Abstract
We present a geometric surface parameterization algorithm and several visualization techniques adapted to the problem of understanding the 4D peristaltic-like motion of the outflow tract (OFT) in an embryonic chick heart. We illustrated the techniques using data from hearts under normal conditions (four embryos), and hearts in which blood flow conditions are altered through OFT banding (four embryos). The overall goal is to create quantitative measures of the temporal heart-shape change both within a single subject and between multiple subjects. These measures will help elucidate how altering hemodynamic conditions changes the shape and motion of the OFT walls, which in turn influence the stresses and strains on the developing heart, causing it to develop differently. We take advantage of the tubular shape and periodic motion of the OFT to produce successively lower dimensional visualizations and quantifications of the cardiac motion.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Cardiac development depends on genetic programs that are modulated by hemodynamic forces and environmental factors. Since the heart starts pumping blood early during embryonic development [10, 20] cardiogenesis mainly occurs under blood flow conditions. Blood flow is essential for proper cardiac development, with altered flow or absence of flow leading to cardiac malformations [13, 24, 26].
At early stages of embryonic development the heart has a tubular structure that soon begins pumping blood, and then bends and loops to form a looping tubular heart, and eventually the four-chambered heart. Although it is known that altering blood flow causes heart defects [9, 21, 23] exactly how this happens is not well understood. The focus of the analysis techniques in this paper is understanding how the pumping tubular structure’s motion, specifically the Out Flow Tract (see Fig. 1), is altered by changing blood flow conditions, for example, by comparing normal embryos to ones with bands placed around them.
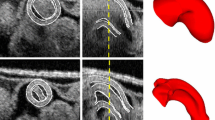
Heart outflow tract (OFT) surfaces reconstructed from OCT data: (a) Original OCT data with contours to illustrate the myocardial walls (external ring-like structure, with the outer and inner myocardium surfaces delineating the myocardium) and lumen-wall interface (bluish line). OFT surfaces depicted when the ventricular end of the OFT is (b) fully closed, (c) opening, (d) fully opened, and (e) closing (Color figure online).
The motion exhibited by the OF is both peristaltic-like (successive cross-sectional contraction) and longitudinal (the tube lengthens and shrinks). The aortic sac end of the OFT is relatively fixed in space, but the ventricular end moves — and, unfortunately, it moves in and out of the segmented volume. This motion makes consistent parameterization of the tube over time non-trivial. For this reason, we first present an algorithm (Sect. 4) that defines the parameterizations of the surfaces over time using domain-specific knowledge about how the tissue is deforming and the desired down-stream analysis.
Next, we extend the algorithm to support temporal alignment of multiple chick heart data sets. Once data sets are consistently parameterized, we apply data reduction and analysis techniques to examine OFT motion. Specifically, we employ area and volume analysis to produce both qualitative and quantitative measurements of the OFT contraction rate and how contraction travels along the OFT tube and derivative analysis to generate qualitative visualizations of how the OFT contraction and expansion occur.
Contributions: We present a consistent parameterization algorithm suited for analysis of cylindrical-like biological surfaces. Using these parameterizations, we develop analysis techniques which map complex surface data to 2D images and video streams, and then use standard image processing techniques to generate quantitative plots capturing global behavior.
2 Related Work
Previous work has been performed in the areas of parameterization techniques, and visualization approaches for biological data. Here we summarize previous works and their relevance to our study.
2.1 Parameterization Approaches
There are a wide variety of techniques for creating consistent parameterizations of a related collection of surfaces. The simplest approach is to map all the surfaces to a common domain or base mesh [12]. Some parameterization techniques are ad-hoc, but most are based on the minimization of a 2D surface metric, for example, conformal or area measures [2, 4–7, 17, 25], geodesics on a shape-space manifold [3, 16], or strain [22]. Alternatively, one can create a deformation of one shape to the other and minimize the 3D deformation energy [14, 28] or a statistical measure [1]. The techniques that come from the computer graphics or vision literature primarily deal with surfaces that have distinctive features, and aim to map those features on each surface [15]; unfortunately, we have no such features.
