Abstract
Donor oocyte IVF is regarded as an effective treatment for diminished ovarian reserve, because oocyte donors are usually young and believed to be a source of highly competent gametes. The aneuploidy rate for embryos derived from anonymously donated oocytes remains poorly characterized, however. This investigation reviewed comprehensive chromosomal screening results on embryos obtained from anonymous donor-egg IVF cycles to determine both the aneuploidy rate and parental source of the genetic error.
Methods
Parental DNA data obtained prior to IVF were correlated with embryo PGD results for chromosome-specific assessments. This approach permitted mitotic and meiotic copy errors to be differentiated for each chromosome among all embryos tested, thus providing information on the specific parental source of embryo aneuploidy (i.e., from the anonymous egg donor vs. the partner’s husband).
Results
This retrospective review identified 305 embryos generated from 23 patients who began IVF treatment in 2013. For oocyte donors (n = 23), mean (±SD) age was 24 ± 2.7 years. For embryos derived from oocyte donation with full chromosomal reporting (n = 284), euploidy was present in 133 (46.8 %). Considering all embryo chromosomes, the average error rate was 18 %. 133 of 155 observed embryo aneuploidies (85.8 %) were attributable to a maternal (oocyte donor) source. Among all aneuploid embryos (n = 155), chromosomal errors from both genetic parents (i.e., oocyte donor and partner’s husband) were present in 52.9 %. While oocyte donor age ranged from 20 to 29 years, some genetically abnormal embryos were produced from donors of each age, and there was no correlation between oocyte donor age and embryo aneuploidy. Moreover, a high-average correlation coefficient across all pairs of chromosomal abnormalities (r = 0.60) suggests that chromosomes tend to have multiple and simultaneous errors (complex aneuploidy) even when oocytes from young donors are used.
Conclusion
These data show that even when young, healthy donors provide oocytes for IVF, the probability of embryo aneuploidy remains high. The oocyte donor appears to make an important contribution to embryo aneuploidy even when her age is <30 years. If these findings are confirmed with larger, prospective studies, the integration of routine preimplantation testing with donor oocyte IVF cycles to identify single euploid embryos for transfer should be considered.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Aneuploidy
- Donor oocyte
- Implantation failure
- Parental genotype
- IVF
- Parental
- Gamete
- Segmental copy imbalance
Introduction
Preimplantation genetic diagnosis (PGD) and preimplantation genetic screening (PGS) are techniques for genetic assessment of embryos prior to transfer into the uterus. These tests offer “at-risk” individuals a greatly improved chance to have an unaffected child. A component of in vitro fertilization (IVF), each is associated with a growing range of uses in clinical fertility practice. Of note, in Europe PGD/PGS is variously prohibited, allowed, or practiced in the absence of legislation, depending on national statues [1]. There are no regulations addressing the provision of PGD or PGS in the United States [2].
In the early 1990s, PGD was first successfully applied to sex determination of embryos to reduce the likelihood of transmitting sex-linked conditions to offspring. In the setting of a family history of any recessive X-linked disease predominantly affecting males (i.e., glucose-6-phosphate dehydrogenase deficiency, Duchenne muscular dystrophy, hemophilia A and B, Wiskott–Aldrich syndrome, etc.), parents might elect to undergo embryo screening to identify female vs. male embryos. Then, only an unaffected female embryo would be transferred [3].
From that early success, reproductive medicine has embraced a substantial expansion of applications for preimplantation embryo assessment in IVF. This technology is currently used to identify embryos with hundreds of very serious single-gene disorders like Huntington’s disease, as well as to permit embryo sex selection on an elective basis [4]. Moreover, because poor IVF outcomes are often related to embryonic chromosomal abnormalities [5], PGS is increasingly used to screen for aneuploid embryos to optimize pregnancy rates and attenuate the miscarriage rate after in vitro fertilization procedures [6].
