Abstract
Microalgal lipid metabolism is of broad interest because microalgae accumulate large amounts of triacylglycerols (TAGs) that can be used for biodiesel production (Durrett et al Plant J 54(4):593–607, 2008; Hu et al Plant J 54(4):621–639, 2008). Additionally, green algae are close relatives of land plants and serve as models to understand conserved lipid metabolism pathways in the green lineage. The green alga Chlamydomonas reinhardtii (Chlamydomonas hereafter) is a powerful model organism for understanding algal lipid metabolism. Various methods have been used to screen Chlamydomonas mutants for lipid amount or composition, and for identification of the mutated loci in mutants of interest. In this chapter, we summarize the advantages and caveats for each of these methods with a focus on screens for mutants with perturbed TAG content. We also discuss technical opportunities and new tools that are becoming available for screens of mutants altered in TAG content or perturbed in other processes in Chlamydomonas.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Triacylglycerol
- Lipids
- High-throughput genetics
- Chlamydomonas
- Algae
- Nitrogen deprivation
- Lipid analysis
- Fluorescence-activated cell sorting
- Lipid droplets
Chlamydomonas Is a Powerful Model Organism for Answering Key Questions in Microalgal Lipid Metabolism
Chlamydomonas is a unicellular green alga that can be used to study a broad spectrum of biological processes, including photosynthesis, the algal carbon concentrating mechanism, cell motility, light reception, phototaxis, nutrient stress responses and the cell cycle. Chlamydomonas has a number of useful features that make it convenient for genetic characterization of these processes. It is haploid during vegetative growth, so phenotypes of mutants can be observed without the need to generate homozygous lines (Harris et al. 2009). The nuclear (Merchant et al. 2007), chloroplast (Maul et al. 2002) and mitochondrial (GenBank accession number U03843) genomes have all been sequenced and can be transformed (Boynton et al. 1988; Kindle et al. 1989; Randolph-Anderson et al. 1993). A collection of selection markers, expression cassettes and fluorescent tags has been developed (Harris et al. 2009; Rasala et al. 2014). Large numbers of nuclear mutants can be generated by chemical or random insertional mutagenesis (Tam and Lefebvre 1993). Chlamydomonas has a well-characterized and rapid mating cycle, allowing combination of mutations and the analysis of segregation patterns by tetrad and random spore analyses (Harris et al. 2009).
Chlamydomonas is also a powerful model system for studies of lipid metabolism. Four qualities make it particularly useful for this application: (1) Chlamydomonas can be induced to accumulate TAGs in lipid droplets under nutrient-deprived conditions, for example under nitrogen (N) deprivation (Moellering and Benning 2010; Wang et al. 2009). Thus, Chlamydomonas can be used to understand how TAG is synthesized and assembled into lipid droplets and how algal lipid metabolism is regulated. (2) Its lipid metabolism is primarily regulated by environmental conditions, unlike in land plants, where lipid synthesis and turnover are also tissue-dependent (To et al. 2012). This simplifies the design, analysis and interpretation of experiments. (3) Chlamydomonas is unicellular, which enables higher throughput mutant screens. (4) Chlamydomonas is a member of the green lineage, with thylakoids and a chloroplast similar to those of land plants in structure and composition. Thus, Chlamydomonas can be used to understand processes that are conserved between green algae and land plants, such as the export of fatty acids from the chloroplast (Riekhof et al. 2005).
A significant fraction of current Chlamydomonas lipid studies is focused on the discovery of genes involved in TAG metabolism. Candidate genes with possible functions in TAG metabolism have been found by homology search (Riekhof et al. 2005), transcript profiling under normal growth conditions and N-deprived conditions (Blaby et al. 2013; Boyle et al. 2012; Goodenough et al. 2014; Miller et al. 2010; Schmollinger et al. 2014), and proteomic characterization of lipid droplets (Moellering and Benning 2010; Nguyen et al. 2011). However, the functions of only a handful of these candidate genes have been confirmed experimentally (Boyle et al. 2012; Li et al. 2012c; Yoon et al. 2012). Additionally, mutants with defects in lipids have been identified in screens, but the genes responsible for the lipid phenotypes have only been identified in a small fraction of cases (Cagnon et al. 2013; Xie et al. 2014; Yan et al. 2013; Li et al. 2012c). A comprehensive understanding of Chlamydomonas TAG metabolism will require additional unbiased screens for TAG mutants, combined with efficient methods for identifying the genes disrupted in the hits. Below, we review existing screening and genotyping tools, and present some tools that are in development.
High-Throughput Genetic Analyses of TAG Metabolism Traditionally Include Three Parts
A traditional forward genetic study consists of three major stages: mutagenesis, phenotypic screen and identification of mutated genes. Typically, a screen starts by randomly mutagenizing wild-type cells by radiation, chemicals or DNA insertions (Fig. 10.1). The TAG content in mutants is then analyzed by direct or indirect methods, in single cells or in a clonal population of each mutant. As there is usually a tradeoff between the throughput and the accuracy of phenotypic analyses, phenotyping is often performed in two stages: a high-throughput screen that rapidly identifies candidate mutants (called “primary screen” here) followed by a lower-throughput but higher accuracy analysis of the hits (“secondary screen”). Once mutants with abnormal TAG content are identified, mutated genes can be identified. We will first discuss screen growth conditions and methods of measuring TAG content. We will then discuss available approaches for generating mutants and for identifying mutated genes.
A forward genetic study for mutants with abnormal TAG content consists of three steps. Mutants are generated from wild-type cells using random mutagenesis, followed by high-throughput phenotypic analysis (TAG content analysis in this case). Mutants with higher or lower amounts of TAG than wild-type cells, referred as “hits” in the figure and depicted as two cells with large and small lipid droplets, are isolated. The causative mutation in these mutants is then identified using map-based cloning, whole-genome sequencing or PCR-based methods. An example is given for an insertional mutant where a piece of exogenous DNA inserted into an exon of a gene results in perturbed TAG levels
Growth Conditions Can Affect TAG Content and the Classes of Mutants That Can Be Isolated
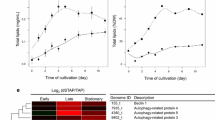
The most common environmental perturbation used for inducing TAG accumulation is N deprivation. Many other stress conditions such as high light and high salinity also induce TAG accumulation, but to a lesser extent (Fan et al. 2011). When N is replete, TAG is detectable but minimal. Under N deprivation, growth stops and TAG is gradually accumulated (Moellering and Benning 2010; Wang et al. 2009). Upon the resupply of N to N-deprived cells, growth resumes and TAG is gradually degraded or diluted by cell division until it reaches a level comparable with the level before N deprivation (Fig. 10.2a) (Cagnon et al. 2013; Li et al. 2012b, 2014; Tsai et al. 2014).
