Abstract
ATP-dependent chromatin remodeling complexes are involved in chromatin remodeling thereby controlling gene expression. These multi-subunit containing complexes contain an ATPase of the SNF2 family that hydrolyzes ATP in order to modify or reshape histone-DNA interactions within nucleosomes. Several subfamilies of the SNF2 family have been identified in different species based on the status of their catalytic ATPase subunit. All of the SNF2 subfamily members, including SWI2/SNF2 (BRG1/BRM), ISWI and CHD/Mi-2β, play critical roles in the maintenance of epidermal homeostasis. The BRG1 chromatin remodeler is required for maintenance of bulge stem cells, hair cycling, normal skin homeostasis, and repair and regeneration processes. ACTL6a (actin-like 6a) modulates the SWI/SNF complex to suppress differentiation in the epidermis. Realignment of the epidermal differentiation complex (EDC) locus, which occurs before activation of EDC genes that drive keratinocyte terminal differentiation, is coordinated by p63 by directly regulating the expression of BRG1. The combined effects of p63 and BRG1 control higher order chromatin remodeling, 3D-genomic organization and efficient expression of EDC genes in epidermal precursor cells during epidermal morphogenesis. BRG1 also suppresses p27kip1, allowing self-renewal of hair follicle bulge cells, and recruits the transcription factor NF-kB, which in turn activates Shh in matrix cells, promoting proliferation. Finally, Shh signaling through Gli activates BRG1 in bulge cells. Hence, BRG1 is necessary for Shh expression in both matrix and bulge compartments, and for hair regeneration and skin repair post-wounding. Mice with deletion of Mi-2β of the SNF2 family in the epidermis die perinatally and display severalphenotypes that differ between dorsal and ventral skin, suggesting spatio-temporal control of gene expression by Mi-2β in the epidermis. Mi-2β is also important for hair follicle morphogenesis and is necessary to reprogram epidermal basal cells to a hair follicle fate. Non-melanoma skin cancers in humans display mutations in the Brm gene following ultraviolet (UV) irradiation, and BRM normally protects epidermal cells from UV irradiation-induced hyper-proliferation, even in the presence of a partial loss of p53, thereby establishing its role as a tumor suppressor.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- ATP-dependent chromatin remodeling complexes
- SWI2/SNF2
- stem cells
- epidermis
- hair follicles
- 3D-genomic organization
- wound healing
- EDC
- BRG1/BRM
6.1 Introduction
Regulation of gene expression requires interplay between the transcription machinery and the multi-subunit protein complexes that are involved in remodeling of chromatin, thereby controlling the spatial and temporal accessibility of sequence specific transcription factors to their corresponding target genes. Such complexes either covalently alter histones or DNA, or dismantle histone-DNA contacts through ATP-dependent nucleosome remodeling. ATP-dependent chromatin remodeling (ATP-DCR) complexes contain multiple subunits, including an ATPase of the SNF2 family that hydrolyzes ATP in order to modify or reshape the histone-DNA interaction within the nucleosomes, leading to nucleosome sliding, removal of histones, and/or exchange of histone variants [1]. The first chromatin remodeling complex was discovered during screens in yeast for molecules involved in the signal transduction responsible for mating type switching, and is also known as Switch or SWI [2].
In different species, several subfamilies of the SNF2 family, including SWI2/SNF2, ISWI and CHD/Mi-2, have been identified based on the status of their catalytic ATPase subunit. SWI2/SNF2, also known as the BRG1/BRM-associated factor (BAF) complex, is one of the best characterized nucleosome remodeling complex subfamilies, and is highly conserved among eukaryotes. Animal SWI2/SNF2 complexes contain one of the two catalytic ATPase subunits, BRG1 (SNF2α) or BRM (SNF2β) , and a variable, eight to fourteen regulatory subunit configuration of BAFs [3]. These subunits can be encoded by at least 20 different genes, resulting in over 200 amalgamations of complexes. These target different DNA sequences and may have different functions [1, 4]. All of these alterations to nucleosome architecture result in an open chromatin structure and transcriptional activation, leading to gene expression.
The following Chapter focuses on the in vivo role of ATP-dependent chromatin remodeling in the control of epidermal differentiation and skin stem cell activity during hair follicle morphogenesis, hair cycling and wound healing in mammals such as mice and humans.
