Abstract
microRNAs (miRNAs) are a small class of ~22 nucleotide long RNAs, which control gene expression through repression of mRNA translation and induction of mRNA decay. One miRNA can potentially target up to several hundred mRNAs, which makes miRNAs powerful regulators of gene expression patterns rather than single genes. miRNAs are involved in almost every biological process, including self-renewal, pluripotency, reprogramming, and differentiation, and are therefore proposed to represent useful tools for regeneration. In this chapter, the biogenesis and function of miRNAs will be described, as well as their role in maintenance of self-renewal and pluripotency, reprogramming of somatic cells, and cardiac differentiation. Understanding the role of miRNAs during generation of iPS cells, cardiac differentiation of cardiac progenitor cells and pluripotent stem cells, and reprogramming of somatic cells, will help develop safe and efficient therapies for cardiac regeneration.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
microRNAs (miRNAs), ~22 nucleotide noncoding RNAs , are post-transcriptional regulators of gene expression that are involved in the regulation of almost every biological process, including self-renewal, pluripotency, and differentiation (Tüfekci et al. 2014). By inhibiting mRNA translation or inducing mRNA degradation, these small noncoding RNA molecules modulate gene expression. There are almost 2000 miRNAs encoded in the human genome. Each miRNA can target up to several hundred complementary mRNAs, giving miRNAs the ability to control gene expression patterns rather than single genes (Friedman et al. 2009). miRNA expression profiling studies have demonstrated that each cell type possesses a unique miRNA expression pattern, which results in different cellular characteristics (Liang et al. 2007). Because miRNAs control gene expression networks, they might represent a useful strategy to coordinate proliferation and differentiation of cells that are able to contribute to cardiac repair and regeneration.
This chapter will describe the mechanism of miRNA-mediated gene silencing and the role of miRNAs in the regulation of different processes, including pluripotency and self-renewal of stem cells, reprogramming of somatic cells, and differentiation of stem cells and progenitor cells into cardiovascular cell types. Altogether, this chapter will give insight into the potential of miRNAs for cardiac regeneration.
1 The Cellular Function of microRNAs
1.1 Introduction
The discovery of microRNAs (miRNAs) led to a radical change in the field of RNA molecular biology . Until 1993, there was no knowledge of one of the main mechanisms of gene regulation. Today, miRNAs are acknowledged as major regulators in many physiological processes (Tüfekci et al. 2014). The growing awareness of the importance of miRNAs in different biological processes has been followed by an explosion of scientific publications. In these publications, it has been estimated that over 60 % of the human protein-coding genes are regulated by miRNAs (Friedman et al. 2009). miRNAs were first discovered by Victor Ambros and colleagues who performed a genetic screen to investigate defects during Caenorhabditis elegans development . They discovered that the gene lin-4, a repressor of lin-14, did not encode a protein. Instead, lin-4 encoded a pair of small RNAs of ~22 and ~61 nucleotides in length. The longer RNA was proposed to be the precursor of the shorter RNA molecule. Moreover, lin-4 RNA exhibited antisense complementarity to sites in the 3′-untranslated region (UTR) of the lin-14 gene. This suggested an antisense regulatory mechanism (Lee et al. 1993). The shorter lin-4 RNA is now recognized as the first member of an elaborate group of small RNAs involved in gene suppression (Lee and Ambros 2001). However, before the turn of the century, there was no evidence for other small non-coding RNAs similar to lin-4. Fortunately, the discovery of Let-7 gave insight into the big world of these small RNAs. Let-7 encodes a 21 nucleotide small RNA involved in the transition to the adult stage of C. elegans (Reinhart et al. 2000), and also inhibits translation of its target by binding to the 3’-UTR (Abrahante et al. 2003). In contrast to lin-4, orthologs of Let-7 were found in different species, including molluscs, sea urchins, flies and humans (Hertel et al. 2012). This suggested a general and widespread role for small RNAs in the regulation of gene expression during development. Further studies have shown that small RNAs perform a regulatory function in almost every biological process (Tüfekci et al. 2014). In 2001, Lagos-Quintana et al. proposed the term ‘microRNAs’ for this still growing family of small RNAs (Lagos-Quintana et al. 2001). Currently, the latest version of miRBase (mirbase.org) contains 1881 mature human miRNAs (Kozomara and Griffiths-Jones 2014). Therefore, miRNAs compose one of the largest families of gene regulatory molecules in the human genome.
1.2 Biogenesis of microRNAs
In the most basic definition, miRNAs are small non-coding RNAs of approximately 22 nucleotides long, which control gene expression by inhibiting messenger RNA (mRNA) translation and inducing mRNA instability and degradation (Huntzinger and Izaurralde 2011). Computational predictions of microRNA targets estimate that each human miRNA can control up to hundreds of different mRNAs (Xu et al. 2014). In the human genome, approximately one third of the miRNAs have their own promoter. Other microRNAs are found in exonic or intronic region s of either non-coding or coding genes (Saini et al. 2007). The majority of intronic microRNAs are transcribed from the same promoter as their host gene. However, approximately one third of the intronic miRNAs are transcribed from an independent promoter, allowing for more regulation of their transcription (Ozsolak et al. 2008).
miRNA biogenesis starts with the synthesis of a primary miRNA (pri-miRNA ) transcript, consisting of up to several thousand nucleotides that contain stem-loop structures and flanking single strand segments. Pri-miRNAs are largely transcribed by RNA polymerase II, as capped and polyadenylated transcripts (Cai et al. 2004). Pri-miRNAs can contain one stem loop structure (pre-miRNA) or a cluster of several stem loop structures. Pri-miRNAs are processed by the microprocessor-complex into −70 nucleotide pre-miRNA within the nucleus. The major components of the microprocessor-complex are the RNase type III endonuclease Drosha and cofactor Di George syndrome critical region 8 (DGCR8 ). DGCR8 interacts with the pri-miRNA at the junction of the stem loop and the single strand segments, and directs Drosha to cleave approximately 11 base pairs (bp) downstream of this junction (Han et al. 2006). In turn, Drosha cleaves the hairpin secondary structure out of the pri-miRNA, generating pre-miRNA with 3′ overhangs (Han et al. 2004).
After pri-miRNA processing by the microprocessor, the resulting ~70 nucleotide hairpin pre-miRNA is transported out of the nucleus by Exportin 5, a RAN-GTP dependent nucleo/cytoplasmatic cargo transporter (Lund et al. 2004). A small subclass of miRNAs, located in intronic regions (mirtrons), can bypass processing by the microprocessor and are directly transported to the cytoplasm by Exportin 5 (Westholm and Lai 2011). In the cytoplasm, the pre-miRNA is again processed by an RNase type III enzyme, called Dicer, which recognizes the 3′ overhang of the pre-miRNA and cleaves within the stem loop, generating an imperfect ~22 nucleotide miRNA-duplex. Dicer associates with dsRNA binding domain proteins, transactivating response RNA-binding protein (TRBP) and protein activator of PKR (PACT), which assist with Dicer’s cleavage precision, facilitating accurate cleavage of miRNA duplexes (Wilson et al. 2014). Subsequently, this complex recruits one of the Argonaute proteins (AGO1-4 ) and a GW182 protein (Pfaff et al. 2013), which form the key components of the multi-protein miRNA-induced silencing complex (miRISC) (Chendrimada et al. 2005). The function of the miRISC is to select and recruit one of the strands of the miRNA duplex, which guides the catalytic complex to its complementary mRNA. In result, the miRISC induces inhibition of translation and/or instability and subsequent degradation of the mRNA (Huntzinger and Izaurralde 2011). Fig. 6.1 displays the biogenesis of miRNAs.
The miRNA biogenesis pathway. In the nucleus, pri-miRNAs are transcribed by RNA polymerase II as capped (m7G, 7-methylguanosine-cap) and polyadenylated transcripts. The pri-miRNA can consist of one pre-miRNA or a cluster of pre-miRNAs. Processing of the pri-miRNA is catalyzed by the micro-processor complex (Drosha and DGCR8) inside the nucleus. Following micro-processing, the pre-miRNA is transported out of the nucleus by RAN-GTP dependent transporter Exportin 5. In the cytoplasm, the pre-miRNA is processed by Dicer, assisted by TRBP and PACT. The generated miRNA duplex is loaded into the miRISC, composed of an Ago protein (AGO1-4) and GW182. This is followed by guide strand selection. The mature miRNA inside the miRISC (red) guides the complex to the target mRNA
1.3 microRNA Target Recognition
The miRNA within the miRISC targets mRNA molecules to induce miRNA-mediated translational repression as well as miRNA-mediated mRNA decay. Target interaction between mRNAs and miRNAs involves seed-pairing between nucleotides of both strands. Recognition of the mRNA target does not require perfect complementarity. In mammals, mRNA target recognition generally involves mRNA sites in the 3′-UTR that base-pair with approximately 7 nucleotides near the 5′-end of the miRNA. These miRNA nucleotides are termed ‘the seed sequence’ (Lewis et al. 2005). However, miRNA target sites in the coding sequence of mRNAs have also been discovered and miRNA target sites that cannot be explained by this canonical seed-pairing model also exist. This makes miRNA target identification a very difficult task (Thomas et al. 2010). Even though the miRNA guides the miRISC, the protein components of the miRISC execute the silencing of target mRNAs.
1.4 microRNA-Induced Silencing Complex
The first miRNA, lin-4, was reported to inhibit translation of lin-14 mRNA without destabilizing the mRNA molecule (Lee et al. 1993). There have been several studies using C. elegans and mammalian cell cultures that only observed translational repression at the initiation of translation (Ding and Grosshans 2009; Bhattacharyya et al. 2006). In contrast, others reported miRNA-mediated deadenylation, decapping, and subsequent decay of miRNA-target molecules (Eulalio et al. 2009). Over the past few years, there has been growing evidence for translational repression as well as mRNA decay by miRNAs (Huntzinger and Izaurralde 2011).
