Abstract
Bacteria associated with marine organisms have received major attention worldwide due to their potential in pharmaceutical industries. A total of 32 bacterial strains were isolated and identified, 22 from Tachypleus gigas and 10 from Carcinoscorpius rotundicauda from Balok Beach, Malaysia. Overall, Gram-negative bacteria showed dominancy on both species. A combination of biochemical tests, macro morphology, and micro morphology was used to roughly determine genus of bacterial isolates; where results from 16S rDNA sequencing were taken as the end results due to its high reliability. Results from the molecular approach showed only Pseudoalteromonas sp. (38.46 %), Vibrio sp. (46.15 %), and Photobacterium sp. (15.38 %) might exhibit mutualism with horseshoe crabs, T. gigas and C. rotundicauda. Study on the diversity of bacteria associated with horseshoe crabs can enhance the understanding of these animals and the potential sustainability of producing useful compounds for defense mechanisms.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Horseshoe crabs are an ancient, unique group of marine arthropods that have contributed to many clinical research studies, such as the discovery of Limulus amoebocyte lysate (LAL), a lipopolysaccharide (endotoxin) detection compound used to test for the presence of Gram-negative bacteria (Manco et al. 2010). The associations of microbes and marine organisms have received major attention in the past few decades due to the high potential for these interactions to yield pharmaceutical and medical products. Bacteria have been proven to associate with many marine organisms such as sponges, soft corals, and mollusks, which synthesize useful bioactive compounds (Armstrong et al. 2001; Lee et al. 2001; Vasanthabharathi and Jayalakshmi 2012). The association of the marine bacteria with the marine host organism is typically mutualistic, which benefits both the host and the bacteria (Lopanik 2014). Currently, information on the association of bacteria with horseshoe crab species, especially in Malaysia, is still very limited where only a few studies have been reported on the eggs, juvenile and adult horseshoe crabs. This study was conducted to identify and characterize marine bacteria associated with horseshoe crabs.
2 Methodology
2.1 Sample Collection and Isolation of Bacteria
Bacteria samples were derived from the book gills and mouth of horseshoe crabs from Balok Beach, Malaysia. The exterior of the horseshoe crabs were cleaned by using sterile distilled water several times to avoid contamination by bacteria from the environment. A cotton swab was used to take the sample, which was incubated overnight at 37 °C on marine agar to isolate single colonies.
2.2 Phenotypic Characterization
Gram-negative and positive bacteria were identified by using heat to fix the smears and a simple staining procedure using crystal violet, iodine and safranin (Black 2008). The isolates were routinely grown on marine agar and incubated at 37 °C for biochemical tests. Catalase existence was determined by the method of Reiner (2010), and an oxidase test was used to detect cytochrome oxidase (Shields and Cathcart 2010).
The production of sulphide, formation of indole, and motility of the bacteria were detected by a Sulfur, Indole, and Motility (SIM) Test (Vasanthabharathi and Jayalakshmi 2012). A Triple-Sugar-Iron (TSI) Test was used by the method of Ateba and Marumo (2014) to differentiate Gram-negative enteric bacilli based on carbohydrate fermentation and the production of hydrogen sulfide.
2.3 Extraction of DNA
Total genomic DNA was extracted by using DNeasy Blood and Tissue Kit (Qiagen, USA). Total DNA was isolated from 1 ml of culture after overnight incubation at 37 °C. The total DNA extracts were electrophoresed in an Agarose gel electrophoresis system. The total DNA extracted from the isolates were amplified by 16S rDNA sequence followed by PCR amplification universal primers 16rDNA primer pair 27F (5′-GAGTTTGATCMTGGCTCAG-3′) and eubacteria-specific primer 1492R (5′-TACGGTTACCTTGTTACGACTT-3′) (Weisburg et al. 1991).
The PCR amplification was conducted in 25 μl containing 2.5 μl of 10× Taq buffer, 1.5 μl of MgCl2 (25 mM), 0.5 μl of dNTP mix (10 mM each), 0.5 μl of each primer (10 μM), 0.2 μl of 1 U Taq DNA polymerase, 1 μl template DNA and deionized water. The PCR cycling program started with one initial denaturation step of 95 °C for 30 s; 35 cycles consisting of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, and elongation at 72 °C for 60 s. The final elongation step was extended at 72 °C for 7 min.
The amplification reaction was performed by using Master-Cycler gradient (Eppendorf, Germany). All purified products, using reverse and forward primers, were sent for DNA sequencing (First BASE Laboratories Sdn. Bhd, Selangor, Malaysia). The resulting gene fragments were aligned using BLAST (Basic Local Alignment Search Tool) search (http://www.ncbi.nml.nih.gov/BLAST) to investigate the region of similarity between sequences, including nearest neighbors (Ismail and Sarijan 2011).
