Abstract
In this paper, we present a new approach for segmentation of left and right ventricles from cardiac MR images. A two-level-set formulation is proposed which is the extension of distance regularized level set evolution (DRLSE) model in [1], with the 0-level set and k-level set representing the endocardium and epicardium, respectively. The extraction of endocardium and epicardium is obtained as a result of the interactive curve evolution of the 0 and k level sets derived from the proposed variational level set formulation. The initialization of the proposed two-level-set DRLSE model is generated by performing the original DRLSE from roughly located endocardium. Experimental results have demonstrated the effectiveness of the proposed two-level-set DRLSE model.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Distance Regularized Level Set Evolution (DRLSE)
- Right Ventricle Segmentation
- Epicardial Contours
- Desired Object Boundary
- Endocardial Contours
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Currently, cardiovascular disease is becoming one of the leading causes to death. Cine-magnetic resonance imaging (cine-MRI) is an important modality which contains accurate information for non-invasively diagnosing and treating cardiac disease. To quantify cardiac function through ventricle volumes, masses, and cavity ejection function (EF), segmentation of left (LV) and right (RV) ventricles from cine-MRI is crucial for physicians and radiologists [2–4]. Manual segmentation of LV and RV which requires delineation slice by slice is a time consuming and error-prone task. Although various automatic segmentation techniques have been proposed to segment the cardiac structure, the LV and RV segmentation is still an open problem because of the poor image contrast across the desired ventricle boundaries [5, 6].
Active contour models and level set methods have been widely implemented to segment different biological structures from medical images [7–10]. Several desirable advantages exist for active contour models over other classical image segmentation methods, such as thresholding, edge detection and region grow. Firstly, active contour models have capable to achieve sub-pixel accuracy of object boundaries. Second, these models can be easily formulated in a principled energy minimization framework and facilitate incorporation of various prior knowledge, such as shape or intensity distribution, for robust image segmentation [11, 12]. Third, active contour models can provide smooth and closed contours as segmentation outcomes, which are essential for segmenting most of the biological structures and can be readily used for further applications, such as shape analysis and recognition [13].
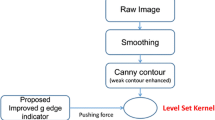
In order to segment the inner and outer contours of both ventricles from cine-MRI, we propose a two-level-set approach, which based on the strengths from DRLSE in [1]. In this new method, endocardial and epicardial contours are mathematically represented by two specified level contours of a level set function. Biventricular segmentation is expressed as an optimization problem of the level set function such that both level set contours best capture the biological structures of epicardium and endocardium.
This paper is further structured as follows: Sect. 2 describes the details of proposed algorithm, Sect. 3 proposes a two-step approach for segmentation of left and right ventricles and Sect. 4 presents the implementation details as well as segmentation results, which is followed by the concluding remarks in Sect. 5.
2 Two-Level-Set Approach
2.1 Anatomical Knowledge for Left and Right Ventricle Segmentation
First, we take account of the anatomy of the both ventricles in the formulation of proposed model. Endocardium is the innermost contour of the ventricle, which is a smooth membrane of endothelial cells that lines the cavities of the heart and the valves [14]. Myocardium is a thick layer of cardiac muscle which is responsible for the contraction and relaxation of the ventricles and atria, and this layer is composed almost completely of cardiomyocytes [3]. The outside of the myocardium is covered with a thin layer called the epicardium, which consists mostly of connective tissue and fat [3, 14].
Based on several informative observations of the cardiac anatomy, the desired segmentation outputs should fulfill the following two criteria: firstly the endocardial and epicardial contours should be smooth contours and secondly the interval between endocardial and epicardial contours should vary smoothly. In particular, the first criterion will be introduced to smooth the epicardial and endocardial contours individually, while the second criterion will be introduced to provide an interaction between the two contours such that the distance between them is gradually varying.
2.2 Edge Based Segmentation of LV and RV Using Two Distance Regularized Level Sets
We formulate the segmentation of both ventricles as an problem of seeking an optimal level set function such that its 0-level and k-level contours best fit the epicardial and endocardial contours respectively. What is more, according to the anatomical properties of endocardium and epicardium, as discussed in Sect. 2.1, the two-level-set function should satisfy the following two properties: first, the 0-level and k-level contours are smooth; and second, the distance between the 0-level and k-level contours is smoothly changing. For the above considerations, we propose a variational framework with an energy functional in the following form:
where \({\mathcal{L}}(\phi )\) is an energy functional defined from edge-based image information, such that it is minimized when the 0-level and k-level contours of the function \(\phi \) are on the endocardium and epicardium; \({\mathcal{A}}(\phi )\) is weighted area term and introduced to speed up the motion of the 0-level and k-level contour in the level set evolution process, which is necessary when the initial contour is placed far away from the desired object boundaries. Energy \({\mathcal{R}_p}(\phi )\) is the double-well potential defined in [1],
In this paper, we let the level set function \(\phi \) take positive values inside the 0-level contour \(C_0\) and negative values outside \(C_0\).