6.2 Control of Epidermal Differentiation
Skin is the largest organ of the body and protects us from toxic and arid external hazards by establishing and maintaining the epidermal permeability barrier (EPB). Proliferating cells in the epidermal basal layer undergo stepwise terminal differentiation, giving rise to a stratified epithelium. Cells in the outermost layer of the epidermis become enucleated and form the stratum corneum, whose cells are sloughed off periodically and are replenished from the proliferative basal layer [5].
Prior in vitro studies suggested that BRM and BRG1, the two ATPases of the SWI2/SNF2 family, regulate the expression of independent sets of genes [6]. However, viability of Brm-null mice indicated that these two factors are functionally redundant in vivo and that BRG1 can compensate for loss of BRM [7]. In contrast, as fibroblasts lacking Brg1 are viable but Brg1-null embryos die very early during development (during the peri-implantation stage), BRG1 might exert cell-specific functions in early development [8].
In order to elucidate the in vivo role of these factors in the later stages of development, including in skin development and maintenance of skin homeostasis, mice bearing LoxP-flanked (floxed) Brg1-alleles were established. Brg1/SNF2α was ablated in the forming epidermis using K14-Cre [9] or K14-Cre-ERT2 [10] transgenic mice that express either the bacteriophage P1 Cre-recombinase or the ligand-dependent Cre-ERT2 recombinase driven by the human K14 promoter, which is active in the surface ectoderm and the basal layer of the epidermis [11]. BRG1 is expressed in the surface ectoderm including that of the outgrowing limbs as early as embryonic day 10 (E10) of development [12]. At E18.5, BRG1 is strongly expressed in most, if not all, basal cells, as well as in about 70% of the spinous and 30% of the granular cells of the developing epidermis. Using constitutively active Cre recombinase, Brg1 was efficiently ablated in epidermal keratinocytes and in the ectodermal layer of the limbs before E12 [12, 13]. Ablation of Brg1 in the surface ectoderm induced severe hindlimb defects due to lack of maintenance of the apical ectodermal ridge (AER). The absence of forelimb defects in constitutive Cre mutants most likely reflects the earlier development of the forelimb, which occurs before efficient expression of the Cre recombinase [14], rather than differential participation of BRG1 in fore- and hindlimb development. Hindlimb defects can be avoided by inducing Brg1 ablation in postnatal skin. Indra and co-workers demonstrated that BRG1 is dispensable for formation of embryonic epidermis, but is essential for establishment of the epidermal permeability barrier (EPB). Interestingly, temporal ablation of Brg1 in the epidermis of mice lacking BRM showed that the BRM/BRG1 ATP-dependent chromatin remodeling complex is not required for epidermal proliferation and “early” differentiation, and revealed partial redundancy between BRM and BRG1 in regulating “late” terminal differentiation of the epidermal keratinocytes E12 [13].
Taken together, these results suggested that BRG1 selectively controls the expression of genes involved in the epithelial mesenchymal interactions required for limb patterning [15] and in terminal differentiation of keratinocytes during development. Similar to the situation in undifferentiated F9 embryonal carcinoma cells and in peri-implantation embryos [8, 16], BRM, which is dispensable for epidermis and limb formation, cannot functionally replace BRG1 in these processes. However, BRM can partially substitute for BRG1 in keratinocytes undergoing terminal differentiation.
It was recently shown that ACTL6a (actin-like 6a), a protein also known as BAF53a/INO80K/Arp4, modulates the SWI/SNF complex to suppress differentiation in the epidermis [17]. ACTL6 expression is downregulated during epidermal differentiation and is most strongly expressed in the less differentiated cells close to the epidermal basement membrane [17]. Spatio-temporal ablation of the epidermal ACTL6a resulted in reduced progenitor functions, premature terminal differentiation and epidermal thinning (hypoplasia) during epidermal development and also in adult tissue homeostasis [17]. Significant derepression of specific well-characterized differentiation related genes at the mRNA and protein levels was observed upon loss of ACTL6a [17]. ACTL6a target gene characterization identified KLF4 (Kruppel-like factor 4), a known activator of epidermal differentiation, as a key target of ACTL6a repression. A large number of genes that are regulated by ACTL6a were also identified as targets of KLF4. In line with this, KLF4 loss together with deletion of ACTL6a significantly compensated for the defects caused by ACTL6a depletion in progenitors [17].