Recent studies on translational repression of miRNAs propose that the miRISC interferes with the function of the cap-binding complex. Mammalian mRNAs exist as capped and polyadenylated transcripts that are circularized by protein complexes interacting with the 5′-cap structure (comprising the cap-binding complex) and the 3′-poly(A)-tail. Circularization of mRNAs is necessary for efficient translation and interfering with the cap-binding complex can result in loss of circularization and subsequent translational repression (Mathonnet et al. 2007; Fukao et al. 2014). In addition, GW182 proteins are known to interact with poly(A) binding proteins (PABP ). Recently, it was found that interaction between GW182 and PABP leads to disassociation of PABP from the mRNA molecule. This can cause disruption of mRNA circularization and inhibit efficient translation, hereby facilitating translational repression (Zekri et al. 2013).
A second mechanism by which miRNAs regulate target mRNAs is mRNA destabilization and subsequent mRNA decay. This process is initiated by deadenylation of the mRNA molecule by one of the deadenylase complexes recruited by GW182. GW182 interacts directly with two deadenylase complexes, CCR4-NOT and, to a lesser extent, PAN2-PAN3 (Braun et al. 2011). The deadenylase complexes produce an unstable mRNA molecule without a poly(A)-tail. Subsequently, the miRISC proteins initiate removal of the 5′-cap (m7G) of the target mRNA. This process is called decapping and requires activators mRNA-decapping enzyme 1 (DCP1), Me31B, and HPat, which are recruited by the miRISC. These factors attract mRNA-decapping enzyme 2 (DCP2), which catalyzes decapping of the target mRNA (Nishihara et al. 2013). This is an irreversible process that commits the mRNA to full degradation by the major cytoplasmic 5′–3′ exonuclease XRN1 (Braun et al. 2012).
Studies on the relationship between translational repression and mRNA decay have demonstrated that translational repression contributed little to the repression of endogenous mRNAs in comparison with mRNA decay (10–25 % and 66–90 %, respectively) (Eichhorn et al. 2014; Guo et al. 2010; Hendrickson et al. 2009). However, these global measurements of translational efficiency and mRNA levels were often made at relatively late time points after introducing miRNAs. Therefore, they are thought to reflect the long-term steady-state effects of miRNAs. Two recent studies examined the initial effect of miRNAs on inducible reporter genes. They concluded that miRNAs predominantly exert their effects through translational repression on newly synthesized targets. This step is followed by mRNA deadenylation, decapping and subsequent mRNA decay, which is the dominant effect of miRNAs at steady state conditions (Djuranovic et al. 2012; Béthune et al. 2012).
Abovementioned mechanism enables miRNAs to regulate gene expression at a post-transcriptional level. As earlier mentioned, miRNAs are differentially expressed in different cells and cell types, including embryonic stem cells (ESCs) , induced pluripotent stem (iPS ) cells , and fully differentiated cells, resulting in different gene expression profiles and cellular properties (Wilson et al. 2009; Houbaviy et al. 2003). Many studies have examined genome-wide miRNA expression profiles of stem cells during differentiation or self-renewal to improve understanding of miRNAs involved in the regulatory networks of these cellular processes (Mallon et al. 2014; Razak et al. 2013).
2 The Role of microRNAs in Pluripotent Stem Cells
2.1 Introduction
Stem cells are undifferentiated cells that are able renew themselves, and differentiate into specialized cells to regenerate tissues . The potency of stem cells describes the potential to differentiate into different cell types. Totipotent stem cells can differentiate into every embryonic and extra embryonic cell type, and are able to generate an entire organism. However, only cells produced by the first few divisions of a fertilized egg are totipotent. Pluripotent cells are descendants of totipotent cells and can differentiate into cells of all embryonic cell types, but not extra embryonic cell types. Furthermore, adult multipotent stem cells, often termed progenitor cells, can only differentiate into several closely related cell types (Mitalipov and Wolf 2009). Pluripotent stem cells hold significant potential for clinical therapies because they theoretically possess the capacity to regenerate all types of tissue. However, these characteristics are only displayed by cells of the inner cell mass (ICM) from early embryos. Fortunately, early studies demonstrated that the pluripotent state of stem cells can be captured by placing pre-implantation blastocysts in culture, leading to the generation of pluripotent embryonic stem cells (ESCs) (Evans and Kaufman 1981).
Furthermore, the derivation of complete adult and sexually mature animals by transplantation of somatic nuclei into enucleated oöcytes demonstrated that somatic nuclei also withhold the information to generate a complete adult organism (Gurdon et al. 1958). Further studies demonstrated that differentiated cells can also be reprogrammed to pluripotent stem cells by nuclear transfer or cell fusion (Campbell et al. 1996; Blau et al. 1983). A real breakthrough in the reprogramming of somatic cells was made when Shinya Yamanaka invented the induced pluripotent stem (iPS ) cell technology in 2006. Yamanaka and co-workers demonstrated that pluripotency can be induced by introducing four transcription factors, Oct4, Sox2, Klf4, and c-Myc (OSKM), in mouse adult fibroblasts by retroviral transfection (Takahashi and Yamanaka 2006). This approach offers the opportunity to generate pluripotent stem cells from differentiated cells of an individual, which opened the field of personalized regenerative medicine. Unfortunately, reprogramming with OSKM has remained slow (2–4 weeks) and inefficient (0.01–0.1 % of total cells) (Stadtfeld and Hochedlinger 2010).
In the last few years, a variety of pluripotent and adult stem cells have been proposed to promote cardiac regeneration . The heart itself possesses little regenerative capacity, since cardiomyocytes exhibit a very slow turnover and the heart lacks a sufficient stem cell reservoir to regenerate itself. In this regard, pluripotent stem cells and progenitor cells have been proposed to replenish the loss of cardiomyocytes or to induce proliferation and differentiation of resident stem cells in the heart. Several subsets of stem cells and cardiac progenitor cells have shown experimental and clinical benefits. Alas, the effects of cell-based therapy have been modest, often due to low engraftment of applied cells and the limited differentiation into cardiovascular cell types of adult progenitor cells (Li et al. 2012; Balsam et al. 2004). In contrast, ESCs and iPS cells are able to differentiate into all cardiovascular lineages. However, efficient differentiation before transplantation is necessary since pluripotent cells may lead to teratomas after delivery in patients (Sun et al. 2010).
Because miRNAs are powerful regulators that control gene expression networks, they might represent useful tools to promote reprogramming of somatic cells into iPS cells or direct somatic cells to specific lineages in vivo and in vitro . Understanding miRNAs involved in self-renewal, reprogramming and differentiation may enable the development of safer and more efficient cell therapies or lead to the development of in vivo reprogramming strategies to regenerate the human heart.
2.2 microRNAs in Regulation of Pluripotency
The importance of miRNAs in the regulation of self-renewal and pluripotency came to light in studies with Dicer- and DGCR8-deficient mouse ESCs (mESCs) (Wang et al. 2007; Murchison et al. 2005) that lacked canonical miRNA processing. Disruption of both Dicer alleles resulted in embryonic lethality, indicating the importance of miRNAs in development. However, it is possible to generate proliferating Dicer-deficient mES cell lines, which indicates that canonical miRNA processing is not required for the maintenance of self-renewal. Nevertheless, Dicer-deficient mESCs do show severe defects in cell cycle progression and differentiation . This might be expected, since almost 50 % of all present miRNA molecules in mESCs, are produced from four loci (miR-21, miR-17–92 cluster, miR-15b-16 cluster and miR-290 cluster) involved in cell cycle regulation and oncogenesis (Calabrese et al. 2007). Similar to Dicer disruption, deletion of both DGCR8 alleles resulted in mouse embryonic lethality early in development, again indicating the importance of canonical miRNA processing in development. Furthermore, DGCR8-deficient mES cell lines also displayed defects in differentiation and cell cycle progression (Wang et al. 2007). In general, loss of miRNAs in mESCs compromises the exit of self-renewal and progression of the cell cycle.
Further studies have focussed on miRNA profiling of mESCs and human ESCs (hESCs ), which have revealed specific miRNA expression profiles. Interestingly, only a limited number of miRNAs are transcribed in a pluripotent stem cell state, and these are immediately silenced once cells receive differentiation signals (Houbaviy et al. 2003). Furthermore, reprogramming factors Oct4, Sox2, Nanog and c-Myc, which are able to induce pluripotency in somatic cells, occupy promoters of these miRNA families, hereby regulating their expression (Marson et al. 2008). The most abundant and ESC-specific miRNAs are transcribed from two miRNA clusters: the miR-290 (human homologues miR-371–373) cluster and the miR-302–367 cluster .
Transcription of miR-302–367 is regulated by Oct4 and Sox2 in hESCs and mESCs (Marson et al. 2008; Card et al. 2008). This miRNA cluster encodes miR-302a, miR302b, miR302c, miR-302d, and miR-367 (Suh et al. 2004), and promotes pluripotency by targeting several pathways. Lipchina et al. (2011) identified a list of 146 validated miR-302–367 targets through complementary experimental and computational approaches (Lipchina et al. 2011). The miR-302–367 cluster members silence inhibitors of cell survival and G1-S transition of the cell cycle. Repression of these targets contributes to the unusual short G1 cell cycle of ESCs, necessary to maintain rapid self-renewal of pluripotent stem cells (Wang et al. 2008a). Furthermore, miR-302–367 targets genes that promote heterochromatin formation to ensure an open chromatin formation, which is characteristic for pluripotent cells. Other targets of miR-302–367 include genes involved in metabolic regulation to promote glycolytic metabolism. By downregulating genes involved in oxidative phosphorylation and mitochondrial biogenesis, miR-302–367 promotes a metabolic state characteristic for ESCs (Lipchina et al. 2012). Another set of target genes of miR-302–367 is involved in intracellular vesicle transport and endocytosis. Although the exact role of intracellular vesicles and endocytosis in maintenance of self-renewal and pluripotency is unclear, studies have indicated that these processes play a role in proliferation (Szczyrba et al. 2011) and signalling pathways involved in development (Weigert et al. 2004).
