Abstract
DBSCAN is a well-known density based clustering algorithm, which can discover clusters of different shapes and sizes along with outliers. However, it suffers from major drawbacks like high computational cost, inability to find varied density clusters and dependency on user provided input density parameters. To address these issues, we propose a novel density based clustering algorithm titled, VDMR-DBSCAN (Varied Density MapReduce DBSCAN), a scalable DBSCAN algorithm using MapReduce which can detect varied density clusters with automatic computation of input density parameters. VDMR-DBSCAN divides the data into small partitions which are parallely processed on Hadoop platform. Thereafter, density variations in a partition are analyzed statistically to divide the data into groups of similar density called Density level sets (DLS). Input density parameters are estimated for each DLS, later DBSCAN is applied on each DLS using its corresponding density parameters. Most importantly, we propose a novel merging technique, which merges the similar density clusters present in different partitions and produces meaningful and compact clusters of varied density. We experimented on large and small synthetic datasets which well confirms the efficacy of our algorithm in terms of scalability and ability to find varied density clusters.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In today’s era, the amount of data produced by different applications is increasing at a very fast pace. As of 2012, \(90\,\%\) of the data in the world has been generated in the last two years alone [1]. There exists an immediate need to convert this huge data into some useful information and knowledge. Clustering is an important unsupervised learning technique in data mining which partitions the data objects into class of similar objects, based on the principle of maximizing intraclass similarity and minimizing interclass similarity [2]. For mining of huge data, clustering is well suited because unlike classification, it does not require labeling of large number of test tuples which is a very costly task. Clustering is broadly used for many applications like pattern recognition, data analysis, market research, image processing etc.
DBSCAN [3] is one of the popular density based clustering algorithm and has been widely used in past for various applications like land use detection, anomaly detection, spam identification, medical imaging, weather forecasting etc [4, 5]. Unlike partitioning based clustering algorithms, DBSCAN does not require apriori knowledge regarding number of clusters to be formed. It has good noise handling abilities and can discover non-convex clusters of arbitrary shapes and sizes. However, DBSCAN has three major drawbacks. First, it is computationally very expensive due to iterative neighborhood querying and faces scalability problems while processing large data. Second, the input density parameters \( \varepsilon \), MinPts, which directly influence the clustering results are specified by the user. Third, due to adoption of global \( \varepsilon \) value, it is unable to detect clusters of varied density.
To address these problems, many enhancements over DBSCAN have been proposed in literature, but they address only some of the limitations of DBSCAN, as discussed above. Today, the size of data is too huge that even a simple data analysis task faces scalability issues. Being no exception, DBSCAN also faces the scalability problem due to its high computational cost. Therefore, a need arises for parallelization of DBSCAN algorithm to reduce its processing time. Also, in huge data, it becomes a very challenging task to find good quality clusters with minimal domain knowledge. Poor quality clusters, even obtained in a very less time, are meaningless. Therefore, quality of clusters produced by a clustering algorithm are equally important as its processing time. Some parallel and distributed solutions for DBSCAN have been proposed in literature, but they are inefficient in dealing with varied density clusters. Hence, there arises a need of a density based clustering algorithm which is highly scalable and discovers varied density clusters with automatic computation of input parameters.
In this paper, we propose a density based clustering algorithm, titled VDMR-DBSCAN (Varied Density Map Reduce DBSCAN) which overcomes the major drawbacks of traditional DBSCAN and its various enhancements. VDMR-DBSCAN is designed on top of Hadoop [7] Platform and uses Google’s MapReduce [6] paradigm for parallelization. Varied density clusters are obtained using the concept of Density Level Partitioning. Most importantly, we have proposed a novel merging technique, which merges the similar density clusters present in different partitions and tends to produce meaningful and compact clusters of varied density. We evaluated VDMR-DBSCAN on large and small synthetic datasets. The results reveal that the proposed algorithm is highly scalable and detects varied density clusters with minimal requirement of domain knowledge.
The rest of the paper is organized as follows. Section 2 presents related work, which discusses the traditional DBSCAN and its various enhancements. In Sect. 3, our proposed algorithm VDMR-DBSCAN is discussed in detail. Section 4 presents experimental settings and results, followed by Sect. 5 which concludes the paper.