Indeed, evidence is accumulating that implantation and pregnancy rates may remain encouraging even for IVF patients using native oocytes up to age 42, with the proviso that only euploid embryos (verified by PGS) are transferred [7]. Such results are consistent with the observation that advancing maternal age is directly correlated with an increasing frequency of chromosomal aberration in embryos [8, 9]. Since up to 60 % of all conceptions (unassisted) result in miscarriage before 12 weeks’ gestation irrespective of age [10], it seems likely that ploidy error in human embryos is not a challenge confined only to oocyte sources of advanced age. For example, when selected chromosomes were studied in embryos obtained from donor-egg IVF treatment, the aneuploidy rate in this partial genomic assessment was higher than expected, particularly considering the egg donors themselves had no infertility diagnosis [11].
Building on this earlier pioneering work, our study reviewed use of comprehensive chromosomal screening in the context of anonymous donor-egg IVF. Using increased bandwidth to capture comprehensive screening data on all 23 pairs of chromosomes, this investigation aimed to answer two unresolved questions: (1) What is the true incidence of genetic abnormality in embryos produced from anonymously donated oocytes and (2) what is the gametic source of embryo aneuploidy observed in donor oocyte IVF?
Methods
Study Design
This retrospective investigation reviewed selected data from all in vitro fertilization (IVF) cases from a single institution in California in 2013 to identify the subset of patients where PGS was performed on embryos derived exclusively from anonymous oocyte donors. IRB approval was sought although the proposal was classified as exempt, since the study design reviewed already collected data and no specific patient identifiers were recorded. For our study, 23 cases meeting the eligibility criteria were identified; these patients produced 305 embryos for full molecular karyotyping. This information was collated with parental DNA obtained immediately before IVF (i.e., from the anonymous egg donor and the partner’s husband) for chromosome-specific assessments. This approach permitted mitotic and meiotic copy errors to be differentiated for each chromosome among all embryos tested, thus providing information on the specific parental source of embryo aneuploidy.
Oocyte Donor and Patient Selection
Anonymous oocyte donors had completed comprehensive medical and psychological evaluation as described previously [12]. Additionally, donors underwent a genetic evaluation and were required to have a normal result (no mutations) on an expanded carrier test [13] before enrollment. Recipients had their initial reproductive endocrinology consultation and monitoring at our facility, and all baseline laboratory tests were within normal limits. Anonymous oocyte donor counseling was provided by an accredited psychologist before starting gonadotropins. Each recipient selected her anonymous oocyte donor via secure Internet portal with an electronic lockout mechanism to prohibit multiple recipients from accessing the aggregate donor pool at the same time. A dedicated nurse coordinator was available to facilitate oocyte donor selection in all cases. Following registration of each provisional donor–recipient match, the corresponding anonymous oocyte donor entry was deleted from the donor library, thus creating a 1:1 ratio for each recipient and their anonymous oocyte donor (i.e., no two IVF recipients utilized oocytes from the same anonymous donor for this analysis).
The anonymous oocyte donor commenced controlled ovarian hyperstimulation, and transvaginal ultrasound-guided oocyte collection followed 36 h after s.c. hCG administration as previously described [14]. Sperm from the recipient’s partner was used to fertilize all freshly retrieved eggs obtained from the anonymous oocyte donor; intracytoplasmic sperm injection (ICSI) was performed in all cases.
For all records reviewed for this study, recipient and partner/husband ages were tabulated, as was age of the anonymous oocyte donor. Husband’s sperm concentration and sperm motility were calculated as an average of two semen analyses performed no more than 6 months before treatment. The following laboratory parameters were also evaluated: number of oocytes fertilized (via ICSI), number of 2pn zygotes produced, number of embryos biopsied, day of biopsy, and number of euploid embryos. In addition, the number and frequency of error observed in each chromosome was recorded, with reference to the (genetic) parental origin of the abnormality, as described previously [15].