3 Results and Discussion
3.1 Macro Morphology and Micro Morphology
A total of 32 bacterial strains were isolated from horseshoe crabs; 22 were from T. gigas, and the remaining 10 were from C. rotundicauda. The macro morphology (shape, elevation and color of colonies) of isolated bacteria from both species was recorded. Based on macro morphology, most isolated bacteria were narrowed down to a few genera or species.
Gram-staining of 22 isolated bacteria from T. gigas showed dominancy of Gram-negative bacteria, which included 21 strains (95 %), and only 1 Gram-positive strain (5 %) was observed. Three out of ten bacterial isolates (30 %) from C. rotundicauda were Gram-positive, while the remaining seven colonies (70 %) were Gram-negative. This result was in agreement with previous studies by Kaiser and Benner (2008) in which the marine environment was dominated by Gram-negative microorganisms, yet Gram-positive microbes were still present. Our study solely depended on bacteria cultured from the horseshoe crabs, T. gigas and C. rotundicauda.
3.2 Identification of Marine Bacteria Isolates
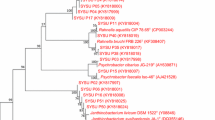
Identification of bacteria was done with reference to Bergey’s Manual of Determinative Bacteriology by Holt et al. (1994). The combination of macro morphology, micro morphology, and biochemical tests enabled identification of bacteria to the genus level. Among the 32 isolates, only 13 were sent for sequencing because some isolates belonged to the same genus or species (Tables 18.1 and 18.2). Of the isolates sequenced, nine were from T. gigas and four were from C. rotundicauda.
Sequencing results showed that the 13 isolates belong to three different genera, which were Pseudoalteromonas, Vibrio, and Photobacterium from the Gram-negative bacteria group. Therefore, with these sequencing results, 22 isolates from T. gigas and 10 isolates from C. rotundicauda were grouped into families. In T. gigas samples, 13 strains (13/22 = 59 %) were in the Pseudoalteromonadaceae family and 9 strains (9/22 = 41 %) were in the Vibrionaceae family.
On the other hand, among the ten C. rotundicauda isolates, six strains (6/10 = 60 %) belonged to the Vibrionaceae family, while the remaining four strains (4/10 = 40 %) were in the Pseudoalteromonadaceae family. According to Drancourt et al. (2000), almost 90 % of identifications from the overall performance of 16S rDNA sequence analysis were outstanding, compared to isolates identified by biochemical profile and Gram-staining, which failed to produce an accurate result. In this study, the sequencing results proved that some biochemical tests were less reliable compared to the 16S rDNA sequencing method.
Pseudoalteromonas sp. is Gram-negative bacillus bacteria. They are motile by a single polar flagellum. Species in this genus are reported to have association with marine invertebrates such as sponges (Vasanthabharathi and Jayalakshmi 2012). Members of Pseudoalteromonas are known to produce bioactive substances. For example, P. phenolica are antibiotic-producing bacteria when they are associated with their hosts (Isnansetyo and Kamei 2003). This genus is also reported to have anti-bacterial, anti-fungal, and anti-fouling activities (Berborn et al. 2011). The dominancy of Pseudoalteromonas sp. showed that they might have a strong association with the horseshoe crab. However, no previous studies have reported that bacteria from this genus showed association with horseshoe crabs.
Vibrio sp. is a genus of Gram-negative bacteria with a curved rod shape, facultative anaerobes, motile by polar flagella with sheaths. This genus was reported to be the most commonly associated with crustaceans, demonstrating pathogenic effects, while at the same time producing bioactive compounds (Jayasinghe et al. 2008). According to Bell et al. (1994), V. parahaemolyticus associated with marine organisms showed anti-microbial and hemolytic activities. Isolated bacteria from T. gigas and C. rotundicauda showed the presence of Vibrio sp., suggesting a potential source of bioactive compounds.
Photobacterium sp. is Gram-negative bacteria in the Vibrionaceae family. They are morphologically rod, motile by one to three polar flagella, facultative aerobes, and some strains have the ability to emit light. Many species of Photobacterium were proven to live in symbiosis with marine organisms (Chimetto et al. 2010). Many species of marine fish form bioluminescent symbioses with Photobacterium spp., including P. kishitanii, P. leiognathi, and P. mandapamensis (Urbanczyk et al. 2011). So far, other species of Photobacterium, P fischeri were first reported and identified to live symbiotically with light organs of luminous fishes (Ruby and Nealson 1976).