The miR-290 cluster comprises the most highly expressed miRNAs in mESCs (>70 %) and possesses similar or identical seed sequences as the miR-302–367 cluster (Houbaviy et al. 2003). The human genome contains homologues of the miR-290 cluster, namely the miR-371–373 family (Wang et al. 2013). Besides the miR-302–367 cluster, certain members of the miR-290 cluster (miR-291a, miR-294, and miR-295) also regulate the G1-S phase transition during the cell cycle by primarily targeting cyclin-Cdk inhibitors (Wang et al. 2008a). Furthermore, Oct4 physically occupies the promoter of the miR-290 cluster, regulating its expression (Marson et al. 2008). MiR-290 targets include epigenetic repressive DNA methyltransferases, such as retinoblastoma-like protein 2 (Rbl2 ) (Sinkkonen et al. 2008). By downregulating Rbl2, this miRNA cluster promotes expression of Oct4, which in return promotes expression of the miR-290 cluster. Other targets of miR-290 include the NF-κB subunit p65, which normally promotes differentiation of ESCs (Lüningschrör et al. 2012). According to a recent study, the cluster also promotes glycolytic ESC metabolism by targeting Mbd2, which normally represses glycolytic metabolism (Cao et al. 2015). Though the miR-290 cluster is not expressed in hESCs, the homologous miR-371–373 family is predicted to target the same pathways in hESCs since almost all members from both clusters share the same seed sequence.
Interestingly, the seed sequence (AAGUGCU) of members of miR-290 and miR-302–367 clusters is shared by other miRNAs highly expressed in ESCs, including members of the miR-17–92 cluster, miR-106a–363 cluster, and miR-106b–25 cluster (Fig. 6.2) (Li and He 2012). Conservation of seed sequences between miRNA clusters expressed in ESCs indicates common mechanistic roles and partial redundancy of miRNAs in maintenance of self-renewal and pluripotency. However, these clusters also contain members with different seed sequences that might attribute to distinct characteristics of pluripotent stem cells of different species (Table 6.1) (Nichols and Smith 2009).
MicroRNAs in the maintenance of pluripotency. The figure displays miRNA clusters that are involved in the maintenance of self-renewal and pluripotency. These miRNA clusters posses several members with similar or identical seed sequences (AAGUGCU), indicating common mechanistic roles of miRNAs in these processes
2.3 microRNAs in Alternate States of Pluripotency
Pluripotent stem cells of different species exhibit different characteristics and cellular responses. The ‘naïve’ pluripotent stem cell state is only represented by mESCs derived from the ICM of murine blastocysts. This state is characterized by expression of Oct4 by a distal enhancer, global reduction of DNA methylation, and repressive chromatin marks on developmental regulatory gene promoters (Marks et al. 2012). In contrast, hESCs are termed ‘primed’ pluripotent cells, and display more similarity to cells derived from the post-implantation murine embryonic epiblast with regard to gene expression patterns, epigenetic state, and signalling response to maintain an undifferentiated state (Tesar et al. 2007). These epiblast cells are called epiblast stem cells (EpiSCs ) and are thought to represent a less pluripotent state than ‘naïve’ pluripotent cells, since they cannot contribute to blastocyst chimeras (Tesar et al. 2007). Properties of ‘naïve’ and ‘primed’ pluripotent stem cells are elaborately reviewed elsewhere (Nichols and Smith 2009).
A limited number of studies have investigated differences in miRNA expression profiles between alternate states of pluripotency. Nevertheless, these studies have demonstrated that mESCs predominantly express the miR-290 cluster, preceding miR-302–367 expression at the early epiblast stage later in development (Spruce et al. 2010). EpiSCs and hESCs predominantly express the miR-302–367 cluster (Kim et al. 2011). These findings indicate that miR-302–367 expression corresponds with a slightly less pluripotent cell state.
Several studies have suggested that human pluripotent stem cell lines (hESCs and human iPS cells) exhibit different developmental potential, which translates into varying differentiation propensity (Osafune et al. 2008), especially concerning neural cell types (Hu et al. 2010) or hemangioblastic lineages (Feng et al. 2010). Differentiation propensity is very important to consider when using cell sources for cardiac regeneration. In a study by Kim et al. (2011), hESCs and iPS cells that expressed high levels of the miR-371–373 cluster demonstrated higher level of ‘naïve’ ESC markers and greater neural differentiation propensity. Their results suggest that human pluripotent cell lines that highly express the miR-371–373 cluster may represent a more ‘naïve’ pluripotent state, corresponding with greater differentiation propensity (Kim et al. 2011).
Due to their high pluripotent state, self-renewal, and ability to differentiate into all cells of the cardiovascular lineage, ESCs and iPS cells appear to be the ideal cell source for cardiac regeneration. However, clinical use of pluripotent stem cells is hampered by significant obstacles, including safety concerns, immune rejection, and low efficiency of iPS cell generation (Nussbaum et al. 2007; Okano et al. 2013). Since miRNAs perform an important regulatory function in self-renewal and pluripotency, they may improve reprogramming efficiency or function as a strategy for iPS cell generation without the use of viral vectors.
2.4 microRNAs in Reprogramming
Transduction of fibroblasts with transcription factors OSKM directly affects expression of several miRNA clusters, including miR-290 (and human homolog miR-371–373) (Judson et al. 2009; Subramanyam et al. 2011), miR-302–367 (Subramanyam et al. 2011), and paralogous clusters miR-17–92, miR106b–25, and miR106a–363 (Li et al. 2011). Since c-Myc and Klf4 are involved in oncogenesis, they are best avoided during iPS cell generation when considering clinical applications (Dang 2012; Yu et al. 2011). Therefore, researchers have attempted to generate iPS cells and enhance reprogramming efficiency using aforementioned miRNAs in combination with reduced numbers of pluripotency factors.
Based on the high expression levels of the miR-290 cluster in mESCs, first attempts of miRNA-mediated reprogramming involved overexpression of different members of this cluster in mouse embryonic fibroblasts (Wang et al. 2008a). Addition of miR-291-3p, miR-294, and miR-295 increased the efficiency of reprogramming in combination with retroviral transfection of Oct4, Sox2, and Klf4 (OSK) (Judson et al. 2009). miR-294 demonstrated greatest increased efficiency of up to 75 % of that achieved with OSKM (Judson et al. 2009). Therefore, it was suggested that miR-294 can efficiently substitute for c-Myc when reprogramming fibroblasts into iPS cells .
Furthermore, members of the miR-302–367 cluster that share the same seed sequence with members of the miR-290 cluster, also demonstrated the ability to improve reprogramming efficiency (Judson et al. 2009). Recent studies suggest that miR-302 improves reprogramming efficiency by suppressing several targets, including a repressor of Oct3/4 (NR2F2) (Hu et al. 2013) and several epigenetic regulators (AOF2, AOF1, MECP1-p66, and MECP2) to promote Oct3/4 expression and open chromatin formation, respectively (Lin et al. 2011).
Three other miRNA clusters (miR-17–92 cluster, miR-106b–25 cluster, and the miR-106a–363 cluster), were shown to be highly expressed during early stages of reprogramming with OSKM (Li et al. 2011). Overexpression of miR-106b–25 members, miR-93 and miR-106b, increased reprogramming efficiency fourfold in fibroblasts transfected with OSK. Li et al. (2011) have shown that miR-93 and miR-106b target the TGFβ-receptor 2 (TGFbr2) and cyclin-Cdk inhibitor p21 (Li et al. 2011). TGFbr2 is a receptor kinase that induces epithelial-to-mesenchymal transition (EMT) upon TGF-β signalling, a process were epithelial cells lose their cell-cell contacts and gain migratory and invasive properties (Thiery et al. 2009). Fibroblasts , a product of EMT, can only be reverted to a pluripotent state by repressing pro-EMT signalling (i.e. TGF-β signalling), hereby initiating the reverse process, mesenchymal-to-epithelial transition (MET) (Li et al. 2010). Consistent to these findings, embryonic stem cells are morphologically similar to epithelial cells and express E-cadherin, an epithelial marker (Baum et al. 2008). Moreover, human miR-302b and human miR-372 were also found to inhibit TGFβ-signalling by targeting TGFbr2. Addition of these miRNAs to OSK or OSKM resulted in a 10- to 15-fold increase of reprogramming efficiency, respectively (Subramanyam et al. 2011).
In addition to miR-290, miR-302–367, miR-17–92, and miR-106b–25 cluster members, a recent library screen of 379 miRNAs observed improved reprogramming efficiency during reprogramming with OSK in combination with the miR-130–301–721 family (Pfaff et al. 2011). This newly identified miR-family is indirectly involved in cell cycle regulation by targeting transcription factor Meox2, which normally promotes expression of cell cycle inhibitor p21 (Pfaff et al. 2011).