Ovarian Stimulation and Fertilization
Before commencing gonadotropin therapy, oocyte donors underwent transvaginal ultrasound evaluation with remeasurement of serum FSH, LH, and estradiol on day 3 of the index cycle. Pituitary downregulation was achieved with a GnRH agonist administered on day 21 of the cycle immediately preceding treatment, as previously described [14]. Periodic transvaginal ultrasound and serum estradiol measurements were used to track follicular growth and thickness of the endometrial lining. When ≥3 follicles reached the 19 mm mean diameter, periovulatory hCG was administered by subcutaneous injection of recombinant hCG (250 μg Ovidrel®, Merck Serono; Geneva, Switzerland) with oocyte retrieval performed under transvaginal ultrasound guidance 35–36 h later. Following removal of all cumulus cells, ICSI was performed, and normal fertilization was verified 16–18 h after injection by the presence of two pronuclei and two polar bodies.
Embryology Protocol
Embryo biopsy was performed either on the morning of day 3 or on day 5 (blastocyst stage). Biopsy at day 3 was completed after laser-assisted hatching followed by removal of a single blastomere. Extended embryo culture occurred in Global single-step medium (IVF on Line; Guilford, CT) to blastocyst stage. On day 3 when embryos were at the 6–8 cell stage, a laser (Lycos, Hamilton Thorne; Beverly, MA) was used to create a 6–9 μ circular lacuna in the zona pellucida. This enabled rapid biopsy of trophectoderm (TE) on day 5. Between 3 and 5 herniated TE cells were gently aspirated by a pipette and, when necessary, freed from the blastocyst by application of laser pulses. Harvested TE cells were washed in PBS and placed within a PCR tube with 2.5 μL 1× PBS.
Cell Isolation, DNA Amplification, and Genotyping
Genetic material was obtained from oocyte donors via buccal swabs, from the recipient’s husband by peripheral venipuncture, and from the embryos by either single-cell day 3 blastomere biopsy or multicell day 5 trophectoderm biopsy. Single tissue culture (PMNs) and egg donor buccal cells were isolated using a sterile tip attached to a pipette and stereomicroscope (Leica; Wetzlar, Germany). For fresh day 3 embryo biopsy, individual blastomeres were separated via a micromanipulator after laser-facilitated zona hatching as described above; a micromanipulator was also used to isolate individual sperm cells. Except for sperm, single cells for analysis were washed ×4 with buffer (PBS buffer, pH 7.2; Life Technologies, Carlsbad, CA). Multiple displacement amplification (MDA) with proteinase K buffer (PKB) was used for this procedure; cells were placed in 5 μL PKB (Arcturus PicoPure Lysis Buffer, 100 mM DTT, 187.5 mM KCl, 3.75 mM MgCl2, 3.75 mM Tris-HCl) incubated at 56 °C × 1 h, followed by heat inactivation at 95 °C × 10 min, and held at 25 °C × 15 min. MDA reactions were incubated at 30 °C × 2.5 h and then 65 °C × 10 min.
Genomic DNA from buccal tissue was isolated using the QuickExtract DNA Extraction Solution (Epicentre; Madison, WI). Template controls were included for the amplification method. Amplified single cells and bulk parental tissue were genotyped using the Infinium II (Illumina; San Diego, CA) genome-wide single nucleotide polymorphism (SNP) arrays (CytoSNP 12 chip). The standard Infinium II protocol was used for parent samples (bulk tissue), and Genome Studio was used for allele calling. For single cells, genotyping was accomplished using an Infinium II genotyping protocol.
Copy Number and Haplotype Phasing
Because some commercial software packages use heterozygosity to determine copy number and high rates of ADO with preferential amplification in single-cell measurements can cause unpredictable heterozygosity (regardless of chromosome copy number), performance is poor when calling copy number on noisy single-cell data. Accordingly, previous investigators [9] developed a chromosome copy number classification algorithm in MATLAB (MathWorks; Natick, MA), predicated on parental genotypes and the observed distribution of unprocessed single-cell microarray channel intensities collated by parental origin [16, 17].
In brief, this approach is based on prior work [15] whereby the statistical behavior of each parental group differs as a function of the underlying chromosome copy number of the embryo. These changes are predictable and derive from additional allelic content that is contributed by (or missing from) each parent [15]. Moreover, rank statistics are examined for each parental context and compared to the expected orderings under the various chromosome copy number possibilities. Next, the probability is examined for each parental context that could have swapped rank by random chance to establish copy number and calculate confidences [15, 18].