2 Related Work
In 1996, Ester et al. proposed DBSCAN [3], a well known density based clustering algorithm. It defines the density in terms of two user provided input parameters: \( \varepsilon \), MinPts. To find clusters, DBSCAN starts with an arbitrary unclassified point \(p_{i}\) and finds the points present in \( \varepsilon \)-neighborhood of \( p_{i} \) (\( NEps(p_{i})\)). If \(\vert NEps(p_{i}) \vert \) is greater than MinPts, then a cluster C is formed which includes all the points in \(NEps(p_{i})\), with \(p_{i}\) as a core point, otherwise \(p_{i}\) will be marked as a noise point. DBSCAN then iterates to expand the cluster C by collecting the density-reachable points from all core points present in C. This step repeats until no more points can be added to the cluster. The algorithm will continue until all the points in dataset are assigned to a cluster or labeled as noise point. DBSCAN has some drawbacks like unscalability, inability to find varied density clusters and dependency on user provided input density parameters.
In 1999, Ankerst et al. proposed OPTICS [8] to attack the varied density problem with DBSCAN by creating an augmented ordering of the data points, which represents its density based clustering structure. OPTICS does not produce clustering results explicitly and experimentally its runtime is almost constantly 1.6 times the run time of DBSCAN. Like DBSCAN, it also suffers from scalability problems. In 1999, Xu et al. proposed PDBSCAN [9], a master-slave-mode parallel implementation of DBSCAN. It works on shared nothing architecture where communication between nodes occur through message passing, thereby it leads to a large communication overhead while dealing with huge data. Also, it is unable to deal with clusters of varied density. In 2006, Uncu et al. proposed GRIDBSCAN [10] to solve the varied density problem with DBSCAN. Empirically, GRIDBSCAN is better than DBSCAN in terms of accuracy however, in terms of computational complexity, it is very expensive due to pairwise distance computations which makes it unsuitable for clustering of large data. In 2007, Liu et al. proposed VDBSCAN [11] to attack the varied density problem of DBSCAN. VDBSCAN is good at finding varied density clusters but it is not scalable and parameter k has to be subjectively chosen. In 2008, Mahran et al. proposed GriDBSCAN [12] to solve the scalability issues with DBSCAN. Data partitioning in GriDBSCAN generates a large number of redundant boundary points which degrades the execution efficiency of parallel algorithm and increases the merging time. Furthermore, it is unable to find varied density clusters and the input parameters for DBSCAN execution are provided by the user. In 2012, Xiong et al. proposed DBSCAN-DLP [13] to solve the varied density problem with DBSCAN using the concept of Density level partitioning. DBSCAN-DLP is computationally very expensive due to the computation of \(k^{th}\) nearest neighbor distances and density variations values, which make it unsuitable for large datasets. To address the scalability problem of DBSCAN, Dai et al. proposed DBSCAN-MR [14] in 2012, which is a MapReduce based parallel DBSCAN and uses Hadoop platform for parallel processing. DBSCAN-MR is highly scalable, but it is unable to find varied density clusters and input density parameters are need to be provided by user.
None of the above stated clustering techniques, is efficient enough to tackle all the three issues of DBSCAN. Therefore, we attempt to provide a solution by proposing a novel density based clustering algorithm VDMR-DBSCAN, which overcomes all these limitations.
3 Proposed Work
VDMR-DBSCAN is designed on top of Hadoop platform and uses MapReduce paradigm for parallelization. MapReduce improves the scalability of our algorithm by dividing large data into small partitions and sending those partitions to different nodes in the Hadoop cluster, where they can be processed independently. Later, the results from different nodes are aggregated to obtain final results.
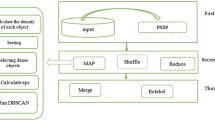
VDMR-DBSCAN framework consists of five different stages: Data Partitioning, Map, Reduce, Merge & Relabeling. In the first phase, input data is partitioned using PRBP (Partition with reduced boundary points) [14] partitioning, with an objective to minimize the boundary points. The data partitions are stored in HDFS (Hadoop Distributed File System) [7]. In the second phase, the partitions obtained are clustered independently using density level partitioning [13], to obtain clusters of varied density. Then, the local clusters present in different partitions are merged to obtain global clusters, followed by relabeling of data points. In the following subsections, we discuss different phases of VDMR-DBSCAN which is also shown in Fig. 1.