2.2 Visualization Approaches
For visualization purposes, the embryonic heart OFT is essentially a deforming tube. Visualizing characteristics of the OFT tube (e.g. curvature or some measure of texture) can be done by color-coding the tube surface and rotating the tube in order to view the tube from all sides. An obvious approach for visualizing the whole tube in a single image is to cut the tube surface open and lay it out flat in the plane, resulting in a 2D image that is easy to inspect and understand. This technique has been used, for example, in virtual colonoscopies [11, 29], in which the primary challenge is to effectively recreate the relatively complex geometry of the interior of the colon (e.g., polyps) while avoiding topology inaccuracies that arise from the scanning process. To better visualize the OFT tube, we employ the same unfolding technique.
Because our data has a temporal element we need to extend this approach A simple technique is to select several cross-sectional planes along the heart tube and plot either cross-sectional contours or areas within the contours versus time [8, 19]. This allows examination of the wall motion along the heart tube and how this motion changes over time and space. We extend this approach to visualize the entire temporal tube.
3 Data Acquisition
The data sets we use come from chick embryonic OFTs imaged using in-vivo OCT imaging (fully described in [18, 19, 27]). The final output from these procedures, which is used as input for this study, is a set of three 3D surface meshes (outer and inner myocardium surfaces and the lumen surface) at 195 time points over the entire cardiac cycle, which gives a total of \(3 \times 195=585\) surface meshes per embryo. At the embryonic stage studied here (approximately 3 days of incubation, HH18), the period of the cardiac cycle is about 400 ms, and thus the time span between consecutive meshes is about 2 ms. Note that these meshes are not in correspondence. We have eight data sets (four normal and four banded) which we use to validate our algorithms; we do not attempt to make strong biological claims from this small sample size.
4 Consistent Parameterization
In this section we describe how we generate a consistent mesh parameterization of the heart surfaces both temporally within one chicken embryonic OFT, and across multiple chicken embryos. We are interested in tracking the motion of the OFT over time (Fig. 2).
Producing a consistent parameterization of the OFT tube at four time points (\(T_A \) - minimum, \(T_C\) - maximum, \(T_B\) and \(T_D\) are mid-way between the two). The geodesic constraint results in a relatively uniform geodesic spacing of the grid lines. The inner curve of the tube (darkest line) is used to align the grid radially between time points and to prevent “twisting” of the parameterization.
Motivations and observations: While during the cardiac cycle the OFT tube moves in space, it remains roughly C-shaped. The longitudinal geodesics (traced from one end of the tube to the other) have a distinct minimum length along the inside of the C-shape (see Fig. 1 for side view). Further, the myocardium can be assumed to be incompressible, which enables estimation of tissue motion given that the aortic sac side of the OFT is fixed in space. Let \({M_t}\) represent the set of surfaces extracted from the 4D images by the segmentation algorithm. Informally, we want (A) a geodesic parameterization of these surfaces, (B) to use the curve on the inside of the tube (shortest geodesic length) as a stable feature for alignment, and (C) cross sections along the tube that do not intersect.
-
A
Let \(u,v \in [0,1] X [0,1]\) be the parameterization of the surface at each time, and let \(G_t\) be the mapping from the domain [0, 1] X [0, 1] to the surface \(M_t\) at time \(t \in [0,1]\). Specifically, u is the circumferential parameter (around the tube) and v is the longitudinal parameter (along the tube). Then even increments in u and v result in even (geodesic) increments along the surface.
-
B
For all time t, the curve \(G_t(0,v)\), \(v \in [0,1]\), corresponds to the shortest longitudinal geodesic on the surface \(G_t\). By shortest longitudinal geodesic we mean the shortest geodesic from the class of geodesics that connect a point on the inlet boundary to a point on the outlet boundary.
-
C
Define a cross section at \(V(v), v \in [0,1],\) by intersecting \(G_t\) with the plane defined by the three points \(G_t(0,v)\), \(G_t(1/3,v)\), and \(G_t(2/3,v)\) (essentially three equally-spaced points on the cross-section contour at v). Further we impose that the family of cross sections V for all v do not intersect inside of \(G_t\) (see Fig. 3).