Detection of three unmatched haplotypes adds additional confidence to a trisomy call, as many chromosome copy number errors are meiotic and will be associated with this configuration. Accordingly, this method included parental information with high-confidence disomic single-cell measurements on offspring and recombination probabilities to determine the parental chromosome phase. A maximum likelihood estimator algorithm was used to phase full chromosomes for all parental genotype contexts. Possible haplotypes in single-cell measurements are then evaluated to detect meiotic trisomies.
Segmental copy imbalances were detected by dividing each chromosome into five segments, with the aforementioned algorithm applied to each section independently. If any segments differ in copy number with high confidence, then the corresponding chromosome is flagged. Note that the reported copy number for chromosomes with a segmental imbalance is reflective of the call on the majority of the chromosome, even if part of the chromosome shows gain or loss. Thus, depending on size, segmental copy imbalances may reduce composite confidence of the complete chromosome call. However, confidences on chromosomes with segmental imbalances may still be high if the deletion is relatively small and/or the remainder of the chromosome is called with very high confidence [15].
Individual chromosome means and standard deviations of normalized microarray probe intensities were used to call chromosome copy number. For each single-cell measurement, a training set of single-cell amplification microarray measurements was used to normalize probe intensities across each chromosome. An algorithm was next used to compute the most likely chromosome state for all the single-cell amplification microarray data.
Statistical Analysis
Data were aggregated, analyzed, and visualized with Tableau 8.2 (Tableau Software; Seattle, WA). To estimate a reference population’s aneuploidy rate and the donor (maternal genetic) aneuploidy contribution, a binomial proportion confidence interval was used on each proportion estimate using the Wald test. When sample size was small (defined as min[np, n(1 − p)] < 5), an adjusted Wald method [19] was used to improve estimate accuracy. For this analysis, the confidence level was set at 95 % by default (90 % for aneuploidy rate comparisons). To compare two sample ratios, the 2-proportion z-test was used for large samples (defined as min[np, n(1 − p)] ≥ 5); Fisher’s exact test was used when sample size was small.
Results
A total of 676 IVF cases proceeded to oocyte retrieval during the 12-month review period ending December 2013. Of these, 50 were anonymous oocyte donors undergoing ovum pickup. The male partners of the intended parents had a mean (±SD) age of 44.3 ± 7.1 (range 25–58 years). Average sperm concentration and motility were 52.8 M/mL (range 2.4–135 M/mL) and 40.8 % (range 2–81 %), respectively.
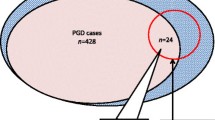
A total of 428 patients requested PGS during the study interval. Intersecting these two patient subsets identified 24 IVF cases which included both anonymous oocyte donation and PGS (see Fig. 17.1). Analysis of this group revealed that 305 embryos were subjected to biopsy and full molecular karyotyping. The mean (±SD) age of recipient females in this study population was 42.5 ± 4.0 (range 35–52) years. Mean (±SD) age was 24.0 ± 2.7 (range 20–29) years for oocyte donors (n = 24).
In this study group, the mean (±SD) number of oocytes which underwent fertilization by ICSI was 17.7 ± 7.8 (range = 6–35), and this yielded an average of 15.1 ± 6.7 2pn zygotes per patient (range = 6–32). Most embryos (86 %) were biopsied on day 3, while the remainder (14 %) were biopsied on day 5. Although the number of blastocyst biopsies was relatively small (n = 44), it was possible to record embryo ploidy as a function of biopsy timing. Using this approach, we found the incidence of missed calls (“no signal”) on chromosomes to be significantly higher among embryos biopsied at day 3, resulting in reduced reporting efficiency for this group compared to the blastocyst biopsy group (92 % vs. 100 %; p = 0.05).