The energy \(\mathcal{L}(\phi )\) is defined by,
where g is an edge indicator function, which is defined as,
The above defined energy \(\mathcal{L}(\phi )\) computes the line integral of the function g along the 0-level and k-level contours. Obviously, this energy is minimized when the 0-level and k-level contours of the level set function \(\phi \) are located on desired object boundaries, where the function g takes smaller values than other non-edge locations. The weighted area term is defined by,
The minimization of the energy \(\mathcal{A}_g\) is achieved by shrinking and expanding the 0-level and k-level contours, depending on the sign of \(\alpha _0\) and \(\alpha _k\) of the banded region between the 0-level and k-level contour when they arrive at object boundaries where take larger values.
2.3 Energy Minimization
With the energy terms including \({\mathcal{L}}(\phi )\), \({\mathcal{A}}(\phi )\) and \({\mathcal{R}_p}(\phi )\) defined above, we propose to minimize the following energy functional:
This energy functional can be minimized by alternately minimizing \({\mathcal{F}}\) with respect to each of its variables. The energy minimization process starts with an initialization of the level set function \(\phi \) and the smooth function \(\alpha \). We minimise the function \({\mathcal{F}}\) with respect to \(\phi \) applying gradient flow method and get,
where \(d_p\) is the double-well potential defined in [1].
3 Application of Proposed Method to Segmentation of Left and Right Ventricles
In this section, we describe a two-step approach for segmentation of LV and RV. In the first step, we use the DRLSE to perform a preliminary segmentation of LV and RV to roughly locate the endocardial contours of the LV and RV. The level set function obtained in the first step is used as the initial level set function of the proposed level set evolution in the second step, with the 0-level and k-level contours representing the initial endocardial and epicardial contours, respectively. The final endocardial and epicardial contours of LV and RV is then obtained as the result of the level set evolution in the proposed model. The details of this two-step approach are described below.
3.1 Roughly Locate Endocardial Contours of LV and RV Using DRLSE
In the first step, we use distance regularized level set evolution (DRLSE) model to obtain a preliminary segmentation of left and right ventricles, which is then applied to define the initial level set function for the distance regularized two-level-set model described in Sect. 2. We put two square blocks inside LV and RV as the initialization of DRLSE. Then, the final zero level contour in DRLSE is evolved to capture the inner contours of both LV and RV.
3.2 Extraction of Both Endocardial and Epicardial Contours Using Proposed Model
The second step of our method aims to accurately capture both the endocardial contours as well as the epicardial contours of both LV and RV at the same time, based on the previously initialized results. With the above two-level-set representation of endocardial and epicardial contours, we implement the mathematical model discussed in Sect. 2 to accurately segment left and right ventricles from cine-MRI based on previously initialized contours from DRLSE method.
4 Experimental Results
In this section, we will demonstrate the implementation details as well as the segmentation results of the proposed two-step approach with the application to segment ventricles from cardiac cine-MRI data, where the data are obtained from MICCAI 2012 right ventricle segmentation challenge.
4.1 Parameters Selection
In first step, we set the parameters for the DRLSE model with \(\varDelta t=1\), \(\mu =0.2\), and \(\lambda =10\). In the second step, we numerically solve the level set evolution equation of proposed model presented in Eq. 1 by following a standard finite difference scheme proposed in [1]. The details of the selected parameters for the proposed model are disclosed in the following. The time step \(\varDelta t\) used in the approximation of temporal derivative is set to \(\varDelta t = 0.1\) in our implementation. For the datasets used in this chapter, we set the other parameters \(\alpha _0=-3\), \(\alpha _k=-3\), \(\rho = 3\), \(\mu = 1\), \(\omega = 0.5\), \(\lambda _1 = 0.002\), \(\lambda _2 = 0.05\), and \(\nu _1 = 0.001 \times 255 \times 255\), \(\nu _2 = 0.001 \times 255 \times 255\). The choice of the level \(k = 140\).
4.2 Segmentation Results
Results for MICCAI 2012 Right Ventricle Segmentation Challenge. Our two-step approach has been tested on the dataset of MICCAI 2012 right ventricle segmentation challenge (http://www.litislab.eu/rvsc), and this two-step approach has already showed the promising capability over other existing segmentation algorithms.
Figure 1 shows the illustrative segmentation results of both left and right ventricles from a selected patient cine-MRI using proposed method after applying DRLSE initialization, where each of these figures corresponds to one of the middle steps within iteration (iterates from left to right and from top to bottom). It is easy to find out the output of the proposed segmentation model (as shown in the bottom right figure in Fig. 1) has capable to accurately capture both the endocardial and epicardial contours, even in presence of the intensity inhomogeneity and image noises. Figure 2 shows the results and ground truth of the subject named P15 in the training set. We choose two slices at end-diastole and end-systole and we can reach promising results through our proposed method.