Instead of resembling miRNA expression profiles of pluripotent cells, an alternative approach to promote reprogramming with miRNAs might be to reduce expression of miRNAs that are highly expressed in differentiated fibroblasts. Let-7 family miRNAs, which are specifically depleted in ESCs, repress pluripotency factors and are highly expressed during differentiation. Interestingly, let-7 and miR-290 cluster miRNAs perform opposite functions in ESC self-renewal and pluripotency (Melton et al. 2010). It has been demonstrated that Let-7 members directly target several ESC-specific genes , including c-Myc, and downregulate important cell cycle molecules such as Cdk4 and CyclinD , to repress the G1-S transition of the cell cycle (Melton et al. 2010; Schultz et al. 2008). Accordingly, let-7 miRNA inhibition resulted in a fourfold increase of reprogramming efficiency compared to three pluripotency factors (OSK) alone (Melton et al. 2010). Repression of tumor suppressor pathways also appears important during the generation of pluripotent cells. For instance, depletion of miR-34 (Choi et al. 2011) and inhibition of miR-199a-3p (Wang et al. 2012), which act downstream of tumor suppressor p53, enhanced OSKM reprogramming efficiency. Moreover, by directly targeting p53 mRNA, ectopic expression of mir-138 (Ye et al. 2012) also significantly improved the efficiency of iPS cell generation. However, p53 is shown to preserve genomic integrity by aborting reprogramming in cells carrying various types of DNA damage, normally resulting in p53-dependent apoptosis (Marión et al. 2009). This reserves a crucial role for p53 in preventing iPS cell generation from suboptimal cells (Marión et al. 2009). Therefore, p53-related miRNAs have to be further investigated before they can be safely used to enhance reprogramming. To date, repression of miRNAs specific for differentiated cells has only slightly increased the efficiency of reprogramming without transcription factor c-Myc (Yang et al. 2011).
Besides stimulating or repressing miRNAs to promote reprogramming efficiency, miRNAs have also been investigated to induce reprogramming without the addition of transcription factors. Only a few studies have reported miRNA-mediated reprogramming of somatic cells to iPS cells without additional reprogramming factors. Reprogramming in the absence of transcription factors was first achieved by expression of the miR-302–367 cluster in cancer cells (Lin et al. 2008). Further investigations of Lin et al. led to the generation of iPS cells through inducible miR-302s (a viral vector containing miR-302a, b, c and d) expression (Lin et al. 2011). Recently, Anokye-Danso et al. (2011) reported that overexpression of miR-302–367 cluster members successfully reprogrammed mouse and human fibroblasts into iPS cells in the absence of additional factors (Anokye-Danso et al. 2011). Direct transfection of mature double-stranded miR-200c in combination with miR-302 and miR-369 was also reported to be sufficient to induce pluripotency in somatic cells (Miyoshi et al. 2011). Lüningschrör et al. (2013) failed to reproduce these miRNA-mediated reprogramming experiments and could not induce pluripotency with miRNAs alone (Lüningschrör et al. 2013).
In summary, it has been demonstrated that miRNAs that target cell cycle inhibitors, epigenetic regulators, and modulators of mesenchymal-to-epithelial transition, clearly enhance reprogramming efficiency of somatic cells into iPS cells. Additional investigations are necessary to reveal if miRNAs alone can be used to effectively reprogram fibroblasts to iPS cells without the use of reprogramming factors (Fig. 6.3).
MicroRNAs involved in reprogramming. The figure displays miRNAs that have demonstrated to promote reprogramming efficiency of somatic cells into iPS cells or were reported to induce reprogramming into iPS cells without additional transcription factors. However, the ability of miRNAs to reprogram cells without additional factors remains questionable
3 The Role of microRNAs in Cardiovascular Lineage Commitment
3.1 Introduction
Considering safety concerns with regard to pluripotent stem cells, differentiation of iPS cells and ESCs into specific lineages before transplantation is necessary. Canonical miRNAs play an essential role in cardiac development, as studies have shown that cardiac-specific knockout of the miRNA-processing enzyme Dicer results in rapid dilated cardiomyopathy, heart failure, and embryonic lethality (Chen et al. 2008). Since miRNAs are powerful regulators of gene expression patterns, they might represent an efficient strategy to modulate commitment of cardiac progenitor cells (CPCs) and pluripotent stem cells to cardiovascular lineages ex vivo or in vivo (summarised in Table 6.2).
3.2 microRNAs in Cardiomyocyte Differentiation and Proliferation
Specific miRNAs involved in cardiac differentiation have been revealed by miRNA expression profiling of hESC-derived cardiomyocytes and during differentiation of CPCs, which highlighted changes in several miRNAs. Especially expression of miR-1, miR-133, miR-208, and miR-499, changed remarkably during cardiac differentiation (Sluijter et al. 2010; Ivey et al. 2008; Wilson et al. 2010).
miR-1 and miR-133 are highly expressed in cardiac and skeletal muscle cells, where they modulate muscle proliferation and differentiation while repressing other lineages (Ivey et al. 2008; Chen et al. 2006). In humans, miR-1 and miR-133 are under regulation of cardiac and muscle specific transcription factors, including serum response factor (SRF ), myocyte enhancer factor 2 (MEF2 ), and myoblast determination protein (MyoD ) (Liu et al. 2007; Zhao et al. 2005). Although miR-1 and miR-133 are transcribed together as a polycystronic cluster, they appear to perform distinct functions during proliferation and differentiation of myoblasts (Chen et al. 2006). miR-1 promotes myogenesis by targeting histone deacetylase 4 (HDAC4), a transcriptional repressor of myogenic gene expression. Furthermore, by directly targeting frizzled family receptor 7 (FZD7) and fibroblast growth factor receptor substrate 2 (FRS2), miR-1 suppresses WNT and FGF signalling, which promotes cardiomyocyte differentiation (Lu et al. 2013). By targeting Hand2, a transcription factor required for ventricular cardiomyocyte proliferation, miR-1 also modulates the growth of the developing heart. Accordingly, miR-1 deficient mice displayed enhanced Hand2 expression, which resulted in an abnormally enlarged heart (Zhao et al. 2007). Consistently, miR-1 overexpression resulted in thin-walled ventricles of the heart in a mouse model, indicating premature cardiomyocytes due to early exit of the cell cycle and hampered cardiomyocyte proliferation (Zhao et al. 2005). Furthermore, miR-1 overexpression promotes cardiomyocyte differentiation from cardiac progenitor cells and ESCs of both mouse and human origin, demonstrated by an increase in cardiac marker expression (Sluijter et al. 2010; Lu et al. 2013).
In contrast to miR-1, which promotes differentiation, overexpression of miR-133 represses cardiac differentiation of hESCs and mESCs (Ivey et al. 2008). Furthermore, miR-133 directly targets muscle transcription factor SRF, suppressing smooth muscle gene expression (Liu et al. 2008). Accordingly, complete deletion of both mir-133 alleles results in ectopic activation of smooth muscle genes in the developing heart, as well as excessive proliferation and apoptosis of cardiomyocytes (Liu et al. 2008). Moreover, miR-133 overexpression in mouse models led to ventricular wall thinning and decreased cardiomyocyte proliferation (Liu et al. 2008; Carè et al. 2007). Altogether, studies have shown that polycystronic transcription of miR-1/miR-133 results in adequate regulation of cardiac proliferation and cardiomyocyte differentiation during cardiac development.
Further studies on miRNAs in cardiac development identified a family of intronic miRNAs, termed ‘myomiRs’, consisting of miR-499, miR-208a and miR-208b, which share many target genes. Although miR-208b and miR-499 are expressed in skeletal and cardiac muscle, miR-208a is only expressed in the heart where it regulates cardiomyocyte hypertrophy (Liu and Olson 2010; Callis et al. 2009). miR-499 is highly expressed in cardiac progenitor cells (Sluijter et al. 2010) and the heart (van Rooij et al. 2009), and in hESCs during cardiac differentiation (Wilson et al. 2010). Overexpression of miR-499 in human cardiac progenitor cells reduced proliferation of these cells and induced differentiation into cardiomyocytes (Sluijter et al. 2010). Furthermore, injection of miR-499 into the border zone of infracted rat hearts enhanced cardiomyogenesis in vivo (Sluijter et al. 2010). Consistent with these findings, expression of miR-499 resulted in upregulation of cardiac markers, such as Nkx2.5 and GATA4, by repression of SOX6 and PTBP3. In contrast, inhibition of miR-499 in mouse and human ESCs inhibits cardiomyocyte differentiation (Sluijter et al. 2010). In the mammalian heart, miR-499 is expressed at very low levels in c-kit positive cardiac stem cells, while it is highly expressed in cardiomyocytes, together with miR-1 and mir-133 (Hosoda et al. 2011). This suggests that these miRNAs are important in the specialization of cells towards a cardiomyocyte cell fate. Interestingly, miR-499 was demonstrated to translocate through gap junctions from cardiomyocytes to c-kit positive cardiac stem cells (CSCs) to promote differentiation into de novo cardiomyocytes (Hosoda 2013).
An alternative strategy to regenerate the heart is to stimulate endogenous cardiomyocyte proliferation. Several studies have shown that zebrafish are able to generate new myocardium from existing cardiomyocytes (Kikuchi et al. 2010; Jopling et al. 2010). During zebrafish heart regeneration, cardiomyocytes around the cardiac injury dedifferentiate into a more immature phenotype and cardiac transcription factors, such as Gata4 and Hand2, are activated (Kikuchi et al. 2010; Schindler et al. 2014). Subsequently, these dedifferentiated cardiomyocytes re-enter the cell cycle and fully repair the injured heart (Jopling et al. 2010). Decreased expression of miR-133 was observed by Yin et al. (2012) during zebrafish heart generation. Accordingly, miR-133 depletion enhanced cardiomyocyte proliferation by elevating repression of cell cycle factors and cell junction components (Yin et al. 2012). More recently, Aguirre et al. (2014) identified two miRNA families, miR-99/100 and let-7a/c, which were also downregulated during zebrafish heart regeneration. Alas, miR-99/100 expression remained unchanged after cardiac injury in mice. Nevertheless, knockdown of miR-99/100 and let-7a/c in mice reduced scar size and improved cardiac function after myocardial injury (Aguirre et al. 2014). Recently, direct injection of viral vectors expressing human miR-590 and miR-199a promoted cardiomyocyte proliferation and restored cardiac function in mice by targeting genes involved in cell cycle and proliferation (Eulalio et al. 2012). Altogether, these results indicate that regenerative mechanisms of the heart may be conserved between species, although they are not sufficiently stimulated upon cardiac injury in mammals.