Assessment of all embryos produced from oocytes contributed by an anonymous donor identified euploidy in 133 of 284 (46.8 %) embryos with full chromosomal reporting (i.e., zero “no calls”). Complete data on all 23 chromosome pairs was reported for 93.1 % of embryos sampled (284 of 305). Considering all embryo chromosomes, mean error rate was 18 %. A chromosome-specific analysis found error present in all chromosomes; chromosome 22 was most often affected, and chromosome 15 was the least likely to have an abnormality (see Fig. 17.2). The relatively high Phi correlation coefficients (see Fig. 17.3) among embryo chromosome pairs with aneuploidy (r = 0.60, range 0.42–0.77; p < 0.01 by chi-square test) indicate that chromosomes tend to have multiple and simultaneous errors (complex aneuploidy).
When analysis was confined only to those embryos with no missed calls for any chromosome, errors attributable to a maternal source (i.e., from the oocyte donor) were noted in 133 of 284 embryos (46.8 %). Conversely, an embryo genetic abnormality of paternal origin was present in 104 of 284 embryos (36.6 %). Among all aneuploid embryos (n = 151), chromosomal errors from both genetic parents (i.e., oocyte donor and partner’s husband) were present in 57.0 % (see Fig. 17.4). While oocyte donor age ranged from 20 to 29 years, some genetically abnormal embryos were produced from donors of each age, and there was no correlation between oocyte donor age and embryo aneuploidy. Likewise, these data did not confirm a correlation between embryo aneuploidy and male partner age or any semen parameter.
Discussion
The role of PGS on the menu of clinical IVF services has evolved substantially in recent years. Although it is tempting to classify PGS applications as simply an accessory to “mainstream” IVF, genetic testing of embryos has been (and will continue to be) a crucial development in the progress of our field. Certainly the successful passage of the world’s first IVF regulatory legislation (Human Fertilisation & Embryology Act, 1990) was strongly influenced and enabled by the arrival of PGD in the United Kingdom [20]; further applications of this technique have continued to push the ethical boundaries for IVF into unfamiliar terrain [21].
In humans, most aneuploidies are triploidies, yet only those involving chromosomes 21, 18, and 13 are compatible with survival to term [22]. Duplication of other autosomes is poorly tolerated and is rarely seen in live births. Viable monosomies are only known to exist for chromosome X, while additional copies of sex chromosomes are developmentally permissive. PGS is a powerful clinical tool to assist in embryo selection to minimize transfer of such embryos, thus improving clinical outcomes.
The arrival of oocyte donation preceded PGS and was originally offered as a treatment for premature ovarian failure or oophorectomy [23]. Egg donation is now commonly in use for many settings besides diminished ovarian reserve, including its use to circumvent transmission of severe genetic disorder(s) in the birth mother to her offspring [24]. While the corrosive effect of age on female infertility can be successfully assuaged for couples using donated oocytes from a younger (presumably more fertile) woman [25], the degree of chromosomal error in embryos derived from such treatment has yielded some unexpected preliminary results [11].
For example, one IVF group recently conducted a 12-year retrospective study on genetic test data collected from anonymous oocyte donor applicants and found that genetic abnormalities caused a significant number of candidates to be excluded from their oocyte donor program [26]. We agree with this approach and, like many institutions, require any potential anonymous oocyte donor to first undergo a careful genetic testing regime before entering the roster of eligible oocyte donors. Indeed, all of the anonymous donors who supplied oocytes for the current study already had been screened for hundreds of genetic disorders in advance of their accession into our egg donor group. However, despite this reassuring clearance (and in the absence of any obvious reproductive pathology in the oocyte donors) the rate of chromosomal error among embryos produced from their eggs was surprisingly high (e.g., 55 % aneuploidy rate).
Previous research attempted to characterize the role of “defective” gametes resulting in the generation of abnormal embryos using an egg-sharing model (where one IVF patient agrees to share her eggs with another IVF patient) [27]. Unfortunately, this can yield an undesirable outcome for the recipient since what she ultimately gets are simply eggs from another infertile patient. Such a study is unsatisfying experimentally because the variable of oocyte pathology cannot be controlled if all the oocytes for study are generated by other patients with manifold infertility diagnoses.