Mutants can be screened for aberrations in the content of TAG or other lipids under several conditions with a variety of approaches. (a) Three examples of mutants with abnormal TAG contents in a certain stage within an N deprivation/resupply cycle are shown. One mutant accumulates a significant amount of TAG without nitrogen deprivation. Another mutant accumulates more TAG than wild-type under nitrogen deprivation. A third mutant has a defect in degrading the TAG accumulated when nitrogen is re-supplied. Note that this is not an exhaustive list of TAG phenotypes, and some mutants exhibit phenotypes under more than one condition. (b–f) Several methods are available for identifying mutants with perturbed lipid content at any timeFig. 10.2 (continued) point. (b) In dot-thinFig. 10.2 (continued) layer chromatography (Dot-TLC), lipids are extracted and resolved radially. Each circle corresponds to a lipid species. A scheme is shown for eight mutants with two mutants depicted in black. Note that one of the two mutants over-accumulates one lipid species. (c) In gas chromatography-flame ionization detector (GC-FID) analysis, whole cell transesterification is performed, followed by gas chromatography separation of fatty acid methyl esters (FAMEs). Each FAME is depicted as one peak. FAME profiles are presented for two strains (the solid line depicts one strain; the dashed line depicts the other). Ratios between different FAMEs can be used to estimate the relative abundance of lipid species. The abundance of total FAMEs can also be obtained. (d) After staining with a lipid sensitive dye, cells of each mutant can be analyzed in bulk with the fluorescence quantified in 96-well format. (e) Flow cytometry allows the analysis of the lipid content of each cell of a mutant. Each dot in the graph represents a single cell. For each cell, the fluorescence emitted from a lipid sensitive dye is typically compared to chlorophyll fluorescence to offset the noise caused by extrinsic factors. Median fluorescence from the lipid sensitive dye across the whole population of cells is used as a metric of the TAG content of each mutant. (f) A mixed population of cells of different mutants is subjected to staining and fluorescence-activated cell sorting. Cells with high-TAG or low-TAG content (in the regions indicated by the red triangles) are isolated
Broadly speaking, a N starvation and re-feeding experiment can reveal mutants that differ from wild-type in up to three stages: (1) Under N-replete conditions, some mutants may already accumulate excess or insufficient TAG (e.g. mutant with high steady state TAG, Fig. 10.2a); such mutants could have mis-regulated signal transduction or metabolism during vegetative growth. Considering the low levels of TAG in wild-type Chlamydomonas cells under N-replete conditions, it would be challenging to screen for mutants with lower steady-state TAG levels. However, this may be possible, as one mutant with such a phenotype was isolated with reverse genetic approaches (Yoon et al. 2012). (2) When N is deprived, mutants with defective or enhanced TAG synthesis or signaling pathways will accumulate less or more TAG, respectively, than wild-type cells (e.g. mutant with high TAG induction, Fig. 10.2a). (3) Finally, one could also identify mutants that degrade TAG faster or slower than wild-type cells (e.g. mutants defective in TAG turnover, Fig. 10.2a). In fact, mutants in all three categories have been identified through forward or reverse genetic studies, demonstrating the power of this N deprivation and re-feeding process for revealing processes affecting TAG metabolism (Li et al. 2012c; Terashima et al. 2014; Tsai et al. 2014; Wang et al. 2009; Work et al. 2010; Boyle et al. 2012; Xie et al. 2014; Yan et al. 2013; Cagnon et al. 2013).
Primary Screens Have Assessed TAG Content by Lipid Stains, Lipid Chromatography or a Protein Bio-marker
The major technical goal in a primary screen is maximizing throughput while enriching for mutants of interest. Biochemical methods can quantify absolute TAG amount but are relatively time-consuming. Staining with lipid-sensitive dyes enables high-throughput analysis with a microtiter plate reader or flow cytometer, although one must be cautious, as artifacts can be caused by factors such as inefficient dye penetration.
Screens Based on Lipid Stains Enable Very High Throughput
Lipid-Binding Dyes Differ in Their Spectral Overlap with Chlorophyll, and Propensity for Photobleaching
Several fluorescent dyes have been used to stain lipid droplets in algae and other organisms. These include Nile Red, BODIPY493/503, BODIPY505/515 and LD540 (Moellering and Benning 2010; Spandl et al. 2009; Wang et al. 2009; Govender et al. 2012). For fluorometric estimation of TAG content in Chlamydomonas, the first concern is the spectral overlap between the dye and chlorophylls. Nile Red is the most frequently used dye for Chlamydomonas because Nile Red fluorescence and chlorophyll fluorescence minimally interfere with each other when certain excitation and emission wavelengths are selected (Terashima et al. 2014).
Another concern for a fluorescent dye is its propensity for photobleaching, which compromises the robustness of the measurement. In an in vitro study, photobleaching of LD540 was determined to be threefold slower than Nile Red and 15-fold slower than BODIPY493/503 (Thiele 2011). BODIPY505/515 was reported to photobleach slower than Nile Red (Govender et al. 2012).
To accurately reflect the amount of TAG in cells, the fluorescence from the dye needs to be linearly correlated with the amount of TAG present. In Chlamydomonas, one staining protocol has been shown to yield a decent correlation (R-squared value of 0.7) between the intensity of Nile Red fluorescence and TAG content (Cagnon et al. 2013).
Staining Can Be Improved by Permeabilizing Cells to Facilitate Dye Entry
One major challenge for quantitative staining of lipid droplets is ensuring that the dye can efficiently enter cells. Several treatments, such as heating by microwaving (Chen et al. 2011), a low amount of detergents (Terashima et al. 2014), or presence of organic solvents (Doan and Obbard 2011) have been reported to enhance the staining efficiency in Chlamydomonas or other algae. The concentration of the dye also needs to be optimized, as a very high concentration can cause fluorescence from dye precipitates in addition to lipid droplets (Cirulis et al. 2012).
Lipid Content Can Be Quantified with a Plate Reader or by Flow Cytometry
Three general formats have been used for screening mutants stained with fluorescent lipid-sensitive dyes: a microtiter plate reader, flow cytometry of individual mutants, and flow cytometry of pools of mutants (Fig. 10.2b). Li et al. and Yan et al. stained mutants in 96-well plates and measured fluorescence emitted by lipid-sensitive dyes with a plate reader (Li et al. 2012c; Yan et al. 2013). Cagnon et al. also stained mutants in 96-well plates, but individually analyzed each mutant by flow cytometry (Cagnon et al. 2013). Deviation of the mutant population from the wild-type population in the lipid-sensitive dye fluorescence dimension was considered a phenotype. As flow cytometry measures the fluorescence of single cells, Xie et al. and Terashima et al. isolated high-TAG or low-TAG mutants from a mixed pool of mutants using fluorescence-activated cell sorting (FACS) (Terashima et al. 2014; Xie et al. 2014). Combined with high-throughput sequencing, FACS of pools of mutants has the potential to reveal all the genes involved in TAG metabolism in a single experiment. The throughput and other features of the above methods are summarized in Table 10.1.
Lipid Species Can Be Separated Physically in Medium-Throughput Screens
In contrast to fluorophore staining, analysis of extracted lipids can reveal differences in the abundances of individual lipid species. Transesterification of lipids into fatty acid methyl esters (FAMEs) followed by gas chromatography-flame ionization detector (GC-FID ) analysis provides the composition as well as the amount of fatty acids contained in the lipid substrates used (Fig. 10.2b). For the purpose of high-throughput screens, direct transesterification with unprocessed algal cells has been developed. Fatty acid profile analysis has enabled the identification of the sole ω-3 fatty acid desaturase in Chlamydomonas (Nguyen et al. 2013; Pflaster et al. 2014). If the starting number of cells is known, this analysis yields the total amount of fatty acids per cell, which is often affected when TAG content is changed (Terashima et al. 2014). Additionally, certain fatty acid species are enriched in one lipid species compared to the others. The ratio of the abundance of a fatty acid species to the total abundance of all fatty acids serves as an indicator of the amount of the corresponding lipid. Because TAG is generally more saturated than membrane lipids, the ratio of C16:0 to C16:4 fatty acid was shown to correlate with TAG content (Liu et al. 2013).