Recent studies also suggest that ACTL6a can associate with different epigenetic regulators, including the Tip60 HAT complexes, the KAT2a HAT complexes, and the SWI/SNF chromatin-remodeling complex [18,19,20]. Depletion of either KAT2a or Tip60 failed to significantly alter expression of differentiation related genes. However, ablation of the largest component of the SWI/SNF complex, BAF250a/ARID1A, but not BAF250b/ARID1B, produced impacts similar to ACTL6a loss: decreased clonogenic growth and premature induction of differentiation. These results indicated that ACTL6a supports the maintenance of the epidermal progenitor state by sequestering BRM1/BRG1 to prevent activation of differentiation programs.
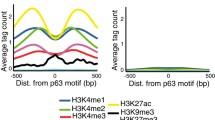
Chromatin immuno-precipitation assays on keratinocytes with functional ACTL6a indicated that compared to undifferentiated cells, differentiated keratinocytes displayed enhanced binding by BRM/BRG1 and RNA polymerase II at the promoters of differentiation genes, including KLF4 as well as KRT10, S100A9, SPRR3, and BMP6. Loss of ACTL6a in undifferentiated progenitor populations enhanced the binding of both BRM and BRG1 as well as RNA polymerase II to differentiation-gene promoters but failed to alter binding to other gene promoters (Fig. 6.1). Thus, ACTL6a helps to maintain the undifferentiated state by inhibiting SWI/SNF chromatin remodeling complex attachment to and activation of KLF4 and other differentiation related gene promoters [17].
Similar to the SWI2/SNF2 chromatin remodeling complex, the ISWI-containing NURF complex and the CHD-containing NuRD complex are important for maintenance of human keratinocyte stem cells in an undifferentiated state [21]. Although there are no other described functions for these complexes in epidermal differentiation and barrier formation, they are required, along with SWI/SNF, for DNA repair. Specifically, the NURF and NuRD complexes are essential for the exposure of damaged DNA bases to the repair machinery and for the repair of double-stranded breaks through recombination [22]. These events are critical for reducing the risk of DNA damage in skin by environmental insults such as solar UV-irradiation, and for diminishing the risk of developing melanoma and non-melanoma skin cancer.
The ATP dependent chromatin remodelers Mi-2α and Mi-2β, alternatively known as CHD3 and CHD4, are expressed in a wide range of developing tissues including skin and mucosal epithelia (Fig. 6.1) [23]. Mi-2β is expressed in epidermis and hair follicle placodes during embryonic development, and in the matrix of mature hair follicles (Fig. 6.1) [24]. Extensive studies of Mi-2β expression at the level of RNA were performed during the epidermal differentiation process. At E10.5, Mi-2β mRNA is uniformly expressed in the single layer ectoderm, and at E14.5 expression is observed in basal and suprabasal layers as well as in the hair peg and matrix of differentiating hair follicles (HF) (Fig. 6.2). By contrast, Mi-2β mRNA expression is very low in mature inter-follicular epithelium (IFE).
In order to study the role of Mi-2β/CHD4 in skin development and homeostasis in detail, Georgopoulos and coworkers generated mice with specific deletion of Mi-2β in keratinocytes using K14-Cre mediated cell-specific recombination [24]. Homozygous mice died within 24 h. of birth with shiny and flaky skin suggesting a possible role for Mi-2β in barrier formation. In addition, the mutants exhibited reduced numbers of HFs, abnormal whisker hairs, and curly tails (to be discussed later). These mice displayed striking difference in the phenotypes between the dorsal and ventral skin in terms of epidermal structure and presence of hair follicles. Early (E10.5) depletion of Mi-2β/CHD4 in the ventral epidermis resulted in reduction of epidermal suprabasal layers and depletion of the basal layers later in embryogenesis. In contrast, later (E13.5) loss of Mi-2β/CHD4 did not affect epidermal differentiation or maintenance of the basal layer, but induction of HFs was blocked. The ventral skin phenotype was caused by deletion of Mi-2β at early stages of epidermal development, whereas the distinct dorsal skin phenotype was due to removal of Mi-2β after initiation of epidermal morphogenesis [24].