Recall that, at each time t, we have three surface meshes representing the OFT wall: the outer and inner myocardial surfaces, and the lumen surface. Our algorithm creates a single, deforming mesh G for each of those three surfaces such that the shape of \(G_t\) matches the original surface \(M_t\) for each time step, but the parameterization of G has the properties given above.
Finding consistent cross-sectional planes that do not intersect. (a) Sketch of the OFT mesh with the geodesics highlighted. (b) Three equally spaced points along a circumferential geodesic, which define a cross-sectional plane. (c) Intersection of cross-sectional planes with the surface mesh at three locations along the OFT. (d) Cross-sectional contours for the three surfaces in a cross-sectional plane. (e) Using the centerline of the mesh results in planes that self-intersect within the OFT.
We represent G using a mesh that has a grid structure, with \(n \times m\) mesh nodes, with n nodes aligned around cross-sectional contours, and m nodes in the longitudinal direction, so that m is also the number of cross-sectional intersecting planes selected for generating the mesh. Note that cross-sectional planes are not necessarily perpendicular to the heart medial axis. The mesh is created by minimizing differences in geodesic distances among neighboring nodes, while ensuring that cross-sectional planes V do not intersect within \(G_t\) (see Fig. 3).
The minimization leading to the mesh parameterization should be done for each of the three OFT surfaces so that the outer and inner myocardial surfaces, and lumen surface, remain in correspondence. In practice, we parameterize the outer myocardial surface and then project the parameterization to the inner myocardial surface (for each vertex we found the closest point on the inner surface). Because the lumen surface folds, lumen parameterization must be done separately (the projection is not unique); we align the end points of the shortest lumen geodesic to the projection of the myocardial end points.
In what follows, we describe in more detail the algorithms for creating a deforming mesh for a single chick heart OFT over time, and for aligning deforming meshes between two (or more) chick heart OFTs. The parameterization itself provides geometric correspondence.
4.1 Single OFT Alignment Algorithm
Our input is a set of 195 surfaces, \(M_t\), consisting of outer myocardial meshes that are not consistently parameterized. For each \(M_t\), we find the shortest geodesic connecting any two end points on the tube boundary (see (B) above).
We cut the mesh along the shortest geodesic and map it to the unit square using Desbrun’s parameterization [2] with area weighting. We place the cut line so that it aligns with \(u=0\), satisfying (B) above. This mapping defines the parameterization \(G_t(u,v)\). It approximately satisfies the desired geodesic constraint (A), in that the mapping attempts to preserve area. (C) is satisfied by the construction of the planes from three equally-spaced points on the contour with constant v, provided the original surface \(M_t\) does not fold back on itself longitudinally in space. Informally, if the curves \(G_t(0,v)\), \(G_t(1/3,v)\), \(G_t(2/3,v)\) do not fold back on themselves then the sequence of triangles defined by those points will not intersect with each other.
To construct our deforming mesh from the parameterization we then place a uniformly-spaced grid (80 nodes in the circumferential direction, 50 in the longitudinal) on the flattened mesh and re-sample the mapped geometry to get the locations for the mesh at time t.
Following this procedure, however, the shortest geodesic can “slide” on the surface between time frames because of slight changes or noise in the tube geometry. To address this, we stabilize the end-points of the \(v=0\) cut line from frame to frame.
The re-parameterization of the existing meshes is then used to create a single, deforming surface that roughly maintains equal geodesic spacing and is temporally smooth. We next discuss finding consistent temporal and spatial alignment between multiple chick OFTs.
4.2 Temporal Alignment (Multiple Hearts)
Our goal is to define a starting point \(t=0\) that is at the same time in the cardiac cycle for all chicks. The heart-cycle has distinct points of maximum contraction (minimum cross-sectional surface area) and maximum expansion (maximum surface area). For each chick heart we find the point of maximal contraction by finding the outer myocardial surface with the minimum volume. We set this to be the \(t=0\) time point. While not perfect, this alignment is enough for our purposes.
4.3 Between Chick Spatial Alignment
Exact structural alignment between multiple chicks is, in general, not possible. This is because imaging varies from chick to chick. Although every effort has been made to consistently image the same portion of the heart, there are no features to align the images. This is compounded by imaging quality, which degrades as the OFT descends into the embryo. Even though we were careful to measure embryos at the same stage of development (HH18), the embryos do differ in size, which also influences how far into the embryo the OFT descends.