Chlamydomonas studies could benefit from the adaptation of other high-throughput biochemical lipid screening techniques developed for other organisms. In Arabidopsis, Xu et al. screened for suppressors of a mutant deficient in synthesis of digalactosyldiacylglycerol (DGDG) by performing lipid extraction and TLC robotically (Xu et al. 2003). While lipids are resolved vertically in a traditional TLC, Xu et al. spotted lipid extracts on a TLC plate and applied a solvent mixture to each spot to separate lipid species radially outwards from the spot (Fig. 10.2b). This approach allows parallel analysis of 384 samples, and could be adapted to TAG mutant screens in algae, which are even more amenable to robotic operations.
Screens Based on Lipid Bio-markers Have Also Been Performed
High-throughput screens for lipid aberrations are also facilitated when a biomarker is available for the lipid of interest. The most abundant constituent of the Chlamydomonas lipid droplet proteome, Major Lipid Droplet Protein (MLDP), is highly correlated with TAG in abundance (Moellering and Benning 2010; Tsai et al. 2014). Tsai et al. screened for mutants with defects in TAG degradation by performing immuno-dot blots against MLDP in mutants arrayed in 96-well format. This allowed the identification of a mutant that exhibits a delay in TAG degradation (Tsai et al. 2014).
Raman Spectroscopy Has Potential for Single Cell-Based Screens
Since staining with fluorescent dyes can be biased by the cell penetration efficiency and provides little composition information on lipids, some groups have sought alternative approaches for rapid, single-cell lipid analysis. Single-cell Raman spectroscopy (SCRS ) is a label-free technique for analyzing the in vivo chemical profiles of single cells (Li et al. 2012a). It detects the emitted light from light-excited molecules, with each wave number shift representing a distinct mode of vibration from a specific molecular structure. In contrast to infrared spectroscopy, Raman spectrometry is not compromised by water molecules thus is suitable for analyzing live biological samples. Recently, several groups have developed techniques based on Raman spectroscopy for analysis of lipids in single algal cells and obtained information on lipid amount and unsaturation levels (Wang et al. 2014; Wu et al. 2011). In addition to lipids, SCRS is also capable of quantitatively measuring the concentration of starch in individual microalgal cells such as Chlamydomonas spp. and Chlorella spp. (Ji et al. 2014). Furthermore, new technologies that isolate and sort single cells based on their Raman spectrum have recently been demonstrated (Wang et al. 2013; Zhang et al. 2014a, 2015). The SCRS approach is label-free, non-invasive, culture-independent, rapid, low-cost, and potentially able to simultaneously track multiple metabolites in individual live cells. As throughput, ease of deployment, cost, and multiplexing are all important considerations in design of assays for detecting and measuring intracellular compounds, such advantages can be exploited for development of many novel applications such as screening of microbial cell factories and optimization of bioprocesses.
Quantitative Analysis and Microscopy Are Used to Confirm TAG Phenotypes
GC-FID Can Be Used to Analyze the Fatty Acid Composition of TAG
The profile of the fatty acids contained in TAG is very useful for determining which pathway is responsible for the extra or missing fatty acids. For example, the pgd1 mutant showed a major deficiency in the 18:1Δ9 fatty acid in TAG and other extraplastidic lipids, indicating a defect in exporting de novo synthesized fatty acids from the chloroplast rather than recycling of membrane lipid species (Li et al. 2012c). To analyze the fatty acid content of TAG specifically, instead of that of whole cells, lipids need to be extracted from cells and resolved, e.g. by TLC. The TAG fraction can then be transesterified and analyzed by GC-FID. In this way, the molar amount of TAG as well as the fatty acid profile of TAG can be determined. A major limitation of GC-FID is that it cannot identify the molecular species of TAG, because the origins of FAMEs are not traceable.
Mass Spectroscopy (MS) Can Be Used to Identify and Quantify TAG Species
MS has been increasingly utilized to quantify TAG content in Chlamydomonas (Liu et al. 2013; Terashima et al. 2014; Wang et al. 2009; Yoon et al. 2012; Vieler et al. 2007). Direct infusion MS minimizes oxidation of polyunsaturated fatty acid species (Danielewicz et al. 2011) while liquid chromatography (LC) prior to MS enables separation and identification of positional isomers of TAG (Kind et al. 2012; MacDougall et al. 2011). With ultrahigh performance liquid chromatography–tandem mass spectrometry (UHPLC–MS/MS), 140 molecular species of TAG were identified for Chlamydomonas. Quantification with this method yielded a consistently lower amount for TAG compared to GC-FID performed on the same samples, possibly due to the difference in ionization efficiency between different TAG species (Liu et al. 2013).
Microscopy Can Be Used to Characterize the Size and Quantity of Lipid Droplets
Hyper or hypo-accumulation of TAG is expected to affect the number or size of lipid droplets. Disruption of different pathways may affect one metric over the other, depending on whether the pathway mainly affects the initial generation or the enlargement of lipid droplets (Wilfling et al. 2013). These parameters can be measured by microscopy on cells stained with lipid sensitive dyes. In a lipid droplet morphology mutant screen performed on a Drosophila cell line, five categories of abnormal lipid droplet morphology were observed (Guo et al. 2008). Microscopy has not yet been used for large-scale primary screens in Chlamydomonas, but it has been employed to visualize the phenotypes of specific mutants. For example, the over-accumulation of TAG in the sta6 starchless mutant was initially observed as an increase in lipid droplet area by microscopy after Nile Red staining (Wang et al. 2009). Electron microscopy revealed a previously unknown association between lipid droplets and the chloroplast and allowed the discovery of large lipid droplets within the chloroplast in the sta6 mutant (Fan et al. 2011; Goodson et al. 2011).
Point Mutations and Insertional Mutations Can Be Used to Disrupt Genes
Point Mutations Can Be Generated by Irradiation or Chemical Mutagens
Point mutations can produce gain-of-function and temperature-sensitive mutants. In Chlamydomonas, point mutations are commonly induced using ultraviolet (UV) radiation or chemical mutagens such as ethyl methane sulfonate (EMS). A general concern with these two approaches is the extent of mutation, which is correlated with the percentage of cells killed during mutagenesis (the “kill rate”). A low kill rate generally means that a larger number of cells need to be mutagenized to cover the genome. A very high kill rate indicates a higher density of point mutations, which may complicate the identification of the mutation causing the phenotype, as many mutations will be genetically linked to the phenotype. EMS mutagenesis has been employed by two groups for the purpose of obtaining high-TAG strains (Xie et al. 2014; Lee et al. 2014).