During the process of epidermal differentiation and permeability barrier formation, it has been suggested that the nucleolus transitions from a state of active transcription in proliferating basal cells to a fully inactive state in the stratum corneum [25]. Current advances in analyses of 3D genomic organization using (a) confocal microscopy after fluorescent in situ hybridization or labeling with transgenic chimeric fluorescent proteins, and (b) chromatin conformation capture (3C, 4C and Hi-C) revealed that the relative positioning of chromosomes in the nucleus is not random, and instead is rather specific to cell type and cell size [26]. In the interphase nucleus, chromosomes reside in defined domain(s) and genes within these regions are non-randomly positioned relative to each other and to nuclear sub-organelles [26, 27]. As an example, in mouse epidermis the position of chromosome 3 in keratinocytes of the basal and suprabasal layers is more peripheral than that of chromosome 11 [25]. Massive remodeling of the higher-order chromatin structure of the epidermal differentiation complex (EDC) on mouse chromosome 3 occurs during epidermal morphogenesis. The locus moves away from the nuclear periphery and towards the nuclear interior into a location that is rich in SC35 (Srsf2)-positive nuclear speckles. This realignment of the EDC locus occurs before transcriptional activation of EDC genes that drive terminal differentiation of keratinocytes. The transcription factor p63, a master regulator of epidermal development [28, 29], orchestrates this lineage-specific, developmentally controlled event in the epidermis. In p63-null mouse skin, significant changes in expression of EDC genes are associated with alteration of the developmentally controlled relocation of the EDC within the nucleus [30]. In epidermal basal cells, p63 directly regulates expression of the ATP-dependent chromatin remodeler Brg1, which binds to distinct domains within the EDC and is required for repositioning of the EDC and Loricrin loci towards the nuclear interior (Fig. 6.1) [30]. This combinatorial effect of p63 and BRG1 drives higher order chromatin remodeling, 3D-genomic organization and efficient gene expression of the EDC genes in epidermal precursor cells during epidermal morphogenesis (Fig. 6.1) [30].
Studies have also shown that transcription factors MAF:MAFB are regulated by lncRNAs TINCR and ANCR, besides p63, to give rise to a complex regulatory gene network for epidermal differentiation [31]. Using DeepCAGE, genome-wide profiling of histone modifications and retroviral integration analysis, Cavazza et al., showed that most of the active promoters are differentially controlled in progenitor and differentiated keratinocytes, while nearly 50% of the enhancers and super-enhancers mediate, in a stage specific manner, the epigenetic changes in differentiated keratinocytes [32]. They also observed that p63 binds to and controls cell specific super-enhancers in both proliferating and differentiating keratinocytes.
6.3 Stem Cell Activity During Hair Follicle Morphogenesis and Cycling
Hair follicle (HF) morphogenesis is initiated during embryonic development and involves epithelial-mesenchymal interactions that require the activity of the Wnt/β-catenin, Shh, Notch, BMP and Edar signaling pathways [33]. In adult life, HFs cycle periodically through anagen (growth), catagen (regression) and telogen (resting) phases [25, 34,35,36]. HF stem cells (HFSCs) in the secondary hair germ and bulge regions are stimulated to proliferate at anagen onset. The secondary hair germ generates the proliferative HF matrix, which produces the hair shaft and its surrounding inner root sheath. Bulge stem cells give rise to the HF outer root sheath, and these contribute to the matrix in the subsequent HF growth cycle [33, 37, 38]. In homeostasis, bulge stem cells contribute only to the HF, but following skin wounding their progeny exit the hair follicle and contribute transiently to epidermal repair [34,35,36]. The ability of HFSCs to self-renew is critical to ensure that HFs can continue to cycle throughout life [33, 39].
HFSCs in the bulge and secondary hair germ are characterized by expression of distinct sets of markers (Krt15+, Lgr5+, CD34+, Sox9+, Lhx2+, Tcf3+, Nfatc1+ for the bulge, and Krt15+, Gli1+, Lgr5+ for the secondary hair germ). Additional stem cell populations reside in the junctional zone (Lrig1+), sebaceous glands (Blimp1+), and isthmus (Lgr6+, Plet1+, Gli1+) [34,35,36, 40,41,42]. Lrig1, Lhx2 and Nfatc1 are thought to play roles in lineage maintenance of these regions [40, 41, 43].
