Abstract
Twitter has been recently used to predict and/or monitor real world outcomes, and this is also true for health related topic. In this work, we extract information about diseases from Twitter with spatio-temporal constraints, i.e. considering a specific geographic area during a given period. We exploit the SNOMED-CT terminology to correctly detect medical terms, using sentiment analysis to assess to what extent each disease is perceived by persons. We show our first results for a monitoring tool that allow to study the dynamic of diseases.
This work was supported by the “Programma Operativo Nazionale Ricerca & Competitivita” 20072013 within the project “PON04a2C - Smart Health 2.0” supported by MIUR (Minister of Education, University and Research).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The amount of digital health related data [6] is becoming more and more huge, being generated both by healthcare industries [23] (e.g. medical records and exams) as well as by social media and virtual networks, where individuals share their experiences and opinions about different topics, including personal health (illnesses, symptoms, treatments, side effects).
While data owned by healthcare industries are often accessible only with restrictions, social media data are generally publicly available, therefore they represent an enormous resource for mining interesting healthcare insights. Among various social networks, the one on-the-edge is Twitter [29], the micro-blogging service whose restriction of 140 characters for post encouraged the development of a kind of shorthand and speed in composing messages.
Twitter has been recently used as an information source to predict and/or monitor real world outcomes [3], from extreme event analysis as the 2013 Syria sarin gas attack [31] or the earthquakes in Japan [24], to more playful scenarios as the inferring of U.S. citizens’ mood during the day [22] or the forecast box-office revenues for movies [2].
Exploiting virtual social networks for healthcare purposes has been recently named with the neologisms Infodemiology and Infoveillance [8], and also Twitter has been exploited, as in [1] the micro-blog is used to detect flu trends, or in [25], where authors tracked and examined disease transmission in particular social contexts via Twitter data, or in [9], where social media improves healthcare delivery by encouraging patient engagement and communication.
In this paper, we monitor health related information using both Twitter data and medical terms present in the SNOMED-CT terminology [15], currently the most comprehensive medical terminology worldwide adopted. Tweets are considered within a specific geographic area, and we extract a (possibily continuous) stream of messages within a given time window, retaining just all those concerning diseases. Then, using natural language processing [12] and sentiment analysis techniques [10, 17], we assess to what extent each disease is present in all tweets over time in that region. Our proposal therefore results in a monitoring tool that allow to study the dynamic of diseases.
Exploiting tweets for health-related issues is not new; in [27] authors present a practical approach for content mining of tweets that is somehow similar to our proposal except for the initial selection of keywords. Indeed, we do not outline in advance a list of significant keywords for tweets extraction, rather we adopt the SNOMED-CT collection to extract any health related tweets. Similarly, in [1] and [16] a predefined list of flu related keywords (e.g. “H1N1”) is considered to accomplish its task, whereas we do not focus on a specific disease. In [13], the temporal diversity of tweets is examined during the known periods of real-world outbreaks for a better understanding of specific events (e.g. diseases). As in our case, time is considered, whereas topic dynamics is inferred using an unsupervised clustering technique (instead of the official SNOMED-CT cited previously); the use of sentiment analysis however is not considered.
The paper is organized as follows. In Sect. 2 we describe the overall architecture of our proposal, and how the data collection and analysis are performed. In Sect. 3 we show an application to a real case, providing concluding remarks and future works in Sect. 4.
2 Architecture
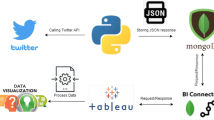
The overall architecture of our proposal is depicted in Fig. 1. As introduced in the previous section, the first step is the extraction of geolocalized tweets; to this purpose, we developed a Python application that extracts a stream of tweets both during a desired time period and within a given region (a box with specified NE and SW coordinates). Note that for better results, only geolocalized tweets have been considered; a less precise solution is to use the user’s provided location but this could lead to misinformation when specified location is not correct.
After having collected tweets, we want to extract only those with health-related content, i.e. where at least a medical term is present. At this step, Natural Language Processing (NLP in Fig. 1) techniques are required to properly filter each tweet by:
-
removing non-English tweets
-
removing irrelevant information, as links, retweet details and usernames
-
applying standard text processing operations as tokenization, stopwords removal, stemming and indexing [4].
2.1 Health-Related Tweets Extraction
In order to discard tweets that do not contain any medical term, we search for index terms in the SNOMED-CT terminology. To better clarify how this search is performed, we briefly cite the SNOMED-CT core components (details can be found in [26]) that are:
-
concepts, that represent all entities that characterize health care processes; they are arranged into acyclic taxonomic hierarchies (according to a is-a semantics)
-
descriptions, explaining concepts in terms of various clinical terms or phrases; these can be of three types, Fully Specified Names (FSNs) that is the main (formal) definition, Preferred Terms (PTs), i.e. the most common way of expressing the meaning of the concept, and Synonyms.