Traditionally, point mutations are identified through map-based cloning, which requires multiple rounds of crossing and a high-density molecular marker system, which is under-developed in Chlamydomonas compared with the plant model A. thaliana (Jander et al. 2002; Rymarquis et al. 2005). In recent years, next-generation sequencing technology has greatly facilitated the mapping of point mutations. Tulin and Cross were able to map 52 mutants by crossing each mutant to wild-type and performing whole-genome sequencing on pools of 10–12 progeny that showed the mutant phenotype. Candidate mutations were identified based on the expectation that only the causative point mutation or tightly linked ones would be uniform in the mutant progeny population (Tulin and Cross 2014).
Insertional Mutagenesis Is Broadly Used in Chlamydomonas
Transformation of an insertion cassette containing a resistance marker into Chlamydomonas cells disrupts the genome at random sites (Tam and Lefebvre 1993; Zhang et al. 2014b). Thus, insertional mutagenesis has been extensively employed to understand many processes including TAG metabolism (Cagnon et al. 2013; Li et al. 2012c; Terashima et al. 2014; Tsai et al. 2014; Yan et al. 2013). General considerations regarding the insertion cassette sequence and transformation method are summarized below.
Selection Is Facilitated with Antibiotic Resistance Markers or Auxotrophy Markers
Commonly used antibiotic resistance markers for Chlamydomonas nuclear genome transformation include ble for resistance to bleomycin, aph7” for resistance to hygromycin B and aphVIII for resistance to paromomycin (Berthold et al. 2002; Sizova et al. 2001; Stevens et al. 1996). aph7” and aphVIII each encode an aminoglycoside phosphotransferase but both can be used in the same strain without interfering with each other (Berthold et al. 2002). Recently, the chloroplast marker AadA, which confers resistance to spectinomycin, has been recoded for the nuclear genome and can be used as a shuttle marker between E. coli and Chlamydomonas (Meslet-Cladiere and Vallon 2012). Other drug-selectable markers include mutant ALS, encoding a mutated form of acetolactate synthase that confers resistance to sulfometuron methyl (Kovar et al. 2002), and CRY1-1, encoding a mutant ribosomal protein that confers resistance to emetine (Nelson et al. 1994).
Besides drug resistance markers, auxotrophy markers are also available for auxotrophic strains. Introduction of the NIT1 gene (encoding the nitrate reductase) into a nit1 background strain enables it to grow on nitrate as the nitrogen source (Kindle et al. 1989); the ARG7 gene (encoding the argininosuccinate lyase) rescues the arginine auxotrophy of arg7 mutants (Debuchy et al. 1989); NIC7 (encoding a quinolinate synthetase) rescues the nicotinamide auxotrophy of nic7 mutants (Ferris 1995); and THI10 (encoding a hydroxyethylthiazole kinase) rescues thiamine auxotrophy of thi10 mutants (Ferris 1995).
Most drug resistance markers are several-fold smaller than NIT1 and ARG7 in size. This makes drug resistance markers easier to manipulate and potentially less vulnerable to endonuclease cleavage during transformation (as discussed later).
Expression of Selection Marker Genes Can Be Enhanced by Strong Promoters or Introns
A strong promoter is desired for abundant production of resistance proteins and optimal transformation efficiency. The Chlamydomonas PSAD promoter (Fischer and Rochaix 2001), RBCS2 promoter (Ribulose Bisphosphate Carboxylase/Oxygenase Small Subunit 2), β-2 tubulin promoter (Brunke et al. 1984; Kozminski et al. 1993) and a hybrid HSP70A-RBCS2 promoter (Schroda et al. 2000) drive high expression of nuclear genes and are extensively used to drive selection markers.
Besides a strong promoter , the presence of the first intron of RBCS2 upstream or downstream of the start codon was demonstrated to enhance the abundance of the marker protein and the transformation efficiency (Berthold et al. 2002; Lumbreras and Purton 1998).
The Extent of Gene Disruption Is Affected by the Gene Feature of the Insertion Site
Insertions into exons typically disrupt gene functions. However, in the absence of a transcriptional terminator, insertions into introns can be spliced out. Commonly, a gene with an insertion in an intron will produce a normal protein but at a lower abundance (Wang 2008).
DNA Can Be Delivered into the Chlamydomonas Genome by Biolistics, Glass Bead Transformation, Electroporation or Agrobacterium-Mediated Gene Transfer
Early transformations of the Chlamydomonas nuclear genome were mostly achieved using biolistic particle delivery . Auxotrophic mutants were successfully complemented with native Chlamydomonas genes using this method (Debuchy et al. 1989; Kindle et al. 1989). However, the transformation efficiency of the biolistic method is typically much lower than the methods developed later. Moreover, it requires a sophisticated particle bombardment system. Soon afterwards, it was observed that exogenous DNA could be delivered into Chlamydomonas genome by agitating the cell/DNA mixture with glass beads . This method does not require specialized equipment but only yielded ~1,000 colonies with 1 μg DNA used in a cell wall-deficient strain (Kindle 1990). A protocol based on electroporation was later developed, enabling transformation efficiencies of up to 100,000 colonies per μg DNA (Shimogawara et al. 1998). More recently, Agrobacterium tumefaciens , which has been primarily used for transferring DNA into dicotyledonous plants, was also shown to introduce DNA into the Chlamydomonas genome (Kumar et al. 2004).
The Number of Insertions Needed to Obtain the Desired Genome Coverage Depends on the Number of Insertions per Mutant
Before mutagenesis, it is important to decide how many mutants are needed to cover a certain fraction of the genome. Results from a simulation suggest that 120,000 insertions are required to cover 80 % of Chlamydomonas genes with one or more allele for each gene (Zhang et al. 2014b). The number of mutants required to cover a certain fraction of the genome can be reduced by increasing the number of insertions per mutant , which can be achieved using a higher amount of transforming DNA per cell (Shimogawara et al. 1998).
Insertion Sites Can Be Identified Using Plasmid Rescue or Various PCR-Based Methods
Several methods have been applied in Chlamydomonas for identification of insertion loci (Table 10.2). If the insertion cassette contains E. coli plasmid sequences, it can be retrieved by restriction digestion of the mutant genome, circularization and transformation back into E. coli (Tam and Lefebvre 1993). Sequencing of the Chlamydomonas genomic regions in the resulting plasmid can reveal the insertion site. This method does not involve PCR and can be used when flanking regions are hard to amplify. However, it requires transformation of an entire plasmid, including bacterial replication origin and bacterial selection marker in addition to the Chlamydomonas selection marker.
Multiple methods developed later use nested PCR to amplify the junction between the insertion and the genome flanking sequence followed by sequencing of the PCR products. Examples include thermal asymmetric interlaced PCR (TAIL-PCR) (Dent et al. 2005), restriction enzyme site-directed amplification PCR (RESDA-PCR) (Gonzalez-Ballester et al. 2005a), and SiteFinding PCR (Li et al. 2012c).
In cases where the insertion cassette lacks a transcriptional terminator, a chimeric mRNA will be produced, containing both the sequence of the selection marker gene and the genomic sequence between the marker gene and the first native transcriptional terminator encountered in the genome. 3′ rapid amplification of cDNA ends (3′ RACE) can be applied to recover the sequence downstream of the marker gene (Meslet-Cladiere and Vallon 2012). Both 3′ RACE and nested PCR methods only require thermal cycling that can be easily performed on multiple microtiter tubes in parallel.
