 (2015)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Li, S., Shi, F., Ma, G., Kim, M., Shen, D. (2015). Improving Accuracy of Automatic Hippocampus Segmentation in Routine MRI by Features Learned from Ultra-High Field MRI. In: Wu, G., Coupé, P., Zhan, Y., Munsell, B., Rueckert, D. (eds) Patch-Based Techniques in Medical Imaging. Patch-MI 2015. Lecture Notes in Computer Science(), vol 9467. Springer, Cham. https://doi.org/10.1007/978-3-319-28194-0_5
Download citation
DOI: https://doi.org/10.1007/978-3-319-28194-0_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-28193-3
Online ISBN: 978-3-319-28194-0
eBook Packages: Computer ScienceComputer Science (R0)