This problem was also addressed when the aneuploidy rate for eight chromosomes in embryos derived from young (<35 years) oocyte donors using fluorescence in situ hybridization analysis was studied. Using this study approach, all oocytes were provided by healthy women who did not have any infertility diagnosis. The authors reported considerable variation between donor cycles with nearly one-third having <30 % genetically normal embryos [11]. Starting from these data where less than half of the embryo’s chromosomes had been evaluated, our work was built on this foundation to screen all 23 pairs of embryo chromosomes in an anonymous donor oocyte IVF setting. Importantly, since the behavior of each parental allelic group is a function of the underlying chromosome copy number of the embryo, and because these modifications may be satisfactorily estimated from additional allelic content contributed by (or omitted from) either the oocyte donor or the recipient’s husband (sperm source), we were able to supply additional information on the parental origin of the genetic problems identified in the embryos derived therefrom.
Earlier research has shown a significantly higher observed pregnancy loss rate among IVF patients with age ≥40 compared to women younger than age 40 [28], establishing that the distribution of genetic error in embryos as a function of maternal age is not stationary. This physiologic process of natural ovarian senescence has been sidestepped for many years by using oocytes provided by younger donors [29]. With further refinement of donor oocyte protocols, acceptance of this treatment in routine IVF practice has increased greatly over the last decade, and when donor oocytes are used, the likelihood of an excellent IVF outcome seems independent of recipient age [30]. In the United States, the incidence of twins is markedly higher among anonymous oocyte donor IVF cycles compared to IVF using native (autologous) oocytes (37 % vs. 29 %, respectively), which provides direct evidence that most clinics are not to following a current ASRM recommendation which encourages single embryo transfers when oocyte donor age is young [31]. Indeed, there now appears to be international consensus that elective single embryo transfers are appropriate for oocyte donor–recipient cycles where the donor has good prognosis and when good quality embryos are available [32].
Of note, comprehensive chromosomal screening has not been applied to embryos of donor oocyte origin to quantify the level of genetic abnormality which persists in such embryos until now. If ever the domain of anonymous donor oocyte IVF were regarded as a realm where the role of genetic error in embryos could be dismissed as unimportant, the current study highlights an important supporting role for PGS in this population of IVF patients, too. Moreover, these data provide some fresh observations on human embryo genetics. Here, we focused on the specific topic of parental origin with respect to chromosomal errors which may be harbored by IVF embryos. Our observation that a high rate of embryonic genetic anomaly could be traced back to the oocyte donor was not anticipated. Thus, it appears that the traditional view that most chromosomal errors are of maternal origin caused by malsegregation in the first meiotic division [33] remains valid, even when the age of the oocyte source is very low.
Our report has some limitations which should be acknowledged. Our data come from a retrospective analysis as an initial step to analyze readily accessible existing data. We aimed to produce a hypothesis about aneuploidy rate in embryos derived from anonymous donor oocytes which could then be tested prospectively [34]. Retrospective work has the potential for incomplete documentation, unrecoverable or unrecorded data, and variance in the quality of information recorded. The reliability of data entry is considered as high for this sample, and the proportion of incomplete records was marginal. Also, because our sample was limited and represented the chance event of an IVF patient using anonymous donor oocytes also incorporating preimplantation testing of embryos produced from this treatment, it is uncertain if these findings can generalize to all anonymous donor-egg IVF cases (it should be noted that a secondary chart review for our study population did not reveal any obvious characteristic which may have influenced the patient’s decision to include PGS in her IVF treatment). Perhaps the high economic cost of IVF in general (and donor oocyte treatment in particular) introduced some selection bias, since only the most affluent IVF patients could have afforded this treatment [35]. It would be interesting to query the remaining donor oocyte IVF patients in this series who declined PGS (n = 27), to understand better why they decided not to request genetic testing of their embryos; this represents an area of future research here. Finally, our analysis of male factor data was confined to the age of the recipient’s husband and only two semen parameters (sperm concentration and motility). We did not include sperm DNA fragmentation data in this study, although this has not yet been correlated with embryo ploidy [36].
In conclusion, although the problem of embryo aneuploidy does diminish somewhat when anonymous donor oocytes are used for IVF, our results show that it does not disappear entirely even when oocytes from donors as young as 20 years of age are used. Prospective investigations utilizing comprehensive chromosomal screening with larger samples will be welcomed for further study of this phenomenon going forward.