One challenge with 3′ RACE and nested PCR approaches is that the distance between the annealing location of the degenerate primer and the insertion junction varies between mutants. In some mutants, the distance may be too large for robust amplification. Additionally, differences in PCR efficiency between different sizes of DNA makes it difficult to adapt these methods to pools of mutants.
Recently, we developed a method for mapping insertion cassettes in pools of tens of thousands of Chlamydomonas mutants (Zhang et al. 2014b). This method, named Chlamydomonas MmeI-based insertion site sequencing (ChlaMmeSeq), relies on the presence of recognition sites for the type IIS restriction enzyme MmeI at the two ends of the cassette. To extract flanking sequences, the genome is digested with MmeI, which binds in the cassette and cleaves 20–21 base pairs into the flanking DNA. This produces a fragment containing 20–21 bases of flanking sequence attached to the cassette, onto which an adapter can be ligated to allow PCR amplification of the flanking sequence. When coupled with high-throughput sequencing, ChlaMmeSeq enables parallel retrieval of 20–21 bp of flanking sequences from more than 10,000 mutants. Approximately 70 % of short flanking sequences obtained can be aligned to a unique locus in the Chlamydomonas genome.
Insertion Sites Are Frequently Messy
The complex nature of many insertions can make it difficult to identify the insertion site and the gene whose disruption causes the observed phenotype. Insertions can contain large deletions, truncated cassettes, and insertions of genomic sequence between the cassette(s) and flanking genomic DNA (Fig. 10.3). In an ideal insertion site, the insertion cassette is inserted without any deletion in the cassette sequence or the genomic sequence at the insertion site (Fig. 10.3a). In this scenario, flanking sequences can be obtained from both sides of the inserted cassette, and a single gene will be directly disrupted by the insertion. However, insertion sites are rarely this clean.
Insertion sites are often complex in Chlamydomonas. Models of cassette insertions in genomic sequence are presented. (a) The cassette is inserted cleanly into the genome. (b) An insertion can be associated with a deletion of genomic DNA. (c) A cassette can be truncated on one or both ends. (d) Genomic DNA from a distant locus (“junk DNA”, shown in yellow) can be inserted next to the cassette. (e) In addition to the mutation caused by cassette insertion, mutations can be present at one or more other loci. Note that a mutant may harbor a combination of these scenarios
Some insertions include a large deletion of genomic DNA that eliminates multiple genes. In such cases, one common strategy to identify the gene whose disruption causes the observed phenotype is to attempt complementation of the mutant with each of the disrupted genes (Fig. 10.3b) (Matsuo et al. 2008; Tsai et al. 2014).
Regardless of the presence of a deletion of genomic sequence, one or both sides of the insertion cassette can be truncated (Fig. 10.3c) (Zhang et al. 2014b). Such truncations can lead to the failure of flanking sequence extraction protocols, due to the loss of the necessary sequences, e.g. primer binding sites or sequences required for plasmid propagation in bacteria.
In addition to possible genomic deletions and cassette truncations, genomic DNA from a distant locus (referred as “junk DNA” here; Fig. 10.3d) can sometimes be found inserted between the cassette and the genomic sequence from the disrupted locus (Zhang et al. 2014b). The presence of junk DNA can make it challenging to identify the insertion site, as the DNA sequence flanking the cassette maps to the genome but does not indicate the true insertion site.
Last but not least, even when the insertion site can be correctly identified, the phenotype of a mutant could be caused by unmarked mutations, such as point mutations that occur during the transformation process (Fig. 10.3e). Thus, the causative relationship between the mutation and the phenotype should always be verified as suggested in the next part.
A Mapped Insertional Mutant Library Will Be a Transformative Tool
Our lab is developing an indexed Chlamydomonas insertional mutant library in which the insertion site is known for each mutant. Mutants are maintained as colony arrays on solid medium, and also preserved cryogenically. This resource will provide mutants in many genes of interest for reverse genetic studies. Additionally, this library will also be usable for various forward genetic screens, eliminating the time-consuming steps of mutant generation and mapping.
One scheme for a genome-saturating TAG mutant screen using this library takes advantage of a feature that enables tracking of the abundance of individual mutants in pools. The mutants in this library are generated with a cassette containing MmeI recognition sites at both ends, allowing the retrieval of a 20–21 bp flanking sequence from each end. If thousands of mutants are pooled together, the flanking sequence of each mutant can serve as a molecular barcode that indicates the abundance of that mutant in the pool (Fig. 10.4). To identify genes with a role in TAG metabolism, mutants can be combined into a single pool, and mutants with unusually high or low TAG content can be enriched by staining with a TAG-binding dye and Fluorescence-Activated Cell Sorting (FACS) (Terashima et al. 2014). Mutants enriched in a high-TAG or low-TAG pool can be identified because their barcodes will be enriched in those pools. Since the physical coordinates of each mutant in the library are already known prior to the screen, mutants in hit genes of interest can then be individually cherry-picked for further characterization.
An indexed library of insertional mutants will facilitate genetic screens for TAG phenotypes. Application of ChlaMmeSeq to each mutant produces a unique 20–21 bp genomic flanking sequence. This unique flanking sequence can serve as a barcode for detecting the presence or quantifying the abundance of the mutant in a pool. For a TAG mutant screen, the colonies from the library can be combined, grown and nitrogen deprived as a pooled sample. Fluorescence activated cell sorting (FACS) can be employed to sort out high-TAG mutants and low-TAG mutants as two pools of hits (depicted as cells with large and small lipid droplets, respectively). ChlaMmeSeq can then be applied to the two pooled samples to obtain insertion flanking sequences/barcodes. Individual mutants for genes of interest can be cherry-picked from the library for further studies
The Causative Relationship between the Mutation and the Phenotype Needs to Be Confirmed by Mutant Complementation
In the prior part, we mentioned the possibility that some mutations can be challenging to locate, and the mutation identified may not always be the one causing the phenotype. Thus, experiments should always be performed to confirm the causative relationship between the mutation and the mutant phenotype.
The Mutation Locus in Insertional Mutants Should Be Characterized by PCR
For insertional mutants, as shown in Fig. 10.3d, the flanking sequence obtained may not reveal the actual insertion site. To gain more confidence on the flanking sequence, one can attempt to retrieve longer flanking sequence beyond a possible junk DNA region. The challenge here is that this method can never tell conclusively that one has read into the actual flanking genomic sequence. A more robust approach is to obtain flanking sequences for both sides of the insertion and see whether they align to the same locus. If they do, it is extremely likely that the insertion is truly at that site. Finally, the most conclusive evidence that a region is disrupted is the absence of a wild-type product when the region spanning the insertion site is amplified by PCR.
Multiple Alleles Increase Confidence in a Genotype-Phenotype Link
When multiple independent mutants in the same gene exhibit the same phenotype, the likelihood that mutation of this gene causes the phenotype is greatly increased. Indeed, this principle is used extensively in Arabidopsis research (Krysan et al. 1999).
Genetic Linkage Analysis Can Be Used to Rule out Non-causative Mutations
In genetic linkage analysis, a mutant is crossed to the wild-type and the phenotypes and genotypes of progeny are determined. A non-causative mutation will typically be found in both progeny with and without the phenotype. However, co-segregation of the phenotype with a mutation of interest does not conclusively establish that the mutation of interest causes the phenotype, as the mutation of interest could merely be at a chromosomal location near the causative mutation.