3.1 Partitioning Phase
VDMR-DBSCAN uses PRBP partitioning [14] to divide the data into small partitions which can be easily handled by a single node in the Hadoop cluster. It partitions the data such that two adjacent partitions share a common region i.e. split region. The points lying in the split region are called boundary points which help in identifying connected clusters present in two adjacent partitions. PRBP mainly focuses on minimizing the boundary points. It works by dividing each dimension in slices of equal width, followed by calculation of data distribution in each slice. Further, the slice ‘s’ with minimum points is selected to partition the data into two partitions. s being the common region (split region), is added to both the partitions. The data space is recursively split until the size of partition fits the node’s memory. We are using PRBP as a data partitioning algorithm for VDMR-DBSCAN with an intuition that reduced number of boundary points will reduce the merging time as well as map reduce time, which in result will improve the execution efficiency of VDMR-DBSCAN. The partitions created by PRBP partitioning are stored in HDFS (Hadoop Distributed File System) [7] from where each partition is read by a mapper in map phase.
3.2 Map Phase
In Map phase, each mapper reads a complete partition from HDFS in the form of (key, value) pair, where key = NULL and value = Partition. Further, each mapper uses Density level partitioning [13] to find varied density clusters, based on statistical characteristics of data points present in a partition. Density level partitioning partitions the data into different density level sets such that points with similar density belongs to same density level set (DLS).
The data is partitioned into different density level sets based on their \(k^{th}\) nearest neighbor distance values (kdist value), where value of k is chosen from the range 3 to 10 [13]. kdist value of a point is the measure of how far is the \(k^{th}\) nearest neighbor of that point, which gives an idea about the density around that point. Algorithm 1, shows the pseudocode for Map phase. Map phase proceeds by finding kdist values for all the points in a partition and stores them in kdistlist. The obtained kdistlist is sorted in ascending order. Further, for each adjacent points \(p_{i}\) and \(p_{j}\) in the kdistlist, \(DenVar( p_{i}, p_{j})\) (density difference between \( p_{i} \)&\( p_{j} \)) is computed using Eq. 1, to obtain DenVarList.
where, \(kdist(p_{j},k)\) is the \( k^{th}\) nearest neighbor distance of point \( p_{j} \).
To obtain the DLSs from DenVarlist, the values in DenVarlist which are larger than a threshold \( \tau \) are separated out and the points corresponding to these separated out DenVar values are put into separate DLS. The value of the threshold \( \tau \) is computed using the statistical characteristics of DenVarlist, as follows:
In Eq. 2, EX is the mathematical expectation, \( \omega \) is the tuning coeffiecient and SD is the standard deviation of the DenVarlist. Value of \( \omega \) is chosen from the range (0,3) [13]. The DLS so obtained are refined further by removing the DLSs of border and noise points and merging of DLSs with similar density levels. Based on the density characteristics of DLS, \( \varepsilon \) values are computed automatically for each DLS using Eq. 3 and stored in EpsList.
Here, maxkdist, meankdist and mediankdist are the maximum, mean and the median kdist values of DLS\( _{i} \) respectively.

Further, DBSCAN is applied on each DLS, using its corresponding \( \varepsilon \) values from EpsList and with \(MinPts = k\). DBSCAN iterates by selecting the initial seed points from the DLS only, but the neighborhood counting is done over all the unprocessed data objects. VDMR-DBSCAN improves the efficiency of traditional DBSCAN from \( O(n^{2})\) to O(n logn) by using KD-Tree [15] spatial index. After all the iterations, final clusters of varied density are obtained and the non-marked points are considered as noise points in the dataset.
The clustering results obtained after applying DBSCAN are divided into two regions: Boundary and local region. The clustering results of boundary region are used in merging of similar density clusters, which are present in adjacent partitions. The results of boundary region are passed to the reducer in the form of (key, value) pairs as (point_index, cluster_id + isCore_point + Eps_value + kdistvalues), where point_index is the index of the point, cluster_id is the cluster identification number for the point, isCore_point is a flag which indicates whether the point is core or not, Eps_value is the \( \varepsilon \) value of the cluster to which the point belongs. kdistvalues is the list of k nearest neighbor distances of a point in the partition. The clustering results of local region are stored in the local disk, in the form of key/value pair as (point_index, cluster_id).