References
Basille C, Frydman R, El Aly A, Hesters L, Fanchin R, Tachdjian G, et al. Preimplantation genetic diagnosis: state of the art. Eur J Obstet Gynecol Reprod Biol. 2009;145(1):9–13.
Baruch S, Kaufman D, Hudson KL. Genetic testing of embryos: practices and perspectives of US in vitro fertilization clinics. Fertil Steril. 2008;89(5):1053–8.
Pray L. Embryo screening and the ethics of human genetic engineering. Nat Educ. 2008;1(1):207.
Sills ES, Palermo GD. Preimplantation genetic diagnosis for elective sex selection, the IVF market economy, and the child–another long day’s journey into night? J Assist Reprod Genet. 2002;19(9):433–7.
Pagidas K, Ying Y, Keefe D. Predictive value of preimplantation genetic diagnosis for aneuploidy screening in repeated IVF-ET cycles among women with recurrent implantation failure. J Assist Reprod Genet. 2008;25(2–3):103–6.
Sills ES, Yang Z, Walsh DJ, Salem SA. Comprehensive genetic assessment of the human embryo: can empiric application of microarray comparative genomic hybridization reduce multiple gestation rate by single fresh blastocyst transfer? Arch Gynecol Obstet. 2012;286(3):755–61.
Harton GL, Munné S, Surrey M, Grifo J, Kaplan B, McCulloh DH, et al. Diminished effect of maternal age on implantation after preimplantation genetic diagnosis with array comparative genomic hybridization. Fertil Steril. 2013;100(6):1695–703.
Schoolcraft WB, Fragouli E, Stevens J, Munne S, Katz-Jaffe MG, Wells D. Clinical application of comprehensive chromosomal screening at the blastocyst stage. Fertil Steril. 2010;94(5):1700–6.
Scott Jr RT, Ferry K, Su J, Tao X, Scott K, Treff NR. Comprehensive chromosome screening is highly predictive of the reproductive potential of human embryos: a prospective, blinded, nonselection study. Fertil Steril. 2012;97(4):870–5.
Fritz MA, Speroff L. Female infertility. In: Clinical gynecologic endocrinology and infertility. 8th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2011. p. 1137–41.
Munné S, Ary J, Zouves C, Escudero T, Barnes F, Cinioglu C, et al. Wide range of chromosome abnormalities in the embryos of young egg donors. Reprod Biomed Online. 2006;12(3):340–6.
Walsh AP, Omar AB, Collins GS, Murray GU, Walsh DJ, Salma U, et al. Application of EU tissue and cell directive screening protocols to anonymous oocyte donors in western Ukraine: data from an Irish IVF programme. J Obstet Gynaecol. 2010;30(6):613–6.
Higgins AS, Flanagan JD, Von Wald T, Hansen KA. Preconception cystic fibrosis screening in infertile couples using an expanded carrier screening test. Obstet Gynecol. 2014;123 Suppl 1:97S.
Sills ES, Schattman GL, Veeck LL, Liu HC, Prasad M, Rosenwaks Z. Characteristics of consecutive in vitro fertilization cycles among patients treated with follicle-stimulating hormone (FSH) and human menopausal gonadotropin versus FSH alone. Fertil Steril. 1998;69(5):831–5.
Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, et al. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod. 2010;25(4):1066–75.
Rabinowitz M, Sweetkind-Singer J, Banjevic M, Johnson DS, Kijacic D, Petrov D, et al. System and method for cleaning noisy genetic data from target individuals using genetic data from genetically related individuals. 2007. U.S. Patent 2007018467A1, 9 August 2007.
Johnson DS, Rabinowitz M, Banjevic M, Singer J, Cinnioglu C, Baner J, et al. Leveraging parental genotypes to increase confidence in genotype calls on single cells. Hum Reprod. 2008;23 Suppl 1:i67.
Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502.
Wu H, Neale MC. Adjusted confidence intervals for a bounded parameter. Behav Genet. 2012;42(6):886–98.