Within these constraints our parameterization can be used to align the OFTs in the radial direction, and provide roughly similar spacing in the longitudinal direction, even if the starting and ending points are not the same. Specifically, we can compare the overall change along and around the tube but we cannot guarantee that these plots image the exact same portion of the tube.
5 Cardiac Motion Visualization and Analysis
In this section we describe several visualization and quantification techniques for analyzing the motion of the heart OFT. OFT wall motion can be described as a peristaltic-like contraction wave that starts at the ventricular end and travels towards the aortic sac. Central to analyzing wall motion along the OFT is the ability to extract cross-sections of the OFT tube as outlined in Sect. 4.
Specific questions we are interested in answering are: Does the contraction wave travel at a uniform speed? Do contraction and later expansion happen in a similar fashion along the tube, or do they vary from one end to the other? How does the band affect contraction waves?
Using the parameterized meshes, we calculate the internal area of each cross-sectional contour (50 cross-sections at each of the 195 time points, using contours from the three OFT surfaces; see Fig. 3). Because for each surface the cross-sectional areas change both in time (t) and space (v), areas can be mapped into a 2D image of v (vertical direction) versus t (horizontal direction), mapping areas to colors (blue is smallest, red is largest) and plotting each contour’s area as a pixel in the 2D image (see Fig. 4A). From these images, we can readily visualize that areas in the ventricle end (bottom) are larger than at the aortic sac end (top), clearly demonstrating that the OFT is a tapered tube. Further, these images enable visualization of the contraction wave that travels through the OFT.
Two-dimensional cross-sectional area images and derived quantifications. (A) Cross-sectional area (color coded) as it changes over the cardiac cycle (horizontal direction) and along the OFT (vertical direction). We overlay on the 2D area image the point of maximum area (maximum OFT expansion) along with the points at which the cross-sectional area reaches 75 % of the maximum (crosses) or one standard deviation from the maximum (stars). (B) 2D plot showing, for each cross-section, maximum area along the OFT. The plot also shows the area that is one standard deviation from the maximum in comparison with 75 % maximal area along the OFT length. (C) 2D plot showing the percentage of time the OFT is expanded, that is OFT cross-sectional area is above 75 % of the maximal area or above one standard deviation from the maximum. (D) The derivative of the cross-sectional area image, with angle mapped to hue and magnitude mapped to intensity.
The 2D area images described above can be analyzed to extract characteristics of the OFT cardiac motion, such as peristaltic wave speed, contraction and expansion rates. Our approach to visualize and quantify cardiac motion is described next.
Peristaltic Wave Visualization: We extracted, for each OFT cross-section, the point at which the area reaches a maximum (the middle black line of Fig. 4A). These data can be further summarized as 2D plots showing the maximal area along the OFT medial axis (Fig. 4B). These plots are useful for comparing OFT sizes and how cross-sectional area changes along the OFT.
Contraction and Expansion Motion: Our next measure examines how long the OFT expansion/contraction lasts, as a percentage of the cardiac cycle. For this measure we tried two approaches, which yielded quantitatively similar results. Our first approach was to use the cross-sectional area data over time (for each position along the OFT), and fit a Gaussian curve centered on the maximum expansion time for that cross-section. We verified that the data was Gaussian-shaped by visually examining a plot of the data with the fitted Gaussian (see supplemental materials). Then, we found the time at which the area was one standard deviation from the maximum area (see Fig. 4A, stars). Our second approach was to use the same data but directly find the time at which the area is 75 % of the maximum (see Fig. 4A, crosses). These data were plotted together with maximum area along the OFT (Fig. 4B). Further, we generated 2D plots showing the time span over which area is expanded, by plotting the percentage of time the area is either above 75 % of maximum or above one standard deviation from the maximum (e.g. Fig. 4C). Of course, we can also chose to plot the percentage time in which the OFT area is contracted. These data are useful for comparing whether contraction/expansion timings change from heart to heart and along the OFT length.