Genetic Complementation Is Critical for Establishing the Causative Relationship between the Mutation and the Phenotype
The strongest evidence that a recessive mutation in a specific gene causes the observed phenotype is the observation that expression of the wild-type gene complements the mutant phenotype. To establish complementation robustly, multiple independent transformants of the wild-type gene into the mutant should be analyzed.
Investigators should be cautious as second site mutations can give the false appearance of complementation. During transformation, suppressor or reversion mutations can arise and revert the phenotype, giving the impression that the mutant has been complemented. To safeguard against this possibility, complementation transformations should include a parallel control transformation with a construct that is not expected to complement. If this latter construct produces colonies that revert the phenotype, reversion mutations are arising, and the investigator should perform multiple independent transformations and test whether the construct of interest yields a significantly greater number of wild-type-like transformants than the control construct.
Concluding Remarks
Algae are a promising platform for producing TAG for biodiesel. Many challenges remain to be overcome before algal biofuels are profitable, including our limited understanding of genes involved in algal TAG metabolism (Chisti 2007). The green alga Chlamydomonas is a powerful model organism for this process. However, due to difficulties in identifying the mutation sites, only a handful of genes have been identified with a role in TAG metabolism. Future efforts should focus on improving the throughput and robustness of screens and the success rate of mutation mapping.
More broadly, TAG serves as a universal storage lipid in eukaryotic organisms (Farese and Walther 2009), while glycerolipids are essential membrane components of all cells. Research on plant and animal lipid metabolism provides important insights into human nutrition and health. Yet, the multicellular nature of land plants and animals limits the throughput of genetic screens. For Chlamydomonas, in contrast, emerging tools including the genome-wide mutant library will enable high-throughput genome-saturating screens. We anticipate that future screens will reveal many conserved aspects of lipid metabolism.
References
Berthold P, Schmitt R, Mages W (2002) An engineered Streptomyces hygroscopicus aph 7″ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist 153(4):401–412. doi:10.1078/14344610260450136
Blaby IK, Glaesener AG, Mettler T, Fitz-Gibbon ST, Gallaher SD, Liu B, Boyle NR, Kropat J, Stitt M, Johnson S, Benning C, Pellegrini M, Casero D, Merchant SS (2013) Systems-level analysis of nitrogen starvation-induced modifications of carbon metabolism in a Chlamydomonas reinhardtii starchless mutant. Plant Cell 25(11):4305–4323. doi:10.1105/tpc.113.117580
Boyle NR, Page MD, Liu B, Blaby IK, Casero D, Kropat J, Cokus SJ, Hong-Hermesdorf A, Shaw J, Karpowicz SJ, Gallaher SD, Johnson S, Benning C, Pellegrini M, Grossman A, Merchant SS (2012) Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J Biol Chem 287(19):15811–15825. doi:10.1074/jbc.M111.334052
Boynton JE, Gillham NW, Harris EH, Hosler JP, Johnson AM, Jones AR, Randolph-Anderson BL, Robertson D, Klein TM, Shark KB et al (1988) Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 240(4858):1534–1538
Brunke KJ, Anthony JG, Sternberg EJ, Weeks DP (1984) Repeated consensus sequence and pseudopromoters in the four coordinately regulated tubulin genes of Chlamydomonas reinhardi. Mol Cell Biol 4(6):1115–1124
Cagnon C, Mirabella B, Nguyen HM, Beyly-Adriano A, Bouvet S, Cuine S, Beisson F, Peltier G, Li-Beisson Y (2013) Development of a forward genetic screen to isolate oil mutants in the green microalga Chlamydomonas reinhardtii. Biotechnol Biofuels 6(1):178. doi:10.1186/1754-6834-6-178
Chen W, Sommerfeld M, Hu Q (2011) Microwave-assisted nile red method for in vivo quantification of neutral lipids in microalgae. Bioresour Technol 102(1):135–141. doi:10.1016/j.biortech.2010.06.076
Chisti Y (2007) Biodiesel from microalgae. Biotechnol Adv 25(3):294–306. doi:10.1016/j.biotechadv.2007.02.001
Cirulis JT, Strasser BC, Scott JA, Ross GM (2012) Optimization of staining conditions for microalgae with three lipophilic dyes to reduce precipitation and fluorescence variability. Cytometry A 81:618–626
Danielewicz MA, Anderson LA, Franz AK (2011) Triacylglycerol profiling of marine microalgae by mass spectrometry. J Lipid Res 52(11):2101–2108. doi:10.1194/jlr.D018408
Debuchy R, Purton S, Rochaix JD (1989) The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the ARG7 locus. EMBO J 8(10):2803–2809
Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK (2005) Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol 137(2):545–556. doi:10.1104/pp. 104.055244
Doan TY, Obbard JP (2011) Improved Nile Red staining of Nannochloropsis sp. J Appl Phycol 23(5):895–901. doi:10.1007/s10811-010-9608-5
Durrett TP, Benning C, Ohlrogge J (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J 54(4):593–607. doi:10.1111/j.1365-313X.2008.03442.x
Fan J, Andre C, Xu C (2011) A chloroplast pathway for the de novo biosynthesis of triacylglycerol in Chlamydomonas reinhardtii. FEBS Lett 585(12):1985–1991. doi:10.1016/j.febslet.2011.05.018
Farese RV Jr, Walther TC (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139(5):855–860. doi:10.1016/j.cell.2009.11.005
Ferris PJ (1995) Localization of the nic-7, ac-29 and thi-10 genes within the mating-type locus of Chlamydomonas reinhardtii. Genetics 141(2):543–549
Fischer N, Rochaix JD (2001) The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics 265(5):888–894
Gonzalez-Ballester D, de Montaigu A, Galvan A, Fernandez E (2005a) Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal Biochem 340(2):330–335. doi:10.1016/j.ab.2005.01.031
González-Ballester D, de Montaigu A, Higuera JJ, Galván A, Fernández E (2005b) Functional genomics of the regulation of the nitrate assimilation pathway in Chlamydomonas. Plant Physiol 137:522–533. doi:10.1104/pp.104.050914
Goodenough U, Blaby I, Casero D, Gallaher SD, Goodson C, Johnson S, Lee JH, Merchant SS, Pellegrini M, Roth R, Rusch J, Singh M, Umen JG, Weiss TL, Wulan T (2014) The path to triacylglyceride obesity in the sta6 strain of Chlamydomonas reinhardtii. Eukaryot Cell 13(5):591–613. doi:10.1128/EC.00013-14
Goodson C, Roth R, Wang ZT, Goodenough U (2011) Structural correlates of cytoplasmic and chloroplast lipid body synthesis in Chlamydomonas reinhardtii and stimulation of lipid body production with acetate boost. Eukaryot Cell 10(12):1592–1606. doi:10.1128/EC.05242-11