3.3 Reduce Phase
Reduce phase finds the pairs of clusters from adjacent partitions, which can be merged in Merge phase. It outputs a Merge_Comb list of mergable cluster pairs and ensures the merging of density similar clusters only. Reduce phase collects the boundary points from Map phase and gathers all the points with same point index from different partitions. Points with same point index (key) are executed at the same reducer. Based on boundary point values, Reduce phase decides whether the two clusters which share a boundary point can be merged or not. However, the final merging will take place in Merge phase only. If two clusters, say, C1 and C2 share a boundary point b, then C1 and C2 forms a merge combination (MC), if they satisfy the following two criterion:
-
Boundary point should be a core point in at least one of the clusters.
-
The difference in the \( \varepsilon \) values of the two clusters should be smaller than a threshold, \( \alpha \).
A core point in the boundary region helps in identifying, if a cluster can be extended upto a different partition. Difference in \( \varepsilon \) values of the two clusters gives a measure of density difference between the two clusters. To ensure merging of similar density clusters, it creates a Merge_Comb list of only those clusters whose \( \varepsilon \) difference is less than \( \alpha \). The value of \( \alpha \) can be controlled, depending upon the quality of clusters required. Algorithm 2, gives a summary of steps involved in Reduce phase.

Non-Merged Clusters: In cases where the clusters are found to be unsuitable for merging then the boundary point which is part of both the clusters, should only be assigned to either of the clusters.
VDMR-DBSCAN solves this issue by using the kdistvalues list which is a part of Mapper’s output. kdistvalues contain the k nearest neighbor distances of a boundary point, in a partition. To find the global value of \(k^{th}\) nearest neighbor distance for the boundary point, kdistvalues of the point are collected from all partitions, in Reducer. The kdistvalues collected for a point, are combined to form a single list ( kdistlist) and sorted in ascending order where \( k^{th} \) value is picked from kdistlist to find \( k^{th} \) nearest neighbor distance for this boundary point. Further, difference between \( \varepsilon \) and kdist value is computed for all the clusters to which the point belongs. The point is assigned to the cluster with minimum absolute difference of \( \varepsilon \) and kdist values. Since, points belonging to same cluster are likely to have similar kdist values therefore, a point is more density similar to the cluster with minimum absolute difference of \( \varepsilon \) and kdist value. The cluster-id of such points are not part of Merge_Comb list and are written to the local disk with the output of local regions. At the end of Reduce phase, Merge_Comb list is written to the HDFS from where it is read by Merge phase for merging of clusters.
3.4 Merge Phase
Merge phase merges the MCs discovered by Reduce phase to identify the clusters which span over multiple partitions. It reads the input from HDFS as (key, Merge_Comb list), written by Reduce phase, where key is the point index and Merge_Comb list contains a list of MCs that can be merged. The output of this phase is a set of MergeList, where each MergeList represents the list of merged clusters. Assume, the output of reducer contains three MCs as \(\lbrace \) P1C1, P2C2 \( \rbrace \), \( \lbrace \) P5C2, P2C2 \(\rbrace \), \( \lbrace \) P4C2, P1C1 \( \rbrace \) which can be further merged, like \( \lbrace \) P1C1, P2C2 \(\rbrace \) can be merged with \( \lbrace \) P2C2, P5C2 \( \rbrace \) to form a MergeList \( \lbrace \) P1C1, P2C2, P5C2 \( \rbrace \), which can be further merged with \( \lbrace \) P4C2, P1C1 \( \rbrace \) to give a final MergeList as \( \lbrace \) P1C1, P2C2, P5C2, P4C2 \( \rbrace \). Merge phase identifies those MCs which can be merged further and merges them to produce a final set of MergeList.