4 Conclusion
This study demonstrates that Pseudoalteromonas sp., Vibrio sp., and Photobacterium sp. can be associated with horseshoe crabs, T. gigas and C. rotundicauda. These marine bacteria might exhibit a mutualistic relationship with the horseshoe crab and may produce useful bioactive compounds. Further study on marine bacteria associated with horseshoe crabs are needed to identify these compounds, which might be important for pharmaceutical industries and contribute to the survival of the horseshoe crab.
References
Armstrong E, Yan L, Boyd KG et al (2001) The symbiotic role of marine microbes on living surfaces. Hydrobilogia 461:37–40
Ateba CN, Marumo BI (2014) Isolation of enterohaemorrhagic Escherichia coli O104 strains from raw meat products in the North West Province, South Africa. J Food Nutr Res 2(6):288–293
Bell R, Carmeli S, Sar N (1994) Vibrindole A, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostacion cubicus. J Nat Prod 57(11):1587–1590
Berborn N, Ng YY, Kjelleberg S et al (2011) Marine bacteria from Danish coastal waters show antifouling activity against the marine fouling bacterium Pseudoalteromonas sp. strain S91 and zoospores of the green alga Ulva australis independent of bacteriocidal activity. Appl Environ Microbiol 77(2):8557–8567
Black JG (2008) Microbiology principles and exploration, 7th edn. Wiley, New York
Chimetto LA, Cleenwerck I, Thompson CC et al (2010) Photobacterium jeanii sp. nov., isolated from corals and zoanthids. Int J Syst Evol Microbiol 60(12):2843–2848
Drancourt M, Bollet C, Carlioz A et al (2000) 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol 38(10):3623–3630
Holt JG, Krieg NR, Sneath PH et al (1994) Bergey’s manual of determinative bacteriology. Williams and Wilkins, Baltimore
Ismail N, Sarijan S (2011) Phylogenetic inference from 18S rRNA gene sequences of horseshoe crabs, Tachypleus gigas between Tanjung Dawai, Kedah and Cherating Pahang. Int J Biol Vet Agric Food Eng 56(8):947–950
Isnansetyo A, Kamei Y (2003) Pseudoalteromonas phenolica sp. nov., a novel marine bacterium that produces phenolic anti-methcilin-resistant Staphylococcus aureus substances. Int J Syst Evol Microbiol 53(2):583–588
Jayasinghe CVL, Ahmed SBN, Kariyawasam MGIU (2008) The isolation and identification of Vibrio species in marine shrimps of Sri Lanka. J Food Agric 1(1):36–44
Kaiser K, Banner R (2008) Major bacterial contribution to the ocean reservoir of detrital organic carbon and nitrogen. Limnol Oceanogr 53(1):99
Lee YK, Lee JH, Lee HKM (2001) Microbial symbiosis in marine sponges. J Microbiol 39:254–264
Lopanik NB (2014) Chemical defensive symbioses in the marine environment. Funct Ecol 28:328–340
Manco M, Putignani L, Bottazzo GF (2010) Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev 31(6):817–844
Reiner K (2010) Catalase test protocol. American Society for Microbiology. http://www.microbelibrary.org/library/laboratory-test/3226-catalase-test-protocol. Accessed 15 June 2014
Ruby EG, Nealson KH (1976) Symbiotic association of Photobacterium fischeri with the marine luminous fish Monocsntris japonica: a model of symbiosis based on bacterial studies. Biol Bull 151(3):574–586
Shields P, Cathcart L (2010) Oxidase test protocol. American Society for Microbiology. http://www.microbelibrary.org/library/laboratory-test/3229-oxidase-test-protocol. Accessed 18 June 2014
Urbanczyk H, Ast JC, Dunlap PV (2011) Phylogeny, genomic and symbiosis of Photobacterium. FEMS Microbiol Rev 35(2):324–342
Vasanthabharathi V, Jayalakshmi S (2012) Bioactive potential of symbiotic bacteria and fungi from marine sponges. Afr J Biotechnol 11(29):7500–7511
Weisburg WG, Barns SM, Pelletier DA et al (1991) 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173(2):697–703
Acknowledgements
We thank Sabah Biodiversity (SaBC), Malaysia and Universiti Malaysia Terengganu (UMT) for funding this research.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Ismail, N. et al. (2015). Marine Bacteria Associated with Horseshoe Crabs, Tachypleus gigas and Carcinoscorpius rotundicauda . In: Carmichael, R., Botton, M., Shin, P., Cheung, S. (eds) Changing Global Perspectives on Horseshoe Crab Biology, Conservation and Management. Springer, Cham. https://doi.org/10.1007/978-3-319-19542-1_18
Download citation
DOI: https://doi.org/10.1007/978-3-319-19542-1_18
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-19541-4
Online ISBN: 978-3-319-19542-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)