Govender T, Ramanna L, Rawat I, Bux F (2012) BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresour Technol 114:507–511. doi:10.1016/j.biortech.2012.03.024
Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV (2008) Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453(7195):657–661. doi:10.1038/nature06928
Harris EH, Stern DB, Witman GB (2009) The Chlamydomonas sourcebook. Academic Press, Oxford
Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A (2008) Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J 54(4):621–639. doi:10.1111/j.1365-313X.2008.03492.x
Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129(2):440–450. doi:10.1104/pp. 003533
Ji Y, He Y, Cui Y, Wang T, Wang Y, Li Y, Huang WE, Xu J (2014) Raman spectroscopy provides a rapid, non-invasive method for quantitation of starch in live, unicellular microalgae. Biotechnol J 9(12):1512–1518. doi:10.1002/biot.201400165
Kind T, Meissen JK, Yang D, Nocito F, Vaniya A, Cheng YS, Vandergheynst JS, Fiehn O (2012) Qualitative analysis of algal secretions with multiple mass spectrometric platforms. J Chromatogr A 1244:139–147. doi:10.1016/j.chroma.2012.04.074
Kindle KL (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A 87(3):1228–1232
Kindle KL, Schnell RA, Fernandez E, Lefebvre PA (1989) Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol 109(6 Pt 1):2589–2601
Kovar JL, Zhang J, Funke RP, Weeks DP (2002) Molecular analysis of the acetolactate synthase gene of Chlamydomonas reinhardtii and development of a genetically engineered gene as a dominant selectable marker for genetic transformation. Plant J 29(1):109–117
Kozminski KG, Diener DR, Rosenbaum JL (1993) High level expression of nonacetylatable alpha-tubulin in Chlamydomonas reinhardtii. Cell Motil Cytoskeleton 25(2):158–170. doi:10.1002/cm.970250205
Krysan PJ, Young JC, Sussman MR (1999) T-DNA as an insertional mutagen in Arabidopsis. Plant Cell 11(12):2283–2290
Kumar SV, Misquitta RW, Reddy VS, Rao BJ, Rajam MV (2004) Genetic transformation of the green alga—Chlamydomonas reinhardtii by Agrobacterium tumefaciens. Plant Sci 166(3):731–738. doi:10.1111/pbi.12190
Lee B, Choi GG, Choi YE, Sung M, Park MS, Yang JW (2014) Enhancement of lipid productivity by ethyl methane sulfonate-mediated random mutagenesis and proteomic analysis in Chlamydomonas reinhardtii. Korean J Chem Eng 31(6):1036–1042. doi:10.1007/s11814-014-0007-5
Li M, Xu J, Romero-Gonzalez M, Banwart SA, Huang WE (2012a) Single cell Raman spectroscopy for cell sorting and imaging. Curr Opin Biotechnol 23(1):56–63. doi:10.1016/j.copbio.2011.11.019
Li X, Benning C, Kuo MH (2012b) Rapid triacylglycerol turnover in Chlamydomonas reinhardtii requires a lipase with broad substrate specificity. Eukaryot Cell 11(12):1451–1462. doi:10.1128/EC.00268-12
Li X, Moellering ER, Liu B, Johnny C, Fedewa M, Sears BB, Kuo MH, Benning C (2012c) A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 24(11):4670–4686. doi:10.1105/tpc.112.105106
Li X, Umen JG, Jonikas MC (2014) Waking sleeping algal cells. Proc Natl Acad Sci U S A 111(44):15610–15611. doi:10.1073/pnas.1418295111
Liu B, Vieler A, Li C, Jones AD, Benning C (2013) Triacylglycerol profiling of microalgae Chlamydomonas reinhardtii and Nannochloropsis oceanica. Bioresour Technol 146:310–316. doi:10.1016/j.biortech.2013.07.088
Lumbreras V, Purton S (1998) Recent advances in chlamydomonas transgenics. Protist 149(1):23–27. doi:10.1016/S1434-4610(98)70006-9
MacDougall KM, McNichol J, McGinn PJ, O’Leary SJ, Melanson JE (2011) Triacylglycerol profiling of microalgae strains for biofuel feedstock by liquid chromatography-high-resolution mass spectrometry. Anal Bioanal Chem 401(8):2609–2616. doi:10.1007/s00216-011-5376-6
Matsuo T, Okamoto K, Onai K, Niwa Y, Shimogawara K, Ishiura M (2008) A systematic forward genetic analysis identified components of the Chlamydomonas circadian system. Genes Dev 22(7):918–930. doi:10.1101/gad.1650408
Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB (2002) The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell 14(11):2659–2679
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren QH, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meir I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riano-Pachon DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan JM, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang PF, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo YG, Martinez D, Ngau WCA, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou KM, Grigoriev IV, Rokhsar DS, Grossman AR (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318(5848):245–251. doi:10.1126/science.1143609
Meslet-Cladiere L, Vallon O (2012) A new method to identify flanking sequence tags in chlamydomonas using 3′-RACE. Plant Methods 8(1):21. doi:10.1186/1746-4811-8-21
Miller R, Wu G, Deshpande RR, Vieler A, Gartner K, Li X, Moellering ER, Zauner S, Cornish AJ, Liu B, Bullard B, Sears BB, Kuo MH, Hegg EL, Shachar-Hill Y, Shiu SH, Benning C (2010) Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol 154(4):1737–1752. doi:10.1104/pp. 110.165159
Moellering ER, Benning C (2010) RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 9(1):97–106. doi:10.1128/EC.00203-09
Nelson JA, Savereide PB, Lefebvre PA (1994) The CRY1 gene in Chlamydomonas reinhardtii: structure and use as a dominant selectable marker for nuclear transformation. Mol Cell Biol 14(6):4011–4019
Nguyen HM, Baudet M, Cuine S, Adriano JM, Barthe D, Billon E, Bruley C, Beisson F, Peltier G, Ferro M, Li-Beisson Y (2011) Proteomic profiling of oil bodies isolated from the unicellular green microalga Chlamydomonas reinhardtii: with focus on proteins involved in lipid metabolism. Proteomics 11(21):4266–4273. doi:10.1002/pmic.201100114
Nguyen HM, Cuine S, Beyly-Adriano A, Legeret B, Billon E, Auroy P, Beisson F, Peltier G, Li-Beisson Y (2013) The green microalga Chlamydomonas reinhardtii has a single omega-3 fatty acid desaturase that localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol 163(2):914–928. doi:10.1104/pp. 113.223941
Pflaster EL, Schwabe MJ, Becker J, Wilkinson MS, Parmer A, Clemente TE, Cahoon EB, Riekhof WR (2014) A high-throughput fatty acid profiling screen reveals novel variations in fatty acid biosynthesis in Chlamydomonas reinhardtii and related algae. Eukaryot Cell 13(11):1431–1438. doi:10.1128/EC.00128-14
Randolph-Anderson BL, Boynton JE, Gillham NW, Harris EH, Johnson AM, Dorthu MP, Matagne RF (1993) Further characterization of the respiratory deficient dum-1 mutation of Chlamydomonas reinhardtii and its use as a recipient for mitochondrial transformation. Mol Gen Genet 236(2–3):235–244