-
relationships between concepts, e.g. the concept (disease) “Staphylococcal eye infection” has “Causative agent” relationship with “Staphylococcus” (different types of relationships exist depending on concepts type)
-
reference sets used to group concepts e.g. for cross-maps to other standard purposes.
In this work, the first two items are considered. In particular, among all concepts hierarchies we focus in the “disorder/disease” since our goal is to detect tweets about diseases; therefore we do not consider other specific hierarchies (e.g. “surgical procedures”). Inside the disorder hierarchy, we search each index term extracted from tweets as a FSN, PT or synonym; if found, that tweet is further processed in order to establish to what extent the specified disorder is present using sentiment analysis (see below).
Note that to guarantee that all medical terms can be successfully detected, a list of additional informal terms is searched if nothing is found within SNOMED-CT. For instance, if the index term is the word “flu”, this has positive match in the synonym list of “influenza” disease (the FSN), but the (also quite common) term “headache” is not explicitly present when browsing SNOMED-CT [11], where this disorder is instead referred as “migraine” both as FSN and its synonym. Including “headache” in an additional list (named “informal terms” in Fig. 1) is the simple solution we adopted; this list is considered just if nothing is found within SNOMED-CT.
Also note that several diseases are defined as a group of words (e.g. “Viral respiratory infection”), therefore during the indexing phase we also retainN-grams with N=2 and 3; diseases with more than three words can be easily disambiguated even with 3 words since not all words are generally significative (e.g. in “Disease due to Orthomyxoviridae” the first and the last words are enough for correct matching).
Finally, detected diseases may be hierarchically related, e.g. “influenza” and “pneumonia” are both children of “Viral respiratory infection” according to the “is-a” semantics. This information could be used for instance by replacing both children with their common parent, in order to build a more generalized, global view of diseases named in the given geographic area during the chosen time period. We choose however to preserve the best level of detail by not using a common ancestor as in the example, while on the other hand we will substitute all terms that represent the same disease with its FSN as indicated in SNOMED-CT. For instance, if different tweets refer to “flu”, “grippe” and “influenza” they will be all considered as tweets about “influenza”.
2.2 Tweets Classification
The next phase is the use of sentiment analysis in order to establish to what extent the disease detected in that tweet is present. Sentiment analysis or opinion mining [20] leverages NLP, text analysis and computational linguistics to extract subjective information, as the mood of the people regarding a particular product or topic; basically, the sentiment analysis can be viewed as a classification problem of labelling a given text (e.g. a statement within a tweet) as positive, negative or neutral.
Opinion mining has been applied to twitter data in several context, e.g. [2], where tweets are used to predict revenues for upcoming movies, or [7], where tweets allow to guess the political election results during U.S. presidential debate in 2008. Several approaches are adopted to perform sentiment analysis; typically, these are (1) machine learning algorithms with supervised models, where training examples labelled by human experts are exploited, or (2) unsupervised models, where classification is performed using proper syntactic patterns used to express opinions.
In the work here described we choose the latter approach. In particular, we first extract main statements from each tweet using the NLTK chunking package [19]; chunking, also called shallow parsing, allows to identify short phrases (clusters) like noun phrases (NP) and verb phrases (VP), thus providing more information than just the parts of speech (POS) of words, but without building the full parse tree of the whole text (tweet). For instance, in the tweet “Last night was too rainy, this morning my headache is stabbing but fortunately my little syster has got over her terrible flu”, the package produces the following chunks:
“Last night”(NP)
“was” (VP)
“too rainy” (NP)
“this morning” (NP)
“my headache” (NP)
“is stabbing” (VP)
“but fortunately” (NP)
“my little syster” (NP)
“has got over” (VP)
“her terrible flu” (NP).
Basically, the sentiment analysis we exploit to discover disease searches for them into NPs chunks (in the example, “headache” and “flu”), while the presence or absence of that diseases can be derived by analyzing VPs chunks. Therefore, in the tweet example the headache is present, while the flu is cited but no more present. We use a proper list of positive and negative verbs to this purpose, obviously taking into account negative verbal forms and propositions to guarantee a correct detection. In addition to the basic mechanism described here, we also estimate to what extent the given disease is present or not combining the linguistical distance (in terms of NP/VP chunks) between the disease and its associated verb and a proper rank we assigned to verbs and disease adjectives. In the example above, “terrible” and “is stabbing” both increase the relevance of their associated disease (details can be found in [5]). We exploit this estimation together with the number of tweets concerning a given disease in order to approximate its impact, e.g. assessing whether few people have terrible flu or many people are few cold in a given area during the monitoring time period.