3.3 microRNAs in Vascular Smooth Muscle Differentiation
The importance of miRNAs in vascular smooth muscle cell (VSMC) development was revealed during experiments with mutant mice that possessed a dysfunctional Dicer allele under a VSMC-specific promoter. Mutant mice died at embryonic day 16 or 17 due to bleeding and impaired VSMC contractility. Interestingly, these vascular defects observed in mutant mice could be partially rescued by overexpression of miR-145, highlighting the importance of this specific miRNA in VSMC development (Albinsson et al. 2010).
miR-145 is the most highly expressed miRNA in VSMCs, and is transcribed together with miR-143. During development, miR-145/143 is expressed in VSMCs under control of Nkx2.5, SRF, and its co-activator, myocardin. These miRNAs target a number of transcription factors, including Klf4 and Elk1 (both involved cellular proliferation), as well as a number of cytoskeletal proteins, to promote differentiation and repress proliferation of VSMCs (Cordes et al. 2009). In addition, miR-145, but not miR-143, is necessary for myocardin-induced reprogramming of fibroblasts into VSMCs and overexpression of miR-145 induced VSMC differentiation from neural crest cells (Cordes et al. 2009). Inhibition of miR-145 did not fully inhibit VSMC differentiation from ESCs but resulted in a more immature SMC phenotype (Cordes et al. 2009). Furthermore, miR-143/145 expression appears necessary for VSMCs to acquire and maintain a contractile phenotype (Boettger et al. 2009). Loss of these miRNAs results in impeded neointima formation because miR-143/145 act downstream of SRF during cytoskeletal remodelling and VSMC migration after vascular injury (Xin et al. 2009). Surprisingly, overexpression of miR-143 and miR-145 also decreased neointima formation in a rat model of acute vascular injury (Elia et al. 2009). These data indicate that miR-143 and miR-145 play crucial roles in modulating VSMC maturation and migration, though they are not essential for VSMC differentiation from stem cells.
Further studies have identified miR-1 , which also mediates cardiomyocyte differentiation, as one of the regulators in VSMC differentiation (Xie et al. 2011). miR-1 is required for VSMC differentiation from ESCs, as inhibition of miR-1 led to a down-regulation of VSMC-specific markers and decreased the number of VSMCs from ESCs. Similar to miR-145, miR-1 also targets transcription factor Klf4 (Xie et al. 2011). In addition to miR-1, miRNA profiling during VSMC differentiation demonstrated an increased expression of miR-10a, which was shown to target HDAC4 (Huang et al. 2010). Accordingly, inhibition of miR-10a blocked VSMC differentiation from ESCs. However, it has not been revealed how repression of HDAC4 by miR-10a is specifically involved in VSMC differentiation. Furthermore, miR-221 and miR-222 were found to be involved in VSMC proliferation by targeting cyclin-dependent kinase inhibitors, p57 and p27 (Davis et al. 2009; Liu et al. 2009). Consistent with these findings, knockdown of miR-221 and miR-222 suppressed VSMC proliferation in vivo (Liu et al. 2009). More recent studies on VSMC differentiation and proliferation have identified other miRNAs that either promote or inhibit VSMC differentiation. miRNAs that promote differentiation include miR-663 (Li et al. 2013) and miR-18a-5p (Kee et al. 2014), while others such as miR-132 (Choe et al. 2013), and miR-365 (Zhang et al. 2014) inhibit VSMC differentiation. Further studies are necessary to discover the potential of these miRNAs with regard to heart regeneration after injury.
3.4 microRNAs in Endothelial Differentiation and Vascular Development
Early studies demonstrated that Dicer-deficient mouse embryos exhibited defects in vasculogenesis and angiogenesis, which was consistent with altered expression of vascular marker genes such as vascular endothelial growth factor (VEGF), vascular endothelial growth factor receptor-1 and receptor-2, and Tie1 (Yang et al. 2005). Later in development, these Dicer-deficient mice had an impaired angiogenic potential after vascular injury (Suárez et al. 2008).
The most extensively studied angiogenic miRNA is miR-126, which was demonstrated to be highly important for vessel integrity and angiogenic signalling by early studies in zebrafish and mice (Fish et al. 2008; Wang et al. 2008b). Targeted deletion of miR-126 resulted in a reduced angiogenic response to endothelial growth factors, loss of vascular integrity, and induced haemorrhages in zebrafish (Fish et al. 2008). In mice, loss of miR-126 also led to leaky blood vessels, emphasising the importance of miR-126 in vascular physiology. As identified by microarray analysis, loss of angiogenic potential and vascular integrity can, at least in part, be attributed to increased levels of Spred1 and Pik3r2, which are repressors of angiogenic signalling and direct targets of miR-126 (Fish et al. 2008; Wang et al. 2008b). Moreover, miR-126 has been demonstrated to modulate expression of stromal cell-derived factor 1 (SDF-1) in ECs, which is proposed to regulate recruitment of vascular progenitor cells to ischemic regions (van Solingen et al. 2011). In addition to pro-angiogenic effects, miR-126 regulates expression of inflammatory mediators in endothelial cells, such as vascular cell adhesion molecule 1 (VCAM-1), hereby limiting leukocyte adhesion and subsequent inflammation (Harris et al. 2008). Although enriched in endothelial progenitor cells (EPCs), miR-126 lacks the potential to induce endothelial cell differentiation in ESCs, as expression of several vascular marker genes was not affected by miR-126 overexpression (Fish et al. 2008). In summary, miR-126 is important for vessel integrity and angiogenesis by modulating angiogenic signalling, vascular progenitor cell recruitment, and inflammation. However, miR-126 is not sufficient to induce EC differentiation in ESCs, indicating a minor role in endothelial lineage commitment.
The miR-17–92 cluster, induced by transcription factor c-Myc, comprises of miR-17, -18a, -19a/b, -20a, and -92a, and is also expressed in ECs. Interestingly, this cluster has shown to perform distinct functions in different contexts. First, the miR-17–92 cluster plays an important role in angiogenesis after vascular injury (O’Donnell et al. 2005; He et al. 2005). One of the clusters members, miR-92a, performs anti-angiogenic functions in ECs by targeting several proangiogenic factors. Accordingly, inhibition of miR-92a enhanced neovascularisation and functional recovery after myocardial infarction (Bonauer et al. 2009). Furthermore, expression of miR-17, miR-18a, miR-19a, and miR-20a, has shown to inhibit angiogenic activity of ECs in vitro, while inhibition of these miRNAs led to stimulation of angiogenic processes (Doebele et al. 2010). In contrast, miR-17–92 cluster appears to enhance neovascularisation in tumour cells. Therefore, it is proposed that the effect of miR-17–92 cluster differs for ischemia-induced angiogenesis and tumour-associated angiogenesis, which is important to consider when developing clinical therapies (Dews et al. 2006).
miRNA profiling during EC differentiation from ESCs further revealed several miRNAs essential for this process (Kane et al. 2010, 2012; Luo et al. 2013). Some miRNAs that significantly change in their expression during EC differentiation either promote angiogenesis (let7b, let7f, miR-126, miR-130a, mir-133/b, miR-210, and miR-296), or impair angiogenesis (miR-20a/b, miR-221, and miR-222), and are reviewed elsewhere (Wu et al. 2009). To date, not all of these miRNAs have been tested for their potential to induce EC differentiation.
Identified as one of the first mammalian miRNAs, miR-21 was found to induce stem cell differentiation by modulation of TGF-β2 signalling (Di Bernardini et al. 2014). In a recent study on EC differentiation from iPS cells, miR-21 overexpression in iPS cells exposed to VEGF induced capillary formation in vitro and in vivo (Di Bernardini et al. 2014). Recently, miRNA gain/loss-of-function analysis identified two other miRNAs involved in EC differentiation from hESCs (Luo et al. 2013). These miRNAs, miR-150 and miR-200c, are proposed to be involved in EC differentiation by repressing transcriptional repressor zinc finger E-box-binding homeobox 1 (ZEB1), which has been identified as a repressor for EC gene expression (Luo et al. 2013). Furthermore, Kane et al. (2012) indicated that miR-99b, -181a, and -181b are also involved in EC differentiation (Kane et al. 2012). Expression levels of these miRNAs increased in a time- and differentiation-dependent manner and peaked in hESC-derived ECs and adult ECs. Lentiviral-mediated transfer of these miRNAs promoted EC-specific markers, as well as neovascularisation after transplantation with hESC-derived ECs in vivo (Kane et al. 2012). However, knockdown of miR-99b, -181a, and -181b did not inhibit EC differentiation, suggesting that these miRNAs are not indispensible in endothelial commitment.
Although several miRNAs in EC differentiation have been described here, many other miRNAs perform regulatory functions in vascular development, angiogenesis and endothelial function, including miR-221 (Nicoli et al. 2012), miR-132 (Anand et al. 2010), miR-218 (Small et al. 2010), miR-23–27–24 cluster (Zhou et al. 2011), and miR-27a/b (Urbich et al. 2012). The role of these miRNAs in vascular development is extensively reviewed elsewhere (Dang et al. 2013).