Rasala BA, Chao SS, Pier M, Barrera DJ, Mayfield SP (2014) Enhanced genetic tools for engineering multigene traits into green algae. PLoS One 9(4):e94028. doi:10.1371/journal.pone.0094028
Riekhof WR, Sears BB, Benning C (2005) Annotation of genes involved in glycerolipid biosynthesis in Chlamydomonas reinhardtii: discovery of the betaine lipid synthase BTA1Cr. Eukaryot Cell 4(2):242–252. doi:10.1128/EC.4.2.242-252.2005
Rymarquis LA, Handley JM, Thomas M, Stern DB (2005) Beyond complementation. Map-based cloning in Chlamydomonas reinhardtii. Plant Physiol 137(2):557–566. doi:10.1104/pp. 104.054221
Schmollinger S, Muhlhaus T, Boyle NR, Blaby IK, Casero D, Mettler T, Moseley JL, Kropat J, Sommer F, Strenkert D, Hemme D, Pellegrini M, Grossman AR, Stitt M, Schroda M, Merchant SS (2014) Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 26(4):1410–1435. doi:10.1105/tpc.113.122523
Schroda M, Blocker D, Beck CF (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J 21(2):121–131
Shimogawara K, Fujiwara S, Grossman A, Usuda H (1998) High-efficiency transformation of Chlamydomonas reinhardtii by electroporation. Genetics 148(4):1821–1828
Sizova I, Fuhrmann M, Hegemann P (2001) A Streptomyces rimosus aphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene 277(1–2):221–229
Spandl J, White DJ, Peychl J, Thiele C (2009) Live cell multicolor imaging of lipid droplets with a new dye, LD540. Traffic 10(11):1579–1584. doi:10.1111/j.1600-0854.2009.00980.x
Stevens DR, Rochaix JD, Purton S (1996) The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol Gen Genet 251(1):23–30
Tam LW, Lefebvre PA (1993) Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135(2):375–384
Terashima M, Freeman ES, Jinkerson RE, Jonikas MC (2014) A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. doi:10.1111/tpj.12682
Thiele C (2011) Fluorescence-based imaging and analysis of cells and cellular components using lipophilic dyes with improved specificity, spectral property and photostability. EP20090010408
To A, Joubes J, Barthole G, Lecureuil A, Scagnelli A, Jasinski S, Lepiniec L, Baud S (2012) WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell 24(12):5007–5023. doi:10.1105/tpc.112.106120
Tsai CH, Warakanont J, Takeuchi T, Sears BB, Moellering ER, Benning C (2014) The protein Compromised Hydrolysis of Triacylglycerols 7 (CHT7) acts as a repressor of cellular quiescence in Chlamydomonas. Proc Natl Acad Sci U S A 111(44):15833–15838. doi:10.1073/pnas.1414567111
Tulin F, Cross FR (2014) A microbial avenue to cell cycle control in the plant superkingdom. Plant Cell 26(10):4019–4038. doi:10.1105/tpc.114.129312
Vieler A, Wilhelm C, Goss R, Suss R, Schiller J (2007) The lipid composition of the unicellular green alga Chlamydomonas reinhardtii and the diatom Cyclotella meneghiniana investigated by MALDI-TOF MS and TLC. Chem Phys Lipids 150(2):143–155. doi:10.1016/j.chemphyslip.2007.06.224
Wang YH (2008) How effective is T-DNA insertional mutagenesis in Arabidopsis? J Biochem Tech 1(1):11–20. doi:10.1111/pbi.12190
Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U (2009) Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell 8(12):1856–1868. doi:10.1128/EC.00272-09
Wang Y, Ji Y, Wharfe ES, Meadows RS, March P, Goodacre R, Xu J, Huang WE (2013) Raman activated cell ejection for isolation of single cells. Anal Chem 85(22):10697–10701. doi:10.1021/ac403107p
Wang T, Ji Y, Wang Y, Jia J, Li J, Huang S, Han D, Hu Q, Huang WE, Xu J (2014) Quantitative dynamics of triacylglycerol accumulation in microalgae populations at single-cell resolution revealed by Raman microspectroscopy. Biotechnol Biofuels 7:58. doi:10.1186/1754-6834-7-58
Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV Jr, Walther TC (2013) Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell 24(4):384–399. doi:10.1016/j.devcel.2013.01.013
Work VH, Radakovits R, Jinkerson RE, Meuser JE, Elliott LG, Vinyard DJ, Laurens LM, Dismukes GC, Posewitz MC (2010) Increased lipid accumulation in the Chlamydomonas reinhardtii sta7-10 starchless isoamylase mutant and increased carbohydrate synthesis in complemented strains. Eukaryot Cell 9(8):1251–1261. doi:10.1128/EC.00075-10
Wu H, Volponi JV, Oliver AE, Parikh AN, Simmons BA, Singh S (2011) In vivo lipidomics using single-cell Raman spectroscopy. Proc Natl Acad Sci U S A 108(9):3809–3814. doi:10.1073/pnas.1009043108
Xie B, Stessman D, Hart JH, Dong H, Wang Y, Wright DA, Nikolau BJ, Spalding MH, Halverson LJ (2014) High-throughput fluorescence-activated cell sorting for lipid hyperaccumulating Chlamydomonas reinhardtii mutants. Plant Biotechnol J 12(7):872–882. doi:10.1111/pbi.12190
Xu C, Fan J, Riekhof W, Froehlich JE, Benning C (2003) A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J 22(10):2370–2379. doi:10.1093/emboj/cdg234
Yan C, Fan J, Xu C (2013) Analysis of oil droplets in microalgae. Methods Cell Biol 116:71–82. doi:10.1016/B978-0-12-408051-5.00005-X
Yoon K, Han D, Li Y, Sommerfeld M, Hu Q (2012) Phospholipid:diacylglycerol acyltransferase is a multifunctional enzyme involved in membrane lipid turnover and degradation while synthesizing triacylglycerol in the unicellular green microalga Chlamydomonas reinhardtii. Plant Cell 24(9):3708–3724. doi:10.1105/tpc.112.100701
Zhang Q, Zhang P, Su Y, Mou C, Zhou T, Yang M, Xu J, Ma B (2014a) On-demand control of microfluidic flow via capillary-tuned solenoid microvalve suction. Lab Chip 14(24):4599–4603. doi:10.1039/c4lc00833b
Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, Jonikas MC (2014b) High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26(4):1398–1409. doi:10.1105/tpc.114.124099
Zhang P, Ren L, Zhang X, Shan Y, Wang Y, Ji Y, Yin H, Huang WE, Xu J, Ma B (2015) Raman-activated cell sorting based on dielectrophoretic single-cell trap and release. Anal Chem. doi:10.1021/ac503974e
Acknowledgements
We thank Arthur Grossman, Robert Jinkerson, Bensheng Liu, Liz Freeman Rosenzweig and Jian Xu for critical reading of the manuscript. This work was supported by the Carnegie Institution for Science and a grant from the National Science Foundation (MCB-1146621).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Li, X., Jonikas, M.C. (2016). High-Throughput Genetics Strategies for Identifying New Components of Lipid Metabolism in the Green Alga Chlamydomonas reinhardtii . In: Nakamura, Y., Li-Beisson, Y. (eds) Lipids in Plant and Algae Development. Subcellular Biochemistry, vol 86. Springer, Cham. https://doi.org/10.1007/978-3-319-25979-6_10
Download citation
DOI: https://doi.org/10.1007/978-3-319-25979-6_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-25977-2
Online ISBN: 978-3-319-25979-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)