Contraction and Expansion Rates: As one final visualization we can show the change in area by taking the image derivative of the 2D area images. The image derivative calculates the gradient of the area, which results in a vector field (vector components are the derivative of area with respect to time, t, and the derivative of area with respect to the circumferential direction, u). We map the angle of the gradient vector to hue and the magnitude of the vector to intensity (Fig. 4D). From these data, maximum cardiac expansion and contraction rates can also be extracted.
6 Conclusion
In this paper we have presented an algorithm for producing consistent spatial correspondence for the deforming heart OFT and extending this correspondence (both temporally and spatially) to multiple embryos, within the limitations of the imaging data. This algorithm is suitable for aligning other tubal surfaces undergoing deformation.
The primary purpose of the correspondence algorithm is to enable comparison within the cardiac cycle (single embryo) and between embryos. We have presented several visualization and analysis approaches that can be used to reduce the dimensionality of the data and quantify some aspects of the overall motion (e.g., peristaltic wave speed and cross sectional shape change).
References
Datar, M., Cates, J., Fletcher, P.T., Gouttard, S., Gerig, G., Whitaker, R.: Particle based shape regression of open surfaces with applications to developmental neuroimaging. In: Yang, G.-Z., Hawkes, D., Rueckert, D., Noble, A., Taylor, C. (eds.) MICCAI 2009, Part II. LNCS, vol. 5762, pp. 167–174. Springer, Heidelberg (2009)
Desbrun, M., Meyer, M., Alliez, P.: Intrinsic parameterizations of surface meshes. CGF 21(3), 209–218 (2002)
Drury, H.A., van Essen, D.C., Joshi, S.C., Miller, M.I.: Analysis and comparison of areal partitioning schemes using two-dimensional fluid deformations. NeuroImage 3, S130 (1996)
Drury, H., Van Essen, D.: Functional specializations in human cerebral cortex analyzed using the visible man surface-based atlas. Hum. Brain Mapp. 5(4), 233–237 (1997)
Eck, M., DeRose, T., Duchamp, T., Hoppe, H., Lounsbery, M., Stuetzle, W.: Multiresolution analysis of arbitrary meshes. In: SIGGRAPH 1995. pp. 173–182. ACM, New York, NY, USA (1995)
Essen, D.C.V., Drury, H.A., Dickson, J., Harwell, J., Hanlon, D., Anderson, C.H.: An integrated software suite for surface-based analyses of cerebral cortex. J. Am. Med. Inform. Assoc. 8(5), 443–459 (2001)
Fischl, B., Sereno, M.I., Tootell, R.B., Dale, A.M.: High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum. Brain Mapp. 8(4), 272–284 (1999)
Garita, B., Jenkins, M.W., Han, M., Zhou, C., Vanauker, M., Rollins, A.M., Watanabe, M., Fujimoto, J.G., Linask, K.K.: Blood flow dynamics of one cardiac cycle and relationship to mechanotransduction and trabeculation during heart looping. Am. J. Physiol. Heart Circulatory physiol. 300(3), H879–H891 (2011)
Goodwin, R.L., Biechler, S.V., Junor, L., Evans, A.N., Eberth, J.F., Price, R.L., Potts, J.D., Yost, M.J.: The impact of flow-induced forces on the morphogenesis of the outflow tract. Front. Physiol. 5(225), 1–9 (2014)
Hamburger, V., Hamilton, H.L.: A series of normal stages in the development of the chick embryo. J. Morphol. 88(1), 49–92 (1951)
Hong, W., Gu, X., Qiu, F., Jin, M., Kaufman, A.: Conformal virtual colon flattening. In: Proceedings of the 2006 ACM Symposium on Solid and Physical Modeling, SPM 2006, pp. 85–93. ACM, New York, NY, USA (2006)
Hormann, K., Polthier, K., Sheffer, A.: Mesh parameterization: theory and practice. In: ACM SIGGRAPH ASIA 2008 courses, pp. 47:1–47:87. SIGGRAPH Asia 2008 (2008)