Merge phase starts with finding difference between \( \varepsilon \) values of clusters, for all MCs in Merge_Comb list and sorts all of them in ascending order based on their \( \varepsilon \) difference value. Then, it first combines the one with the minimum \( \varepsilon \) difference i.e. the most density similar MC to form a MergeList. Further, all the MCs which have an intersection with this MergeList are found, where all MCs are already sorted in ascending order based on their \( \varepsilon \) difference values. The intersection pair with minimum \( \varepsilon \) difference is chosen and its \( \varepsilon \) difference is computed with all the clusters present in this MergeList. If the maximum \( \varepsilon \) difference is found to be less than the threshold \( \alpha \) then this MC is merged with the existing MergeList to form a new MergeList. Further, Merge phase proceeds by iterating on remaining MCs, until no more clusters can be merged.
Mathematically, it can be formulated as,
In Eq. 4, \(eps\_diff(C_{i}, C_{j})\) computes the dissimilarity in the density of clusters \( C _{i} ~ \& ~C_{j}\), where \(eps(C_{i})\) is the \( \varepsilon \) value of cluster \(C_{i}\). After merging of cluster \(C_{i}, C_{j}\), the eps_diff of resulting cluster with another \(C_{k}\) is computed as:
For Merging, \(eps\_diff((C_{i} \cup C_{j}), C_{k}) < \alpha \). Intuitively, this is similar to the complete link hierarchical clustering [2], where the distance between two clusters is determined by the most distant nodes in the two clusters. Similar to complete link hierarchical clustering, merge phase tends to produce compact (density compact) clusters and avoids long chain of clusters which would be meaningless.
For illustration of merging, consider Fig. 2 which represents six MCs namely, \(\lbrace C_{i}, C_{j} \rbrace \), \( \lbrace C_{j}, C_{k} \rbrace \), \( \lbrace C_{k}, C_{m} \rbrace \), \( \lbrace C_{i}, C_{k} \rbrace \), \( \lbrace C_{m}, C_{j} \rbrace \), \( \lbrace C_{i}, C_{m} \rbrace \), arranged in ascending order based on their eps_diff values. Here, lines between the clusters represent their eps_diff. Assuming, \( C_{i} \) and \(C_{j}\) has the least eps_diff, therefore, they are combined to form a MergeList. \(\lbrace C_{j}, C_{k} \rbrace \) is the next most density similar MC (least value of eps_diff), which has an intersection with \(\lbrace C_{i}, C_{j} \rbrace \) and its maximum eps_diff i.e. eps_diff \((C_{i}, C_{k})\) is less than \( \alpha \), therefore it is merged into MergeList. Now, the updated MergeList contains \(\lbrace C_{i}, C_{j}, C_{k} \rbrace \). The next most density similar intersection pair for the MergeList is \(\lbrace C_{k}, C_{m} \rbrace \) whereas, \(eps\_diff(C_{i}, C_{m})\) is greater than \( \alpha \), therefore \( C_{m} \) is not combined with the MergeList.
It can be observed from the above illustration, that a cluster can be combined to a list of clusters only if its eps_diff with all clusters in the list, is less than \( \alpha \), i.e. it is similar in density to all clusters in the list. In absence of this, it may lead to large variations of densities in a cluster and degrade the quality of cluster. Hence, it can be concluded that Merge phase avoids large variations in densities while merging of clusters and tends to provide compact clusters in terms of density.
3.5 Relabeling of Data Points
Once the clusters are combined in Merge phase, then all cluster-ids are sorted in ascending order and all clusters in the list are relabeled as the first one. For example, the clusters in the list \(\lbrace \) P1C1, P2C2, P5C2, P4C2 \(\rbrace \) are relabeled as P1C1. Both boundary points and points in local output are relabeled to correct cluster-ids.
4 Results and Discussions
In this section, we evaluate the performance of VDMR-DBSCAN on synthetic datasets of different densities and sizes and compare its clustering results with DBSCAN-DLP [13] and DBSCAN-MR [14] which are found to be the best versions of DBSCAN clustering algorithm in the current state of art.