Note that for each tweet, a set (generally small due to the limited lenght of tweets) of diseases could be detected. We do not associate however persons (twetter users) with diseases, rather we aim at achieving a “global” vision of the health status in the monitored area; an example of first results is provided in the following section.
3 Results
In this section we show how the approach illustrated in previous sections has been implemented to get first results.
The Python application we developed made use of the Tweepy libraries [28] and Twitter Stream APIs [30] to extracts the stream of tweets on March 2015 (1 month) within the area of New York City, delimited as a box with proper NE and SW coordinates (see Fig. 2); the OAuth APIs [14] has been used for authentication.
The total number of tweets collected was about 178,000 generated by about 60,000 unique users.
Tweets have then been processed with the NLTK python based platform [18] to perform all text-processing operations described in the previous section; SNOMED-CT and the additional informal medical terms allow to isolate health related tweets, while the next phase (i.e. sentiment analysis) classify tweet statements (chunks) to assess whether and how diseases are present.
A list of all diseases extracted can be used to examine each one of them. In Fig. 3 the list of the most relevant diseases detected is shown, each with the number of tweets that contains at least a chunk referring to that disease.
As indicated in previous section, for each disease we also tried to estimate to what extent it is present at a given time. For instance in Fig. 4 we show how influenza is perceived by persons during March in the entire area examined. The two highest value detected from tweets concern the case where people healed from influenza (about 4500 tweets) and the opposite, where people tweet about their serious flu (6420 tweets). We believe that people tend to tweet significant information and probably having just a little bit of influenza is generally considered not so relevant.
Filtering data with space and/or time constraints makes it possible to assess the evolution of that disease, e.g. in Fig. 5 we represent the number of tweets detected across the three 10-day slots of March for “influenza”, showing that there has been an increment of influenza outbreaks during the second decade.
4 Conclusions
We introduced an approach to Tweeter data processing aiming at extracting health related information in a given area during an assigned period; this is achieved by also expoliting the SNOMED-CT medical terminology and sentiment analysis technique. The final goal is to get data for studying the spatio-temporal evolution of a selected disease in the area being considered, and first results are encouraging. We are considering other further questions as:
-
the comparison with other existing proposal/tools, e.g. [21]
-
the contribution that following and followers can provide to improve the accuracy and the meaning of collected data
-
how profiling users (according to age, gender, residence area, device type...) leads to better (targeted) analysis; a related improvement is to address the biased demographic of users that could affect results (e.g. [32]).
-
how to explore other sentiment anaysis methods, for instance combining lexical- and machine learning- based methods as suggested in [10], in order to improve the effectiveness of the proposed approach
-
to gather a larger number of tweets (for instance, over a year or more) even in different geographical areas, to validate our proposal
-
to more deeply explore SNOMED-CT, for instance by exploiting relationships between concepts for a more effective health-related tweets extraction.
References
Achrekar, H., Gandhe, A., Lazarus, R., Yu, S.H., Liu, B.: Predicting flu trends using twitter data. In: 2011 IEEE Conference on Computer Communications Workshops (INFOCOM WKSHPS), pp. 702–707, April 2011
Asur, S., Huberman, B.A.: Predicting the future with social media. CoRR abs/1003.5699 (2010). http://arxiv.org/abs/1003.5699
Atefeh, F., Khreich, W.: A survey of techniques for event detection in twitter. Comput. Intell. 31(1), 132–164 (2015)
Baeza-yates, R., Ribeiro-Neto, B.: Modern Information Retrievial. ACM Press, Seattle (1999)
Carchiolo, V., Longheu, A., Cifalino, S.: Contestualizzazione spaziale di informazioni medico scientifiche tramite sensori sociali. DIEEI - Internal, Report (2015)
Cios, K.J., Moore, W.: Uniqueness of medical data mining. Artif. Intell. Med. 26, 1–24 (2002)