Hove, J.R., Koster, R.W., Forouhar, A.S., Acevedo-Bolton, G., Fraser, S.E., Gharib, M.: Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature 421, 172–7 (2003)
Huang, Q.X., Adams, B., Wicke, M., Guibas, L.J.: Non-rigid registration under isometric deformations. In: SGP, pp. 1449–1457. Eurographics Association (2008)
van Kaick, O., Zhang, H., Hamarneh, G., Cohen-Or, D.: A survey on shape correspondence. In: Proceedings of Eurographics State-of-the-art Report, pp. 1–22 (2010)
Kurtek, S., Srivastava, A., Klassen, E., Laga, H.: Landmark-guided elastic shape analysis of spherically-parameterized surfaces. CGF 32(2.4), 429–438 (2013)
Lévy, B., Petitjean, S., Ray, N., Maillot, J.: Least squares conformal maps for automatic texture atlas generation. ACM TOG 21, 362–371 (2002)
Liu, A., Wang, R., Thornburg, K.L., Rugonyi, S.: Efficient postacquisition synchronization of 4-d nongated cardiac images obtained from optical coherence tomography: application to 4-d reconstruction of the chick embryonic heart. J. Biomed. Opt. 14(4), 044020 (2009)
Liu, A., Yin, X., Shi, L., Li, P., Thornburg, K.L., Wang, R., Rugonyi, S.: Biomechanics of the chick embryonic heart outflow tract at HH18 using 4D optical coherence tomography imaging and computational modeling. PLoS ONE 7(7), e40869 (2012)
Martinsen, B.J.: Reference guide to the stages of chick heart embryology. Dev. Dyn. 233(4), 1217–1237 (2005)
Midgett, M., Rugonyi, S.: Congenital heart malformations induced by hemodynamic altering surgical interventions. Front. Physiol. 5, 287 (2014)
Phan, L., Knutsen, A.K., Bayly, P.V., Rugonyi, S., Grimm, C.: Refining shape correspondence for similar objects using strain. In: EG 3D OR, pp. 17–24. Eurographics Association (2011)
Rothenberg, F., Fisher, S.A., Watanabe, M.: Sculpting the cardiac outflow tract. Birth Defects Res. C 69(1), 38–45 (2003)
Sedmera, D., Pexieder, T., Rychterova, V., Hu, N., Clark, E.B.: Remodeling of chick embryonic ventricular myoarchitecture under experimentally changed loading conditions. Anat. Rec. 254(2), 238–252 (1999)
Thompson, P., Mega, M., Woods, R., Blanton, R., Moussai, J., Zoumalan, C., Aron, J., Cummings, J., Toga, A.: A probabilistic atlas of the human brain in alzheimer’s disease: emerging patterns of variability, asymmetry and degeneration. NeuroImage 9, S597 (1999)
Tobita, K., Garrison, J.B., Liu, L.J., Tinney, J.P., Keller, B.B.: Three-dimensional myofiber architecture of the embryonic left ventricle during normal development and altered mechanical loads. Anat. Rec. Part A Discov. Mol. Cell. Evol. Biol. 283A(1), 193–201 (2005)
Yin, X., Liu, A., Thornburg, K.L., Wang, R.K., Rugonyi, S.: Extracting cardiac shapes and motion of the chick embryo heart outflow tract from four-dimensional optical coherence tomography images. J. Biomed. Opt. 17(9), 1–10 (2012)
Zhang, H., Sheffer, A., Cohen-Or, D., Zhou, Q., van Kaick, O., Tagliasacchi, A.: Deformation-driven shape correspondence. In: SGP, pp. 1431–1439. Eurographics Association (2008)
Zhao, J., Cao, L., Zhuang, T., Wang, G.: Digital eversion of a hollow structure: an application in virtual colonography. J. Biomed. Imag. 2008, 1–6 (2008)
Acknowledgment
This research was funded in part by NSF grants DBI-1052688 and IIS-1302142 and NIH grant R01-HL094570.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Phan, L., Grimm, C., Rugonyi, S. (2015). Visualization Techniques for the Developing Chicken Heart. In: Bebis, G., et al. Advances in Visual Computing. ISVC 2015. Lecture Notes in Computer Science(), vol 9474. Springer, Cham. https://doi.org/10.1007/978-3-319-27857-5_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-27857-5_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27856-8
Online ISBN: 978-3-319-27857-5
eBook Packages: Computer ScienceComputer Science (R0)