4.1 Experimental Settings
We conducted the experiments on Hadoop cluster with 4 DataNodes and 1 NameNode where NameNode contains 8 GB RAM with intel i5 CPU, running Ubuntu-14.04 Linux operating system. DataNodes contain intel i5 CPU and 4 GB RAM with Ubuntu-14.04. For MapReduce platform, we used Hadoop-1.2.1 version on all the nodes. Both JobTracker & NameNode are configured on same node. We have used three synthetic datasets, to illustrate the performance of our proposed algorithm. For intuitive illustration, we have restricted to the datasets in 2 dimensions only. No suitable cluster validity index is found in literature for validating varied density clusters, therefore we have verified the results visually with 2 dimensional data only. We have used two small synthetic datasets: Zahn_compound (DS1) and Spiral dataset (DS2) [16] of varied densities. Figure 3(a) and (b) shows the unclustured data points of dataset DS1 & DS2 respectively. Zahn_compound dataset contains 399 data points whereas spiral dataset contains 312 data points. We have generated, one large 2-dimensional synthetic dataset (DS3) to illustrate the efficiency of proposed algorithm in finding varied density clusters on large data.
4.2 Experimental Results
In this section, the clustering results of VDMR-DBSCAN on above metioned datasets and its comparison with DBSCAN-DLP and DBSCAN-MR are discussed. Clustering results are represented through points in different colors and markers which indicate the different clusters discovered whereas dashed lines represent the data partitions obtained. In VDMR-DBSCAN k, \(\omega \), \(\alpha \) and \(\sigma \) are the user provided input parameters, whereas in DBSCAN-MR k, \( \varepsilon \), MinPts are provided by user. \(\rho \) is the number of partitions created by PRBP partitioning. Experimentally, \(k=4\) and \( \omega = 2.5 \) is found to be an ideal value for multi-density datasets [13].
4.3 Clustering Results on Zahn_compound Dataset (DS1)
Figure 4(a) shows eight varied density clusters obtained after applying VDMR-DBSCAN whereas, Fig. 4(b), shows the clusters obtained by applying DBSCAN-MR, which clearly illustrates the inability of DBSCAN-MR to find varied density clusters. DBSCAN-MR is unable to identify the points around the cyan colored cluster (marked as red colored cross in Fig. 4(b)), as a cluster and treats them as noise points whereas these points are identified as a separate cluster by VDMR-DBSCAN (represented by black dots in Fig. 4(a)). In DBSCAN-MR, clusters in green and red are formed by merging of small density clusters into a single cluster, which is due to relatively larger \( \varepsilon \) value.
Through clustering and merging of two partitions P1 & P2 (Fig. 5), we demostrate the working of VDMR-DBSCAN. Figure 5(a) and (b) show the clusters obtained by VDMR-DBSCAN on P1 and P2 respectively. On applying VDMR-DBSCAN on P1 (for k = 4 & \( \omega \) = 2.5), two \( \varepsilon \) values are generated. Cluster in blue points is generated by \( \varepsilon \) value of 0.40311 whereas red and green colored clusters are obtained by \( \varepsilon \) value of 2.20055 (Fig. 5(a)). In P2, four \( \varepsilon \) values are generated after applying VDMR-DBSCAN. The cluster in green is generated by \( \varepsilon \) value of 1.04209, cluster in magenta has an \( \varepsilon \) value of 2.00381, whereas yellow colored cluster which is formed in boundary region of partition P2 & P3 has an \( \varepsilon \) value of 2.84783 (Fig. 5(b)).
In Fig. 4(a), it is clearly visible that green colored cluster in P2 is merged with green colored cluster in partition P1, this is because the \( \varepsilon \) difference between both the clusters is 1.1585 \((2.20055 - 1.04209)\), which is less than \( \alpha \) (merging threshold), (1.4790). Similarly, red colored cluster in P1 is merged with magenta colored cluster in P2 with an \( \varepsilon \) difference of 0.19674 \((2.20055-2.00381)\) which is also less than \( \alpha \).
Varying Width of Partitioning Slice ( \(\varvec{\upsigma }\) ): Change in \( \sigma \) value, changes the number of partitions, data points in each partition, boundary region and number of boundary points. We have experimented with \( \sigma \) value of 2.2, 3.4 and 3.8 respectively. Figure 6(a), (b) & Fig. 4(a) show the clustering results of VDMR-DBSCAN on partitions created by slice width of 3.4, 2.2 and 3.8 respectively and the results obtained for different slice widths are found to be almost similar.