4 The Role of microRNAs in Direct Reprogramming
As mentioned before, differentiation of pluripotent stem cells before transplantation could represent a safe strategy for cardiac regeneration . However, differentiation protocols often lead to cardiac cells with varying phenotypes, which hampers mechanical and electrical coupling of transplanted cells, especially cardiomyocytes, with the surrounding tissue (Ng et al. 2010). This has forced researchers to develop new strategies for cardiac regeneration, including stimulating cardiac cell populations in situ to promote regeneration. A groundbreaking discovery was made by Ieda et al., who directly converted mouse cardiac fibroblasts into induced cardiomyocyte-like cells (iCMs) with a combination of three cardiac developmental transcription factors, Gata4, Mef2c, and Tbx5 (GMT) (Ieda et al. 2010). By avoiding a pluripotent cell state, some limitations of undifferentiated iPS cells, such as tumorigenicity, can be overcome. Reprogrammed cells expressed cardiac-specific markers, cardiomyocyte gene expression profiles, and exhibited spontaneous contraction. Further studies also used alternative combinations of transcription factors to reprogram cardiac fibroblasts into cardiomyocytes (Song et al. 2012; Protze et al. 2012). In addition, reprogramming efficiency into functional cardiomyocytes could be improved by addition of miR-133 , which generated sevenfold more beating mouse cardiomyocyte-like cells than reprogramming with transcription factors alone (Muraoka et al. 2014).
Recently, others successfully achieved microRNA-mediated reprogramming of cardiac fibroblasts into cardiomyocytes via lentiviral transfection with cardiac-specific miRNAs without the use of additional transcription factors (Jayawardena et al. 2012). Four miRNAs, miR-1, miR-133, miR-208, and miR-499, highly expressed in cardiomyocytes, and tightly involved in cardiac development (Porrello 2013), were used by Jayawardena et al. (2012) to convert mouse cardiac fibroblasts into cardiomyocytes in vitro and in vivo (Jayawardena et al. 2012). Reprogramming of cardiac fibroblasts into cardiomyocytes resulted in more beating iCMs in vivo than in vitro, suggesting that other unknown factors in the heart enhance maturation of the iCMs. Reprogramming efficiency of cardiac fibroblasts into cardiomyocytes remains low (12 %) in this initial phase of miRNA-mediated direct reprogramming (Jayawardena et al. 2015). However, recent studies have shown that miRNAs are a promising alternative for direct reprogramming of somatic cells (Table 6.3).
5 Future Perspectives
Current understanding suggests that the mammalian heart possesses an endogenous regenerative capacity that could be stimulated to regenerate the heart after injury. The ideal cell source for cell-based cardiac regeneration should be able to differentiate into cardiomyocytes that integrate with existing cardiomyocytes inside the heart and differentiate into new blood vessels to supply blood to the infarct zone. Although several stem cell sources have shown little cardiac differentiation potential and predominantly promote cardiac function through paracrine signalling, pluripotent stem cells and cardiac progenitor cells are capable to differentiate into cardiovascular lineages in vitro and in vivo. Despite the enormous potential of autologous iPS cells in personal regenerative medicine, significant safety concerns remain with respect to the generation and transplantation of iPS cells.
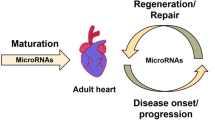
In this chapter, microRNAs have been described as powerful regulators of gene expression patterns, regulating biological processes and cellular behaviour, including stem cell pluripotency, self-renewal, reprogramming, and cardiovascular lineage commitment. Due to increasing knowledge of miRNA expression patterns in different cell types, researchers have identified specific miRNAs that are able to promote reprogramming and induce differentiation of pluripotent stem cells and cardiac progenitor cells ex vivo. Because miRNAs are small, easily synthesized, and administered to cells by lipid-based transfection, they represent useful tools for the development of safe cell-based therapies to regenerate the heart.
Recent studies have reported the possibility to reprogram cardiac fibroblasts directly into beating cardiomyocytes ex vivo and in vivo. Interestingly, the efficiency of this process can be increased by miRNAs and a combination of miR-1, miR-133, miR-208, and miR-499 induced reprogramming of cardiac fibroblasts into functional cardiomyocytes without additional factors. This indicates that miRNAs are able to generate new cardiomyocytes from fibroblasts, hereby promoting in situ regeneration (Fig. 6.4). However, before miRNAs can be used in clinical settings for cardiac regeneration, safe and efficient delivery methods have to be developed (Zhang et al. 2013). By further understanding the role of specific miRNAs in reprogramming, and pluripotent stem cell or cardiac progenitor cell differentiation, clinical therapies could be developed to stimulate endogenous repair mechanisms and in situ regeneration of the human heart.
MiRNA-mediated cardiac regeneration. MiRNAs are involved in maintenance of self-renewal and pluripotency, as well as reprogramming and differentiation. miRNAs control gene expression patterns, enabling the use of miRNAs to direct cellular behaviour. Reprogramming efficiency of fibroblasts into iPS cells can be increased by several miRNAs. Subsequently, pluripotent stem cells can be efficiently differentiated into cardiovascular cell types that can be transplanted to replenish lost cells after myocardial infarction. Furthermore, differentiation of resident cardiac progenitor cells into cardiovascular lineages might also be stimulated by miRNAs to promote cardiac regeneration. Recent studies demonstrated that miRNAs are also sufficient to induce direct reprogramming of cardiac fibroblasts into functional cardiomyocytes
References
Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE (2003) The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell 4(5):625–637
Aguirre A, Montserrat N, Zachiggna S, Nivet E, Hishida T, Krause MN, Kurian L, Ocampo A, Vazquez-Ferrer E, Rodriguez-Esteban C, Kumar S, Moresco JJ, Yates JR, Campistol JM, Sancho-Martinez I, Giacca M, Izpisua Belmonte JC (2014) In vivo activation of a conserved microRNA program induces mammalian heart regeneration. Cell Stem Cell 15(5):589–604
Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC (2010) MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol 30(6):1118–1126
Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA (2010) MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med 16(8):909–914
Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE (2011) Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8(4):376–388
Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC (2004) Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature 428(6983):668–673
Baum B, Settleman J, Quinlan MP (2008) Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol 19(3):294–308
Béthune J, Artus-Revel CG, Filipowicz W (2012) Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep 13(8):716–723
Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W (2006) Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125(6):1111–1124
Blau HM, Chiu CP, Webster C (1983) Cytoplasmic activation of human nuclear genes in stable heterocaryons. Cell 32(4):1171–1180
Boettger T, Beetz N, Kostin S, Schneider J, Krüger M, Hein L, Braun T (2009) Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest 119(9):2634–2647
Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S (2009) MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science 324(5935):1710–1713
Braun JE, Huntzinger E, Fauser M, Izaurralde E (2011) GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44(1):120–133
Braun JE, Truffault V, Boland A, Huntzinger E, Chang C-T, Haas G, Weichenrieder O, Coles M, Izaurralde E (2012) A direct interaction between DCP1 and XRN1 couples mRNA decapping to 5′ exonucleolytic degradation. Nat Struct Mol Biol 19(12):1324–1331
Cai X, Hagedorn CH, Cullen BR (2004) Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10(12):1957–1966
Calabrese JM, Seila AC, Yeo GW, Sharp PA (2007) RNA sequence analysis defines Dicer’s role in mouse embryonic stem cells. Proc Natl Acad Sci U S A 104(46):18097–18102
Callis TE, Pandya K, Seok HY, Tang R-H, Tatsuguchi M, Huang Z-P, Chen J-F, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang D-Z (2009) MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest 119(9):2772–2786
Campbell KH, McWhir J, Ritchie WA, Wilmut I (1996) Sheep cloned by nuclear transfer from a cultured cell line. Nature 380(6569):64–66
Cao Y, Guo W-T, Tian S, He X, Wang X-W, Liu X, Gu K-L, Ma X, Huang D, Hu L, Cai Y, Zhang H, Wang Y, Gao P (2015) miR-290/371-Mbd2-Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J 34(5):609–623
Card DAG, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK (2008) Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol 28(20):6426–6438
Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang M-L, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MVG, Høydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G (2007) MicroRNA-133 controls cardiac hypertrophy. Nat Med 13(5):613–618
Chen J-F, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang D-Z (2006) The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 38(2):228–233
Chen J-F, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH, Meissner G, Patterson C, Hannon GJ, Wang D-Z (2008) Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci U S A 105(6):2111–2116
Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436(7051):740–744
Choe N, Kwon J-S, Kim J-R, Eom GH, Kim Y, Nam K-I, Ahn Y, Kee HJ, Kook H (2013) The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis 229(2):348–355
Choi YJ, Lin C-P, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C, Ozturk A, Hicks GG, Hannon GJ, He L (2011) miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 13(11):1353–1360
Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee T-H, Miano JM, Ivey KN, Srivastava D (2009) miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460(7256):705–710
Dang CV (2012) MYC on the path to cancer. Cell 149(1):22–35
Dang LTH, Lawson ND, Fish JE (2013) MicroRNA control of vascular endothelial growth factor signaling output during vascular development. Arterioscler Thromb Vasc Biol 33(2):193–200
Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A (2009) Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem 284(6):3728–3738
Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A (2006) Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet 38(9):1060–1065