Table 1 shows evaluation results for MICCAI 2012 right ventricle segmentation challenge of our method with others. We use Dice metric (DSC) and Hausdorff distance (HD) assess the RV segmentation results. DSC is a measure of the overlap of two regions and is defined by
where \(|*|\) is the area of region \(*\). Hausdorff distance provides a symmetric distance measure of the maximal discrepancy between two labeled contours, which is defined as,
where d(a, b) donates Euclidean distance. From Table 1, our proposed method shows promising results comparing to the other three methods.
5 Conclusions
In this paper, we present the development of a new segmentation framework for left and right ventricles from cardiac MR short-axis images. This framework contains two key steps: firstly we apply the DRLSE method as initialization of endocardial contour and then we use the two-level-set model to accurately capture both endocardial and epicardial contours for LV and RV, simultaneously. Moreover, this proposed approach based on the strengths from the distance regularized level set evolution (DRLSE) method in [1]. Experimental results have demonstrated the effectiveness of this proposed two-step level set approach for segmenting cardiac left and right ventricles from cine-MRI.
References
Li, C., Xu, C., Gui, C., Fox, M.D.: Distance regularized level set evolution and its application to image segmentation. IEEE Trans. Image Process. 19, 3243–3254 (2010)
Paragios, N.: A variational approach for the segmentation of the left ventricle in cardiac image analysis. Int. J. Comput. Vis. 50, 345–362 (2002)
Petitjean, C., Dacher, J.N.: A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 15, 169–184 (2011)
Kurkure, U., Pednekar, A., Muthupillai, R., Flamm, S.D., Kakadiaris, I.A.: Localization and segmentation of left ventricle in cardiac cine-MR images. IEEE Trans. Biomed. Eng. 56, 1360–1370 (2009)
Qian, X., Lin, Y., Zhao, Y., Wang, J., Liu, J., Zhuang, X.: Segmentation of myocardium from cardiac mr images using a novel dynamic programming based segmentation method. Med. Phys. 42, 1424–1435 (2015)
Frangi, A.F., Niessen, W.J., Viergever, M.A.: Three-dimensional modelling for functional analysis of cardiac images: a review. IEEE Trans. Med. Image 20, 2–5 (2001)
Li, C., Kao, C., Gore, J.C., Ding, Z.: Minimization of region-scalable fitting energy for image segmentation. IEEE Trans. Image Process. 17, 1940–1949 (2008)
Chan, T., Vese, L.: Active contours without edges. IEEE Trans. Image Process. 10, 266–277 (2001)
Qian, X., Wang, J., Guo, S., Li, Q.: An active contour model for medical image segmentation with application to brain ct image. Med. Phys. 40, 8 (2012)
Kass, M., Witkin, A., Terzopoulos, D.: Snakes: Active contour models. Int. J. Comput. Vis. 1, 321–331 (1988)
Chen, Y., Tagare, H.D., Thiruvenkadam, S., Huang, F., Wilson, D., Gopinath, K.S., Richard Briggs, W., Geiser, E.A.: Using prior shapes in geometric active contours in a variational framework. IJCV 50, 315–328 (2002)
Leventon, M., Grimson, W., Faugeras, O.: Statistical shape influence in geodesic active contours. In: 2000 Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 1, pp. 316–323 (2000)
Wu, J., Brigham, K.G., Simon, M.A., Brigham, J.C.: An implementation of independent component analysis for 3d statistical shape analysis. Biomed. Sign. Process. Control 13, 345–356 (2014)
Jolly, M.: Automatic segmentation of the left ventricle in cardiac MR and CT images. Int. J. Comput. Vis. 70, 151–163 (2006)
Zuluaga, M.A., Cardoso, M.J., Ourselin, S.: Automatic right ventricle segmentation using multi-label fusion in cardiac mri. In: Workshop on RV Segmentation Challenge in Cardiac MRI Medical Image Computing and Computer-Assisted Intervention (2012)
Bai, W., Shi, W., O’Regan, D., Tong, T., Wang, H., Jamil-Copley, S., Peters, N., Rueckert, D.: A probabilistic patch-based label fusion model for multi-atlas segmentation with registration refinement: Application to cardiac mr images. IEEE Trans. Med. Imaging 32, 1302–1315 (2013)
Ringenberg, J., Deo, M., Devabhaktuni, V., Berenfeld, O., Boyers, P., Gold, J.: Fast, accurate, and fully automatic segmentation of the right ventricle in short-axis cardiac MRI. Comput. Med. Imaging Graph. 38, 190–201 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Liu, Y., Zhao, Y., Guo, S., Zhang, S., Li, C. (2015). Edge Based Segmentation of Left and Right Ventricles Using Two Distance Regularized Level Sets. In: Bebis, G., et al. Advances in Visual Computing. ISVC 2015. Lecture Notes in Computer Science(), vol 9475. Springer, Cham. https://doi.org/10.1007/978-3-319-27863-6_19
Download citation
DOI: https://doi.org/10.1007/978-3-319-27863-6_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27862-9
Online ISBN: 978-3-319-27863-6
eBook Packages: Computer ScienceComputer Science (R0)