Mulkay M. The embryo research debate: science and the politics of reproduction. Cambridge: Cambridge University Press; 1997. p. 41. 132–3.
Steffann J, Frydman N, Burlet P, Gigarel N, Hesters L, Kerbrat V, et al. Extending preimplantation genetic diagnosis to HLA typing: the French exception. Bull Acad Natl Med. 2011;195(4–5):1015–21.
Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91.
Shulman A, Frenkel Y, Dor J, Levran D, Shiff E, Maschiach S. The best donor. Hum Reprod. 1999;14(10):2493–6.
Dean NL, Edwards RG. Oocyte donation–implications for fertility treatment in the nineties. Curr Opin Obstet Gynecol. 1994;6(2):160–5.
Sauer MV, Paulson RJ, Lobo RA. Reversing the natural decline in human fertility. An extended clinical trial of oocyte donation to women of advanced reproductive age. JAMA. 1992;268(10):1275–9.
Reh A, Amarosa A, Licciardi F, Krey L, Berkeley AS, Kump L. Evaluating the necessity for universal screening of prospective oocyte donors using enhanced genetic and psychological testing. Hum Reprod. 2010;25(9):2298–304.
Katsoff B, Check JH, Mitchell-Williams J. Defective oocytes are not a common cause of unexplained infertility as determined by evaluation of sharing oocytes between infertile donors and recipients. Clin Exp Obstet Gynecol. 2013;40(2):193–5.
Spandorfer SD, Davis OK, Barmat LI, Chung PH, Rosenwaks Z. Relationship between maternal age and aneuploidy in in vitro fertilization pregnancy loss. Fertil Steril. 2004;81(5):1265–9.
Lydic ML, Liu JH, Rebar RW, Thomas MA, Cedars MI. Success of donor oocyte in in vitro fertilization-embryo transfer in recipients with and without premature ovarian failure. Fertil Steril. 1996;65(1):98–102.
Kawwass JF, Monsour M, Crawford S, Kissin DM, Session DR, Kulkarni AD, et al. Trends and outcomes for donor oocyte cycles in the United States, 2000–2010. JAMA. 2013;310(22):2426–34.
Myers ER. Outcomes of donor oocyte cycles in assisted reproduction. JAMA. 2013;310(22):2403–4.
Min JK, Hughes E, Young D, Gysler M, Hemmings R, Cheung AP, et al. Elective single embryo transfer following in vitro fertilization. J Obstet Gynaecol Can. 2010;32(4):363–77.
Larsen EC, Christiansen OB, Kolte AM, Macklon N. New insights into mechanisms behind miscarriage. BMC Med. 2013;11:154.
vonKoss Krowchuk H, Moore ML, Richardson L. Using health care records as sources of data for research. J Nurs Meas. 1995;3(1):3–12.
Sills ES, Collins GS, Salem SA, Jones CA, Peck AC. Balancing selected medication costs with total number of daily injections: a preference analysis of GnRH-agonist and antagonist protocols by IVF patients. Reprod Biol Endocrinol. 2012;10:67.
Bronet F, Martínez E, Gaytán M, Liñán A, Cernuda D, Ariza M, et al. Sperm DNA fragmentation index does not correlate with the sperm or embryo aneuploidy rate in recurrent miscarriage or implantation failure patients. Hum Reprod. 2012;27(7):1922–9.
Conflict of Interest
The authors declare no conflict.
Note A version of this work appeared in the journal Molecular Cytogenetics 2014;7:68.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Sills, E.S., Li, X., Potter, D.A., Frederick, J.L., Khoury, C.D. (2015). Should Molecular Cytogenetic Techniques Be Applied to Facilitate Single Embryo Transfer in Egg Donation Cases? Assessment of Frequency and Distribution of Embryo Aneuploidy After Anonymous Donor Oocyte IVF. In: Sills, E. (eds) Screening the Single Euploid Embryo. Springer, Cham. https://doi.org/10.1007/978-3-319-16892-0_16
Download citation
DOI: https://doi.org/10.1007/978-3-319-16892-0_16
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-16891-3
Online ISBN: 978-3-319-16892-0
eBook Packages: MedicineMedicine (R0)