Chromatin remodeling involving the SWI/SNF complex is important in controlling the activities of genes that regulate stem cell functions. Brahma related gene 1 (BRG1), an ATPase component of the BAF chromatin remodeling structure, plays key roles in normal hair regeneration [44] and is dynamically expressed in HF at different stages of the hair cycle [44]. Low levels of BRG1 expression are observed in late telogen, while early anagen is marked with increased expression mainly in the lower bulge, and at later stages expression declines [44]. To elucidate the physiological role of BRG1 in hair regeneration, Nfatc-Cre mice , which express Cre recombinase specifically in the bulge [43], were combined with a conditional allele of Brg1 [44]. Deletion of Brg1 in the bulge caused decreased matrix cell proliferation, retarded hair growth, and progressive hair loss. BRG1 functions by suppressing p27kip1 and recruiting NF-kB, which in turn activates Shh in matrix cells promoting their proliferation [45, 46] (Fig. 6.2). Shh signaling through Gli activates BRG1 in bulge cells, creating a positive feedback loop. Thus, gene regulation by chromatin restructuring plays a key role in HFSC activation.
The ATP dependent chromatin remodeler Mi-2β and its role in epidermal differentiation have been described in the above section. In addition to its expression in the developing epidermis, Mi-2β is also expressed in the hair placode and matrix of developing HF [24]. Loss of Mi-2β in embryonic dorsal epidermis prevents induction of hair follicle placodes. After initiation of the follicle, markers of follicular morphogenesis such as Edar, Shh, Bmp2 and β-catenin [24, 33, 38] are expressed, and some subsequent morphogenesis of the hair peg proceeds in the absence of Mi-2β; however production of the progenitors that give rise to the inner layers of the hair follicle and hair shaft is impaired [24].
6.4 Stem Cell Activity During Wound Healing
Wound healing is a complex process that requires coordination of inflammatory, proliferative and remodeling mechanisms. HFSCs contribute to tissue repair and regeneration processes, particularly in re-epithelialization and re-establishment of skin homeostasis [36]. For instance, in response to full-thickness wounding, several different HFSC populations are activated, and their progeny migrate out from the HFs to participate in wound re-epithelialization [35, 40, 42, 47,48,49]. Gene regulation mechanisms are controlled by histone modifications, and are essential for normal physiological processes. A recent article by Na et al. (2016) demonstrates that histone modifications are crucial during the wound healing process [50]. Expression of Histone H3K27 demethylase [known as Jumonji domain containing protein D3 (JMJD3)], which has an important role in keratinocyte differentiation, is upregulated in the wound edges and it’s inactivation leads to aberrant wound healing [50]. Specifically, JMJD3 induces keratinocyte activation during the re-epithelialization process by interacting with NF-kB targets on inflammatory, MMP, and growth factor gene promoters [50, 51]. Similarly, expression of the chromatin remodeling protein BRG1 in HF bulge and bulge-derived cells increases following depilation or full thickness wounding in mouse skin [44]. Brg1 functions to facilitate emergence of bulge stem cells from the hair follicle to contribute to wound repair [34, 36, 44, 52].
6.5 Skin Pathophysiology
Exposure to solar UV irradiation can cause sunburn in the short term, and skin photo-aging and skin cancer due to DNA damage in the longer term. Despite these negative effects, controlled UV irradiation can be beneficial in the treatment of skin diseases such as eczema and psoriasis. Brahma (BRM) is a component of the SWI/SNF chromatin remodeling complex. Human non-melanoma skin cancers are reported to have hot spot mutations in the Brm gene, caused by ultraviolet (UV) exposure [53], and recent studies have demonstrated the protective functions of BRM as a tumor suppressor [54]. In line with these findings, irradiation of Brm-null mice with low dose ultraviolet (UV) light revealed their increased susceptibility to skin photocarcinognesis compared with controls, but did not alter the protective apoptotic response to UV-induced sunburn [54]. In the absence of one allele of the tumor suppressor p53, loss of BRM did not further increase tumor incidence, but did result in a higher growth rate of the tumors [54, 55]. Importantly, increased cell division occurred predominantly in the differentiated suprabasal layer of the epidermis, rather than in the basal layer in Brm-/- mice, revealing that BRM protects suprabasal cells from UV induced proliferation (Fig. 6.3) [55]. Suprabasal cells are more exposed to UV than basal cells and are therefore highly susceptible to mutation [56]. Moreover, UV-induced mutation occurs when cells divide without repairing damaged DNA [54, 56]. These observations may help to explain why the authors’ initial studies with Brm-null mice showed increased photo-carcinogenesis, while in later studies where they used a UV dose that simulated chronic sunlight exposure with mild sunburn damage, similar to the effects of natural sun exposure during normal activities in humans, they observed increased UV-induced cellular hyper-proliferation.