Figure 7(a), shows the clustering results of DBSCAN-MR with 5 partitions created by \( \sigma \) = 3.4. In DBSCAN-MR, the \( \varepsilon \) value used should be half of \( \sigma \), so a global \( \varepsilon \) value of 1.7 is used. As compared to the results obtained from VDMR-DBSCAN on the same partitions (Fig. 6(a)), DBSCAN-MR is able to discover only three clusters and rest are flagged as noise points. Due to a global \( \varepsilon \) value, DBSCAN-MR is unable to identify varied density clusters in the dataset. Figure 7(b) shows the clustering results of DBSCAN-MR on six partitions, created by \( \sigma \) = 2.2. It uses a global \( \varepsilon \) value of 1.1. Figure 4(b), shows the clustering results of DBSCAN-MR on three partitions created by \( \sigma \) = 3.8. From the above comparisons, we can clearly observe that VDMR-DBSCAN can efficiently find varied density clusters in a dataset whereas DBSCAN-MR is unable to find, due to single \( \varepsilon \) value. Also, the results of DBSCAN-MR depends on the \( \varepsilon \) value used, as explained above.
4.4 Clustering Results on Spiral Dataset (DS2)
In Fig. 8(a) and (b), clustering results of VDMR-DBSCAN and DBSCAN-MR are compared on DS2 with four partitions, which are created by \( \sigma \) value of 3. It is clearly evident from the two figures, that VDMR-DBSCAN is able to capture the minute density change in yellow and black, magenta and green, also blue and cyan colored clusters, whereas DBSCAN-MR results in noise points and is unable to differentiate between the varied density clusters.
Clustering Results of DBSCAN-DLP on Datasets DS1 & DS2: Results in Fig. 9(a) and (b) show the efficacy of DBSCAN-DLP in discovering varied density clusters. The results are comparable to that of VDMR-DBSCAN but the difference lies in scalability of the algorithm. As VDMR-DBSCAN partitions the data into p partitions and processes each partition parallelly before going for the merge phase, the time complexity is thus reduced by approximately a factor of p. Thus, VDMR-DBSCAN is scalable in comparison to DBSCAN-DLP which is partly evident from Table 1 also.
4.5 Large Synthetic Dataset (DS3)
In this section, clustering results of VDMR-DBSCAN on DS3 are discussed. DS3 consists of 499746 data points. It is synthetically generated to have four clusters of three different densities. Figure 10 shows the data points of DS3 on a two-dimensional plot.
Figure 11(a) shows the result of VDMR-DBSCAN on DS3. Before applying VDMR-DBSCAN the dataset is partitioned using PRBP partitioning. We tried the partitioning with different slice width and out of them slice width of 0.02 is chosen because it produced least number of boundary points. As a result of partitioning, the dataset is divided into 117 partitions with 89459 boundary points. VDMR-DBSCAN is then applied on these 117 partitions and it discovered four varied density clusters in the dataset which are represented by yellow, cyan, red and green colored clusters in Fig. 11(a). Figure 11(b) shows the clusters obtained by DBSCAN-MR on DS3. DBSCAN-MR is run on the partitions obtained, using a global \( \varepsilon \) value of 0.01. As a result, it discovered only three different clusters in the dataset. DBSCAN-MR has merged two clusters of different densities into a single cluster, which is colored as red in Fig. 11(b), whereas VDMR-DBSCAN has discovered it as two clusters (Fig. 11(a)) of red and cyan color.
From Table 1, it can be observed that VDMR-DBSCAN has slightly higher execution time than DBSCAN-MR, which is due to extra computations done by VDMR-DBSCAN during map, reduce and merge phase to discover varied clusters. DBSCAN-DLP has a very high execution time as compared to VDMR-DBSCAN and DBSCAN-MR, because it is not scalable which makes it computationally very expensive while dealing with large datasets. Hence, in terms of execution time, DBSCAN-MR and VDMR-DBSCAN are much more efficient than DBSCAN-DLP. The execution time of DBSCAN-MR and VDMR-DBSCAN are comparable but VDMR-DBSCAN is far better than DBSCAN-MR in terms of finding varied density clusters with automatic computation of input density parameters. Therefore, from above results it can be concluded that VDMR-DBSCAN is better than existing versions of DBSCAN clustering algorithm because it can efficiently find varied density clusters of different shapes and sizes in small as well as large datasets, in a very less time.
