Abstract
This chapter will cover the following topics:
Introduction
-
Why develop PGD?
-
The impetus for its development
-
Research progress that made it possible
-
Legislation—the Warnock report
Early steps towards PGD
-
Sexing of embryos by DNA amplification
-
FISH comes into its own
-
Mosaicism and the extent of aneuploidy—implications for PGD
-
Aneuploidy screening begins
-
First single gene diagnosis
-
First cancer gene PGD
Development of single cell CGH for comprehensive chromosome analysis
-
PGS gets a bad name
-
Application of metaphase CGH
-
Development of array CGH and its application for PGS to select ‘the single euploid embryo’.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Embryo genetics
- Comprehensive chromosomal screening
- History of IVF
- Preimplantation genetic diagnosis
- Preimplantation genetic screening
Introduction
By 1987 early prenatal diagnosis of both chromosomal and single gene defects was possible via chorionic villus sampling, so why was there a perceived need to develop preimplantation genetic diagnosis (PGD)? There were two groups of patients that provided driving forces. Firstly couples known to be at high genetic risk had expressed the fervent wish to be able to start a pregnancy knowing that it would not be affected; many of these couples had experienced the trauma of repeated second trimester terminations of much wanted pregnancies. A second group, for whom PGD would obviously be of great benefit, included couples where the women had been shown from pedigree analysis to be carriers of an X-linked condition for which at the time there was no specific diagnostic test. For this group, the only option was prenatal testing of an established pregnancy to determine the sex. This then led to the termination of all male pregnancies, of which only 50 % were likely to be affected [1]. Various events around this time had made the development of PGD a possible option. In 1983 Trounson and Mohr [2] had shown that it was possible for a normal pregnancy to occur even after the destruction of blastomeres following embryo freezing. This finding indicated that it should be feasible to remove one or two cells from a cleavage stage embryo for diagnosis without compromising its further development. In the UK, Dame Mary Warnock chaired a government committee composed of people from a variety of backgrounds that considered the status of the human embryo before preimplantation with regard to the ethics of research on embryos at this stage of their development. The subsequent report was published in 1984 by the DHSS; it proposed a time limit for research of 14 days after fertilisation, well beyond the time when cells would be removed to allow genetic diagnosis. This paved the way for subsequent government legislation that was in line with the committee’s recommendations.
Early Steps in PGD
With the aim of helping couples at risk of an X-linked disorder, the first approach to PGD was in order to sex the embryo. Handyside and colleagues at the Hammersmith Hospital in London reported in 1989 that they had been able to biopsy single cells from 30 embryos and that the expected proportion had developed to blastocysts after 6 days in culture [3]. Furthermore in all the normally fertilised embryos they were able to determine the sex by DNA amplification of a Y-chromosome-specific repetitive sequence. In 15 cases, the sex was confirmed by means of in situ hybridisation or Y chromosome fluorescence in metaphases. Shortly this was followed by the report from the same group of pregnancies from embryos sexed by Y-specific DNA amplification [4]. However, this approach proved to be error prone since crucially it relied on a negative result to identify the females. The development of the rapid and reliable technique of fluorescence in situ hybridisation (FISH) at the end of the 1980s proved a saviour and was quickly applied to biopsied cells from human embryos with excellent results [5]. The application of FISH for embryo sexing at UCL in London gave reliable diagnostic results but also gave the first indication of the frequency of aneuploidy and chromosomal mosaicism in these embryos created by in vitro fertilisation (IVF) from fertile patients [6]. Prior to this, the IVF specialists were looking forward to treating PGD patients who would be fertile, anticipating that IVF would have a much improved success rate compared with that for infertile couples. Simultaneously, FISH was being applied to biopsied cells from cleavage stage embryos by Munne’s group in the USA and in 1993 they also reported the diagnosis of major aneuploidies in mosaic and full form [7]. Evidently, embryos created by IVF from couples of proven fertility were also prone to mosaic aneuploidy of an extent that was going to affect viability and implantation rates as well as the accuracy of PGD. So it was that an additional aim was added: as well as using PGD to detect heritable genetic disorders, it could be applied to help improve the success rate of IVF for infertile couples by using FISH to detect aneuploidy—this was the birth of PGS—preimplantation genetic screening.
Meanwhile, in 1992 the Hammersmith group reported the first successful PGD for a single gene disorder: the birth of a normal girl, free of cystic fibrosis, after PGD [8]. Within a few years, FISH was being applied in London to biopsied blastomeres to help couples at risk of passing on an unbalanced form of a reciprocal or Robertsonian translocation [9], and in the USA preconception diagnosis was achieved for maternal carriers by testing the first polar body alone while in Chicago it was tested in combination with karyotyping of the second polar body, also only for maternal carriers [10, 11]. While few would dispute on ethical grounds the application of PGD to avoid single gene defects that affect children, its use to avoid passing on genes that predispose to late-onset disorders such as adult cancers provoked more controversy.
Nevertheless, the UK Human Fertilisation and Embryology Authority licensed the procedure in the case of the APC gene that causes familial adenomatous polyposis (and inevitable colorectal cancer) when mutated and the first PGD diagnosis for inherited cancer, and of this condition, was carried out in London and reported in 1998 [12].