Di Bernardini E, Campagnolo P, Margariti A, Zampetaki A, Karamariti E, Hu Y, Xu Q (2014) Endothelial lineage differentiation from induced pluripotent stem cells is regulated by microRNA-21 and transforming growth factor β2 (TGF-β2) pathways. J Biol Chem 289(6):3383–3393
Ding XC, Grosshans H (2009) Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J 28(3):213–222
Djuranovic S, Nahvi A, Green R (2012) miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336(6078):237–240
Doebele C, Bonauer A, Fischer A, Scholz A, Reiss Y, Urbich C, Hofmann W-K, Zeiher AM, Dimmeler S (2010) Members of the microRNA-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood 115(23):4944–4950
Eichhorn SW, Guo H, McGeary SE, Rodriguez-Mias RA, Shin C, Baek D, Hsu S-H, Ghoshal K, Villén J, Bartel DP (2014) mRNA Destabilization Is the Dominant Effect of Mammalian MicroRNAs by the Time Substantial Repression Ensues. Mol Cell 56(1):104–115
Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MVG, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, Condorelli G (2009) The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ 16(12):1590–1598
Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E (2009) Deadenylation is a widespread effect of miRNA regulation. RNA 15(1):21–32
Eulalio A, Mano M, Dal Ferro M, Zentilin L, Sinagra G, Zacchigna S, Giacca M (2012) Functional screening identifies miRNAs inducing cardiac regeneration. Nature 492(7429):376–381
Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292(5819):154–156
Feng Q, Lu S-J, Klimanskaya I, Gomes I, Kim D, Chung Y, Honig GR, Kim K-S, Lanza R (2010) Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28(4):704–712
Fish JE, Santoro MM, Morton SU, Yu S, Yeh R-F, Wythe JD, Ivey KN, Bruneau BG, Stainier DYR, Srivastava D (2008) miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell 15(2):272–284
Friedman RC, Farh KK-H, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Fukao A, Mishima Y, Takizawa N, Oka S, Imataka H, Pelletier J, Sonenberg N, Thoma C, Fujiwara T (2014) MicroRNAs trigger dissociation of eIF4AI and eIF4AII from target mRNAs in humans. Mol Cell 56(1):79–89
Guo H, Ingolia NT, Weissman JS, Bartel DP (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466(7308):835–840
Gurdon JB, Elsdale TR, Fischberg M (1958) Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature 182(4627):64–65
Han J, Lee Y, Yeom K-H, Kim Y-K, Jin H, Kim VN (2004) The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18(24):3016–3027
Han J, Lee Y, Yeom K-H, Nam J-W, Heo I, Rhee J-K, Sohn SY, Cho Y, Zhang B-T, Kim VN (2006) Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 125(5):887–901
Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ (2008) MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci U S A 105(5):1516–1521
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435(7043):828–833
Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO (2009) Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol 7(11), e1000238
Hertel J, Bartschat S, Wintsche A, Otto C, Stadler PF (2012) Evolution of the let-7 microRNA family. RNA Biol 9(3):231–241
Hosoda T (2013) The mircrine mechanism controlling cardiac stem cell fate. Front Genet 4:204
Hosoda T, Zheng H, Cabral-da-Silva M, Sanada F, Ide-Iwata N, Ogorek B, Ferreira-Martins J, Arranto C, D’Amario D, del Monte F, Urbanek K, D’Alessandro DA, Michler RE, Anversa P, Rota M, Kajstura J, Leri A (2011) Human cardiac stem cell differentiation Is regulated by a mircrine mechanism. Circulation 123(12):1287–1296
Houbaviy HB, Murray MF, Sharp PA (2003) Embryonic stem cell-specific MicroRNAs. Dev Cell 5(2):351–358
Hu B-Y, Weick JP, Yu J, Ma L-X, Zhang X-Q, Thomson JA, Zhang S-C (2010) Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A 107(9):4335–4340
Hu S, Wilson KD, Ghosh Z, Han L, Wang Y, Lan F, Ransohoff KJ, Burridge P, Wu JC (2013) MicroRNA-302 Increases Reprogramming Efficiency via Repression of NR2F2. Stem Cells 31(2):259–268
Huang H, Xie C, Sun X, Ritchie RP, Zhang J, Chen YE (2010) miR-10a contributes to retinoid acid-induced smooth muscle cell differentiation. J Biol Chem 285(13):9383–9389
Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12(2):99–110
Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142(3):375–386
Ivey KN, Muth A, Arnold J, King FW, Yeh R-F, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D (2008) MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2(3):219–229
Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ (2012) MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110(11):1465–1473
Jayawardena TM, Finch EA, Zhang L, Zhang H, Hodgkinson CP, Pratt RE, Rosenberg PB, Mirotsou M, Dzau VJ (2015) MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res 116(3):418–424
Jopling C, Sleep E, Raya M, Martí M, Raya A, Izpisúa Belmonte JC (2010) Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 464(7288):606–609
Judson RL, Babiarz JE, Venere M, Blelloch R (2009) Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 27(5):459–461
Kane NM, Meloni M, Spencer HL, Craig MA, Strehl R, Milligan G, Houslay MD, Mountford JC, Emanueli C, Baker AH (2010) Derivation of endothelial cells from human embryonic stem cells by directed differentiation: analysis of microRNA and angiogenesis in vitro and in vivo. Arterioscler Thromb Vasc Biol 30(7):1389–1397
Kane NM, Howard L, Descamps B, Meloni M, McClure J, Lu R, McCahill A, Breen C, Mackenzie RM, Delles C, Mountford JC, Milligan G, Emanueli C, Baker AH (2012) Role of microRNAs 99b, 181a, and 181b in the differentiation of human embryonic stem cells to vascular endothelial cells. Stem Cells 30(4):643–654
Kee HJ, Kim GR, Cho S-N, Kwon J-S, Ahn Y, Kook H, Jeong MH (2014) miR-18a-5p MicroRNA Increases Vascular Smooth Muscle Cell Differentiation by Downregulating Syndecan4. Korean Circ J 44(4):255–263
Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DYR, Poss KD (2010) Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature 464(7288):601–605
Kim H, Lee G, Ganat Y, Papapetrou EP, Lipchina I, Socci ND, Sadelain M, Studer L (2011) miR-371-3 expression predicts neural differentiation propensity in human pluripotent stem cells. Cell Stem Cell 8(6):695–706
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue):D68–D73
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294(5543):853–858
Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans. Science 294(5543):862–864
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
Li MA, He L (2012) microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays 34(8):670–680
Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7(1):51–63
Li Z, Yang C-S, Nakashima K, Rana TM (2011) Small RNA-mediated regulation of iPS cell generation. EMBO J 30(5):823–834
Li T-S, Cheng K, Malliaras K, Smith RR, Zhang Y, Sun B, Matsushita N, Blusztajn A, Terrovitis J, Kusuoka H, Marbán L, Marbán E (2012) Direct comparison of different stem cell types and subpopulations reveals superior paracrine potency and myocardial repair efficacy with cardiosphere-derived cells. J Am Coll Cardiol 59(10):942–953
Li P, Zhu N, Yi B, Wang N, Chen M, You X, Zhao X, Solomides CC, Qin Y, Sun J (2013) MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation. Circ Res 113(10):1117–1127
Liang Y, Ridzon D, Wong L, Chen C (2007) Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8:166
Lin S-L, Chang DC, Chang-Lin S, Lin C-H, Wu DTS, Chen DT, Ying S-Y (2008) Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 14(10):2115–2124
Lin S-L, Chang DC, Lin C-H, Ying S-Y, Leu D, Wu DTS (2011) Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res 39(3):1054–1065
Lipchina I, Elkabetz Y, Hafner M, Sheridan R, Mihailovic A, Tuschl T, Sander C, Studer L, Betel D (2011) Genome-wide identification of microRNA targets in human ES cells reveals a role for miR-302 in modulating BMP response. Genes Dev 25(20):2173–2186
Lipchina I, Studer L, Betel D (2012) The expanding role of miR-302-367 in pluripotency and reprogramming. Cell Cycle 11(8):1517–1523
Liu N, Olson EN (2010) MicroRNA regulatory networks in cardiovascular development. Dev Cell 18(4):510–525
Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN (2007) An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci 104(52):20844–20849
Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN (2008) microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev 22(23):3242–3254
Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C (2009) A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res 104(4):476–487
Lu T-Y, Lin B, Li Y, Arora A, Han L, Cui C, Coronnello C, Sheng Y, Benos PV, Yang L (2013) Overexpression of microRNA-1 promotes cardiomyocyte commitment from human cardiovascular progenitors via suppressing WNT and FGF signaling pathways. J Mol Cell Cardiol 63:146–154
Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303(5654):95–98
Lüningschrör P, Stöcker B, Kaltschmidt B, Kaltschmidt C (2012) miR-290 cluster modulates pluripotency by repressing canonical NF-κB signaling. Stem Cells 30(4):655–664
Lüningschrör P, Hauser S, Kaltschmidt B, Kaltschmidt C (2013) MicroRNAs in pluripotency, reprogramming and cell fate induction. Biochim Biophys Acta 1833(8):1894–1903
Luo Z, Wen G, Wang G, Pu X, Ye S, Xu Q, Wang W, Xiao Q (2013) MicroRNA-200C and -150 play an important role in endothelial cell differentiation and vasculogenesis by targeting transcription repressor ZEB1. Stem Cells 31(9):1749–1762
Mallon BS, Hamilton RS, Kozhich OA, Johnson KR, Fann YC, Rao MS, Robey PG (2014) Comparison of the molecular profiles of human embryonic and induced pluripotent stem cells of isogenic origin. Stem Cell Res 12(2):376–386
Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA (2009) A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 460(7259):1149–1153
Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, Stunnenberg HG (2012) The transcriptional and epigenomic foundations of ground state pluripotency. Cell 149(3):590–604
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134(3):521–533
Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, Filipowicz W, Duchaine TF, Sonenberg N (2007) MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 317(5845):1764–1767
Melton C, Judson RL, Blelloch R (2010) Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463(7281):621–626
Mitalipov S, Wolf D (2009) Totipotency, pluripotency and nuclear reprogramming. Adv Biochem Eng Biotechnol 114:185–199
Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M (2011) Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8(6):633–638
Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R, Inagawa K, Nishiyama T, Kaneda R, Fukuda T, Takeda S, Tohyama S, Hashimoto H, Kawamura Y, Goshima N, Aeba R, Yamagishi H, Fukuda K, Ieda M (2014) MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J 33(14):1565–1581
Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ (2005) Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A 102(34):12135–12140
Ng SY, Wong CK, Tsang SY (2010) Differential gene expressions in atrial and ventricular myocytes: insights into the road of applying embryonic stem cell-derived cardiomyocytes for future therapies. Am J Physiol Cell Physiol 299(6):C1234–C1249
Nichols J, Smith A (2009) Naive and primed pluripotent states. Cell Stem Cell 4(6):487–492
Nicoli S, Knyphausen C-P, Zhu LJ, Lakshmanan A, Lawson ND (2012) miR-221 is required for endothelial tip cell behaviors during vascular development. Dev Cell 22(2):418–429
Nishihara T, Zekri L, Braun JE, Izaurralde E (2013) miRISC recruits decapping factors to miRNA targets to enhance their degradation. Nucleic Acids Res 41(18):8692–8705
Nussbaum J, Minami E, Laflamme MA, Virag JAI, Ware CB, Masino A, Muskheli V, Pabon L, Reinecke H, Murry CE (2007) Transplantation of undifferentiated murine embryonic stem cells in the heart: teratoma formation and immune response. FASEB J 21(7):1345–1357
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435(7043):839–843
Okano H, Nakamura M, Yoshida K, Okada Y, Tsuji O, Nori S, Ikeda E, Yamanaka S, Miura K (2013) Steps toward safe cell therapy using induced pluripotent stem cells. Circ Res 112(3):523–533
Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, Melton DA (2008) Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol 26(3):313–315
Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE (2008) Chromatin structure analyses identify miRNA promoters. Genes Dev 22(22):3172–3183
Pfaff N, Fiedler J, Holzmann A, Schambach A, Moritz T, Cantz T, Thum T (2011) miRNA screening reveals a new miRNA family stimulating iPS cell generation via regulation of Meox2. EMBO Rep 12(11):1153–1159
Pfaff J, Hennig J, Herzog F, Aebersold R, Sattler M, Niessing D, Meister G (2013) Structural features of Argonaute-GW182 protein interactions. Proc Natl Acad Sci U S A 110(40):E3770–E3779
Porrello ER (2013) microRNAs in cardiac development and regeneration. Clin Sci (Lond) 125(4):151–166
Protze S, Khattak S, Poulet C, Lindemann D, Tanaka EM, Ravens U (2012) A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol 53(3):323–332
Razak SRA, Ueno K, Takayama N, Nariai N, Nagasaki M, Saito R, Koso H, Lai C-Y, Murakami M, Tsuji K, Michiue T, Nakauchi H, Otsu M, Watanabe S (2013) Profiling of microRNA in human and mouse ES and iPS cells reveals overlapping but distinct microRNA expression patterns. PLoS One 8(9), e73532
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772):901–906
Saini HK, Griffiths-Jones S, Enright AJ (2007) Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci U S A 104(45):17719–17724
Schindler YL, Garske KM, Wang J, Firulli BA, Firulli AB, Poss KD, Yelon D (2014) Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Development 141(16):3112–3122
Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M (2008) MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res 18(5):549–557
Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W (2008) MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol 15(3):259–267
Sluijter JPG, van Mil A, van Vliet P, Metz CHG, Liu J, Doevendans PA, Goumans M-J (2010) MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler Thromb Vasc Biol 30(4):859–868
Small EM, Sutherland LB, Rajagopalan KN, Wang S, Olson EN (2010) MicroRNA-218 regulates vascular patterning by modulation of Slit-Robo signaling. Circ Res 107(11):1336–1344
Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, Bassel-Duby R, Olson EN (2012) Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 485(7400):599–604
Spruce T, Pernaute B, Di-Gregorio A, Cobb BS, Merkenschlager M, Manzanares M, Rodriguez TA (2010) An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Dev Cell 19(2):207–219
Stadtfeld M, Hochedlinger K (2010) Induced pluripotency: history, mechanisms, and applications. Genes Dev 24(20):2239–2263
Suárez Y, Fernández-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC (2008) Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A 105(37):14082–14087
Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R (2011) Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29(5):443–448
Suh M-R, Lee Y, Kim JY, Kim S-K, Moon S-H, Lee JY, Cha K-Y, Chung HM, Yoon HS, Moon SY, Kim VN, Kim K-S (2004) Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270(2):488–498
Sun N, Longaker MT, Wu JC (2010) Human iPS cell-based therapy: considerations before clinical applications. Cell Cycle 9(5):880–885
Szczyrba J, Nolte E, Wach S, Kremmer E, Stöhr R, Hartmann A, Wieland W, Wullich B, Grässer FA (2011) Downregulation of Sec23A protein by miRNA-375 in prostate carcinoma. Mol Cancer Res 9(6):791–800
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676
Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RDG (2007) New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448(7150):196–199
Thiery JP, Acloque H, Huang RYJ, Nieto MA (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139(5):871–890
Thomas M, Lieberman J, Lal A (2010) Desperately seeking microRNA targets. Nat Struct Mol Biol 17(10):1169–1174
Tüfekci KU, Meuwissen RLJ, Genç S (2014) The role of microRNAs in biological processes. Methods Mol Biol 1107:15–31
Urbich C, Kaluza D, Frömel T, Knau A, Bennewitz K, Boon RA, Bonauer A, Doebele C, Boeckel J-N, Hergenreider E, Zeiher AM, Kroll J, Fleming I, Dimmeler S (2012) MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood 119(6):1607–1616
van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, Richardson JA, Kelm RJ, Olson EN (2009) A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev Cell 17(5):662–673
van Solingen C, de Boer HC, Bijkerk R, Monge M, van Oeveren-Rietdijk AM, Seghers L, de Vries MR, van der Veer EP, Quax PHA, Rabelink TJ, van Zonneveld AJ (2011) MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1(+)/Lin(−) progenitor cells in ischaemia. Cardiovasc Res 92(3):449–455
Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R (2007) DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39(3):380–385
Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R (2008a) Embryonic stem cell-specific microRNAs regulate the G1-S transition and promote rapid proliferation. Nat Genet 40(12):1478–1483
Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN (2008b) The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell 15(2):261–271
Wang J, He Q, Han C, Gu H, Jin L, Li Q, Mei Y, Wu M (2012) p53-facilitated miR-199a-3p regulates somatic cell reprogramming. Stem Cells 30(7):1405–1413
Wang T, Shi S, Sha H (2013) MicroRNAs in regulation of pluripotency and somatic cell reprogramming: small molecule with big impact. RNA Biol 10(8):1255–1261
Weigert R, Yeung AC, Li J, Donaldson JG (2004) Rab22a regulates the recycling of membrane proteins internalized independently of clathrin. Mol Biol Cell 15(8):3758–3770
Westholm JO, Lai EC (2011) Mirtrons: microRNA biogenesis via splicing. Biochimie 93(11):1897–1904
Wilson KD, Venkatasubrahmanyam S, Jia F, Sun N, Butte AJ, Wu JC (2009) MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev 18(5):749–758
Wilson KD, Hu S, Venkatasubrahmanyam S, Fu J-D, Sun N, Abilez OJ, Baugh JJA, Jia F, Ghosh Z, Li RA, Butte AJ, Wu JC (2010) Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: role for miR-499. Circ Cardiovasc Genet 3(5):426–435
Wilson RC, Tambe A, Kidwell MA, Noland CL, Schneider CP, Doudna JA (2014) Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol Cell 57(3):397–407
Wu F, Yang Z, Li G (2009) Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys Res Commun 386(4):549–553
Xie C, Huang H, Sun X, Guo Y, Hamblin M, Ritchie RP, Garcia-Barrio MT, Zhang J, Chen YE (2011) MicroRNA-1 regulates smooth muscle cell differentiation by repressing Kruppel-like factor 4. Stem Cells Dev 20(2):205–210
Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN (2009) MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev 23(18):2166–2178
Xu W, San Lucas A, Wang Z, Liu Y (2014) Identifying microRNA targets in different gene regions. BMC Bioinformatics 15(7):S4
Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G (2005) Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem 280(10):9330–9335
Yang C-S, Li Z, Rana TM (2011) microRNAs modulate iPS cell generation. RNA 17(8):1451–1460
Ye D, Wang G, Liu Y, Huang W, Wu M, Zhu S, Jia W, Deng A-M, Liu H, Kang J (2012) MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells 30(8):1645–1654
Yin VP, Lepilina A, Smith A, Poss KD (2012) Regulation of zebrafish heart regeneration by miR-133. Dev Biol 365(2):319–327
Yu F, Li J, Chen H, Fu J, Ray S, Huang S, Zheng H, Ai W (2011) Kruppel-like factor 4 (KLF4) is required for maintenance of breast cancer stem cells and for cell migration and invasion. Oncogene 30(18):2161–2172
Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E (2013) GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. EMBO J 32(7):1052–1065
Zhang Y, Wang Z, Gemeinhart RA (2013) Progress in microRNA delivery. J Control Release 172(3):962–974
Zhang P, Zheng C, Ye H, Teng Y, Zheng B, Yang X, Zhang J (2014) MicroRNA-365 inhibits vascular smooth muscle cell proliferation through targeting cyclin D1. Int J Med Sci 11(8):765–770
Zhao Y, Samal E, Srivastava D (2005) Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature 436(7048):214–220
Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D (2007) Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell 129(2):303–317
Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S (2011) Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci U S A 108(20):8287–8292
Acknowledgements
This research forms part of the Project P1.05 LUST of the research program of the BioMedical Materials institute, co-funded by the Dutch Ministry of Economic Affairs and the Netherlands CardioVascular Research Initiative (CVON): the Dutch Heart Foundation, Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development, and the Royal Netherlands Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Doeleman, M.J.H., Feyen, D.A.M., de Veij Mestdagh, C.F., Sluijter, J.P.G. (2016). Cardiac Regeneration and microRNAs: Regulators of Pluripotency, Reprogramming, and Cardiovascular Lineage Commitment. In: Madonna, R. (eds) Stem Cells and Cardiac Regeneration. Stem Cell Biology and Regenerative Medicine. Springer, Cham. https://doi.org/10.1007/978-3-319-25427-2_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-25427-2_6
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-25425-8
Online ISBN: 978-3-319-25427-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)