Diakopoulos, N.A., Shamma, D.A.: Characterizing debate performance via aggregated twitter sentiment. In: Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, CHI 2010, pp. 1195–1198. ACM, New York (2010). http://doi.acm.org/10.1145/1753326.1753504
Eysenbach, G.: Infodemiology and Infoveillance. Am. J. Prev. Med. 40(5), S154–S158 (2011). http://dx.doi.org/10.1016/j.amepre.2011.02.006
Fisher, J., Clayton, M.: Who gives a tweet: assessing patients interest in the use of social media for health care. Worldviews Evid.-Based Nurs. 9(2), 100–108 (2012). http://dx.doi.org/10.1111/j.1741-6787.2012.00243.x
Gonçalves, P., Araújo, M., Benevenuto, F., Cha, M.: Comparing and combining sentiment analysis methods. In: Proceedings of the First ACM Conference on Online Social Networks, COSN 2013, pp. 27–38. ACM, New York (2013), http://doi.acm.org/10.1145/2512938.2512951
IHTSDO SNOMED CT Browser. http://browser.ihtsdotools.org/
Jackson, P., Moulinier, I.: Natural Language Processing for Online Applications: Text Retrieval, Extraction and Categorization, 2nd edn. John Benjamins, Amsterdam (2007)
Kanhabua, N., Nejdl, W.: Understanding the diversity of tweets in the time of outbreaks. In: Proceedings of the 22nd International Conference on World Wide Web Companion, WWW 2013 Companion, pp. 1335–1342. International World Wide Web Conferences Steering Committee, Republic and Canton of Geneva, Switzerland (2013). http://dl.acm.org/citation.cfm?id=2487788.2488172
Kumar, S., Morstatter, F., Liu, H.: Twitter Data Analytics. Springer, New York (2013)
Lee, D., Cornet, R., Lau, F., de Keizer, N.: A survey of snomed-ct implementations. J. Biomed. Inform. 46(1), 87–96 (2013). http://www.sciencedirect.com/science/article/pii/S1532046412001530
Lee, K., Agrawal, A., Choudhary, A.: Real-time disease surveillance using twitter data: demonstration on flu and cancer. In: Proceedings of the 19th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, KDD 2013, pp. 1474–1477. ACM, New York (2013). http://doi.acm.org/10.1145/2487575.2487709
Medhat, W., Hassan, A., Korashy, H.: Sentiment analysis algorithms and applications: a survey. Ain Shams Eng. J. 5(4), 1093–1113 (2014). http://www.sciencedirect.com/science/article/pii/S2090447914000550
Natural Language Toolkit. http://www.nltk.org/
Natural Language Toolkit chunk package. http://www.nltk.org/api/nltk.chunk.html
Pang, B., Lee, L.: Opinion mining and sentiment analysis. Found. Trends Inf. Retr. 2(1–2), 1–135 (2008). http://dx.doi.org/10.1561/1500000011
Paul, M.: Discovering health topics in social media using topic models, April 2014. http://dx.doi.org/10.6084/m9.figshare.1007712
Pulse of the Nation. http://www.ccs.neu.edu/home/amislove/twittermood
Raghupathi, W., Raghupathi, V.: Big data analytics in healthcare: promise and potential. Health Inf. Sci. Syst. 2(1), 3 (2014). http://dx.doi.org/10.1186/2047-2501-2-3
Sakaki, T., Okazaki, M., Matsuo, Y.: Earthquake shakes twitter users: real-time event detection by social sensors. In: Proceedings of the 19th International Conference on World Wide Web, WWW 2010, pp. 851–860. ACM, New York (2010). http://doi.acm.org/10.1145/1772690.1772777
Signorini, A., Segre, A.M., Polgreen, P.M.: The use of Twitter to track levels of disease activity and public concern in the US during the influenza A H1N1 pandemic. PLoS One 6(5), e19467 (2011). doi:10.1371/journal.pone.0019467
Snomed, CT. http://www.ihtsdo.org/snomed-ct
Sunmoo Yoon, N.E., Bakken, S.: A practical approach for content mining of tweets. Am. J. Prev. Med. 45(1), S122–S129 (2013)
Tweepy - A Python library for accessing Twitter API. http://www.tweepy.org/
Twitter. http://www.twitter.com/
Twitter Streaming APIs. https://dev.twitter.com/streaming/
Tyshchuk, Y., Wallace, W., Li, H., Ji, H., Kase, S.: The nature of communications and emerging communities on twitter following the 2013 syria sarin gas attacks. In: 2014 IEEE Joint on Intelligence and Security Informatics Conference (JISIC), pp. 41–47, September 2014
When Google got flu wrong. http://www.nature.com/news/when-google-got-flu-wrong-1.12413
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this paper
Cite this paper
Carchiolo, V., Longheu, A., Malgeri, M. (2015). Using Twitter Data and Sentiment Analysis to Study Diseases Dynamics. In: Renda, M., Bursa, M., Holzinger, A., Khuri, S. (eds) Information Technology in Bio- and Medical Informatics. ITBAM 2015. Lecture Notes in Computer Science(), vol 9267. Springer, Cham. https://doi.org/10.1007/978-3-319-22741-2_2
Download citation
DOI: https://doi.org/10.1007/978-3-319-22741-2_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-22740-5
Online ISBN: 978-3-319-22741-2
eBook Packages: Computer ScienceComputer Science (R0)