The Development of Comprehensive Chromosomal Analysis
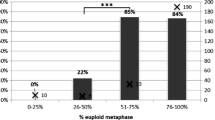
Initially, from 1999 onwards it was reported that the outcome for infertile couples improved significantly after PGS was applied to their embryos, compared with comparable control groups [13]. However, since FISH is perceived as an easy and reliable technique that any laboratory scientist may apply and achieve a successful outcome, it became widely used by IVF centres with no experience of genetic testing. Not surprisingly, the results for the patients were variable and doubts began to be expressed as to the benefits of PGS, mostly applied to older women with fewer embryos for testing. In 2007 a paper was published that reported on the outcome of a randomised clinical trial of PGS that showed a negative effect of screening via PGS [14]; this paper has been widely quoted but also heavily criticised on technical grounds by scientists with extensive experience of the application of FISH to human blastomeres. There are two contributory problems: one that in order to test as many chromosomes as possible, several rounds of hybridisation with FISH probes may be carried out; thus reducing the efficiency and secondly the widespread mosaicism that affects the early human embryo will clearly lead to apparent ‘misdiagnoses’ when testing only a single cell for aneuploidy. In the meantime, research was progressing on methods for comprehensive chromosome testing, based upon analysis of DNA extracted from a single cell. The aim was to be able to apply the technique of comparative genomic hybridisation (CGH) as used in tumour cytogenetics, where karyotyping was not possible. For this to happen, the DNA from each cell had first to be amplified in a manner compatible with analysis via CGH. Two groups from opposite sides of the world (London UK and Melbourne Australia) were successful in achieving single cell CGH analysis of blastomeres and simultaneously both groups published their work in the year 2000 [15, 16]. The results obtained confirmed the early FISH data with respect to the incidence of full and mosaic aneuploidy in apparently normally developing human embryos. These results were achieved by classical metaphase analysis after the combined hybridisation of both test and control DNAs; even with the help of computer software, that analysis required the ability to karyotype and took 72 h for the hybridisation step alone. Although both innovator groups did apply the technique clinically, these factors clearly limited full clinical application. The final step needed was the refinement of array CGH so that it could be applied to the analysis of single cells; early results from this development were described in 2009 [17]. By 2013 it was evident that centres were seeing an improved outcome with regard to both implantation and pregnancy rates compared with those achieved previously by FISH analysis [18]. It may be concluded that the development and application of a reliable aCGH method has made a major contribution to the stated goal of ‘Transferring the single euploid embryo’.
References
Penketh R, McLaren A. Prospects form prenatal diagnosis during preimplantation human development. Baillieres Clin Obstet Gynaecol. 1987;1(3):747–63.
Trounson A, Mohr L. Human pregnancy following cryopreservation, thawing and transfer of an eight cell embryo. Nature. 1983;305:707–9.
Handyside AH, Pattinson JK, Penketh RJA, et al. Biopsy of human preimplantaion embryos and sexing by DNA amplification. Lancet. 1989;18:347–9.
Handyside AH, Kontogianni EH, Hardy K, et al. Pregnancies from biopsied human preimplantation embryos sexed by Y-specific DNA amplification. Nature. 1990;344:768–70.
Griffin DK, Handyside AH, Penketh RJA, et al. Fluorescent in situ hybridization to interphase nuclei of human preimplantation embryos with X and Y chromosome specific probes. Hum Reprod. 1991;6:101–5.
Delhanty JDA, Griffin DK, Handyside AH, et al. Detection of aneuploidy and chromosomal mosaicism in human embryos during preimplantation sex determination. Hum Mol Genet. 1993;2:1183–5.
Munne S, Lee A, Rosenwaks Z. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8:2185–92.
Handyside AH, Lesko JG, Tarin JJ, et al. Birth of a normal girl after in vitro fertilization and preimplantation diagnostic testing for cystic fibrosis. N Engl J Med. 1992;327:905–9.
Conn CM, Harper JC, Winston RML, et al. Preimplantation diagnosis of trisomies 13,14,18 & 21 in translocation carriers using multicolor fluorescent in situ hybridization (FISH). Am J Hum Genet. 1995;57:A277.
Munne S, Scott R, Sable D, et al. First pregnancies after pre-conception diagnosis of translocations of maternal origin. Fertil Steril. 1998;69:675–81.
Verlinsky Y, Eviskov S. Karyotyping of human oocytes by chromosomal analysis of the second polar bodies. Mol Hum Reprod. 1999;5:89–95.
Ao A, Well D, Handyside AH, et al. Preimpantation genetic diagnosis of inherited cancer: familial adenomatous polyposis coli. J Assist Reprod Genet. 1998;15:140–4.
Munne S, Magli C, Cohen J, et al. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Hum Reprod. 1999;14:2191–9.
Mastenbroek S, Twisk M, Van-Echten-Arends J, et al. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357:9–17.
Wells D, Delhanty JDA. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol Hum Reprod. 2000;6:1055–62.
Voullaire L, Salter H, Williamson R, et al. Chromosome analysis of blastomeres from human embryos using comparative genomic hybridization. Hum Genet. 2000;106:210–7.
Wells D, Alfarawati S, Fragouli E. Use of comprehensive chromosome screening for embryo assessment: microarrays and CGH. Mol Hum Reprod. 2009;14:703–10.
Rubio C, Rodrigo L, Mir P, et al. Use of array comparative genomic hybridization (array-CGH) for embryo assessment: clinical results. Fertil Steril. 2013;99:1044–8.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Delhanty, J.D.A. (2015). The Development of PGD. In: Sills, E. (eds) Screening the Single Euploid Embryo. Springer, Cham. https://doi.org/10.1007/978-3-319-16892-0_1
Download citation
DOI: https://doi.org/10.1007/978-3-319-16892-0_1
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-16891-3
Online ISBN: 978-3-319-16892-0
eBook Packages: MedicineMedicine (R0)