5 Conclusions and Future Work
In this paper, we proposed a novel density based clustering algorithm VDMR-DBSCAN, which is highly scalable and finds clusters of varied-density with automatic computation of input density parameters in massive data. VDMR-DBSCAN is designed on top of Hadoop Platform and uses MapReduce paradigm for parallelization. We also proposed a novel merging technique, which merges the similar density clusters present in different partitions and ensures meaningful and compact clusters of varied density. We proved the efficiency of proposed algorithm by experimenting on large and small synthetic datasets. Experimental results revealed that our algorithm is highly scalable and detects varied density clusters with minimal requirement of domain knowledge. One of the future work is to use VDMR-DBSCAN with different existing data partitioning techniques like ESP, CBP [17], to see if they improve the execution efficiency of VDMR-DBSCAN.
References
IBM. http://www-01.ibm.com/software/data/bigdata/what-is-big-data.html
Han, J., Kamber, M., Pei, J.: Data Mining: Concepts and Techniques. Elsevier, Amterdam (2011)
Ester, M., et al.: A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd 96(34), 226–231 (1996)
Birant, D., Kut, A.: ST-DBSCAN: an algorithm for clustering spatial-temporal data. Data Knowl. Eng. 60(1), 208–221 (2007)
Emre, C.M., Aslandogan, Y.A., Bergstresser, P.R.: Mining biomedical images with density-based clustering. In: 2005 International Conference on Information Technology: Coding and Computing, ITCC 2005, vol. 1. IEEE (2005)
Dean, J., Ghemawat, S.: MapReduce: simplified data processing on large clusters. Commun. ACM 51(1), 107–113 (2008)
Hadoop, W.T.: The Definitive Guide, 1st edn. OReilly Media Inc., Sebastopol (2009)
Ankerst, M., et al.: OPTICS: ordering points to identify the clustering structure. ACM Sigmod Rec. 28(2), 49–60 (1999)
Xu, X., Jger, J., Kriegel, H.-P.: A fast parallel clustering algorithm for large spatial databases. In: Guo, Y., Grossman, R. (eds.) High Performance Data Mining, pp. 263–290. Springer US, London (2002)
Uncu, O., et al.: GRIDBSCAN: grid density-based spatial clustering of applications with noise. In: 2006 IEEE International Conference on Systems, Man and Cybernetics, SMC 2006, vol. 4. IEEE (2006)
Liu, P., Zhou, D., Wu, N.: VDBSCAN: varied density based spatial clustering of applications with noise. In: 2007 International Conference on Service Systems and Service Management. IEEE (2007)
Mahran, S., Mahar, K.: Using grid for accelerating density-based clustering. In: 2008 8th IEEE International Conference on Computer and Information Technology, CIT 2008. IEEE (2008)
Xiong, Z., et al.: Multi-density DBSCAN algorithm based on density levels partitioning. J. Inf. Comput. Sci. 9(10), 2739–2749 (2012)
Dai, B.-R., Lin, I.: Efficient map/reduce-based DBSCAN algorithm with optimized data partition. In: 2012 IEEE 5th International Conference on Cloud Computing (CLOUD). IEEE (2012)
Gaede, V., Gnther, O.: Multidimensional access methods. ACM Comput. Surv. (CSUR) 30(2), 170–231 (1998)
University of Eastern Finland. http://cs.joensuu.fi/sipu/datasets/
He, Y., et al.: MR-DBSCAN: a scalable MapReduce-based DBSCAN algorithm for heavily skewed data. Front. Comput. Sci. 8(1), 83–99 (2014)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Bhardwaj, S., Dash, S.K. (2015). VDMR-DBSCAN: Varied Density MapReduce DBSCAN. In: Kumar, N., Bhatnagar, V. (eds) Big Data Analytics. BDA 2015. Lecture Notes in Computer Science(), vol 9498. Springer, Cham. https://doi.org/10.1007/978-3-319-27057-9_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-27057-9_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-27056-2
Online ISBN: 978-3-319-27057-9
eBook Packages: Computer ScienceComputer Science (R0)