By contrast, UVB irradiation of C. elegans nematodes lacking the Brm analog psa-4 caused increased UV-sensitivity and cell death. This finding may reflect differences between the C. elegans and mammalian genomes and/or differences in sensitivity to UVB between C. elegans and mammalian keratinocytes [57].
6.6 Conclusions
The studies summarized above suggest that all subfamilies of the SNF2 family, including SWI2/SNF2 (BRG1/BRM), ISWI and CHD/Mi-2β, play critical roles in the maintenance of epidermal homeostasis by controlling the balance between proliferation and differentiation, and also in controlling epidermal permeability barrier formation. Future studies will further explore the roles of epigenetic mechanisms and their cross-talk with other regulatory pathways in controlling keratinocyte proliferation and the switch to differentiation in healthy and diseased skin.
In particular, the mechanisms underlying targeting of BRG1 to specific domains of the EDC, and its potential interaction with key epidermal transcription factors, such as p63, AP-1, and Klf4, that regulate EDC gene expression in epidermal progenitor cells remain to be determined. The cooperative involvement of BRG1 and SATB1 for establishing the specific EDC configuration in differentiating epidermal keratinocytes needs to be further investigated. Similarly, additional studies to explore the mechanisms by which Mi-2β exerts selective effects on development of the epidermis and related skin appendages will further elucidate the role that Mi-2 β-like chromatin remodelers play in establishing lineage specific stem cell identity. Future studies on the functions of other actin-like proteins, similar to ACTL6a, in multiple tissues will provide insights into developmental regulation by these modulators and the epigenetic regulatory complexes with which they associate.
The BRG1 chromatin remodeler plays crucial roles in maintaining the bulge stem cell pool, controlling hair cycling, permitting normal skin homeostasis, and facilitating repair and regeneration processes (Figs. 6.1 and 6.2). Other factors involved in the process of Gli-mediated activation of BRG1 during hair cycling are currently unknown, and are an important subject for further study (Fig. 6.2). It would be interesting to examine the regulation of BRG1 by other bulge stem cell factors such as Lhx2 and Tcf3 in this context. Improved understanding of BRG1 mediated regulatory controls of other cutaneous stem cell markers such as CD34, CD133 and Lrig1 during hair cycling and wound healing may help identify novel approaches to improve wound repair and tissue regeneration. Regulation of JMJD3 activity may provide a therapeutic approach for treatment of chronic wounds. Similarly, future mechanistic studies will provide more information regarding the role of Mi-2β in mobilization of stem cells during hair follicle morphogenesis and hair cycling, as well as in wound repair.
In the future, a better understanding of the epigenetic control of other key regulators by BRM and/or BRG1 in photo-carcinogenesis, UV-induced DNA damage and UV-induced skin inflammation will be necessary to gain more insights into these processes. Elucidation of detailed molecular mechanisms, together with gene expression profiling of coding and non-coding small and long-RNAs in UVB induced skin, may lead to discovery of potential biomarkers in photocarcinogenesis.
Last but not least, many of the studies discussed here highlight the power of Cre-ERT2-mediated recombination technology for creating spatio-temporally controlled targeted somatic mutations that allow dissection of gene functions throughout skin morphogenesis and in adult life [12, 58, 59].
Abbreviations
- ACTL6a:
-
Actin-like 6a
- AER:
-
Apical ectodermal ridge
- ATP:
-
Adenosine tri-phosphate
- ATP-DCR:
-
ATP-dependent chromatin remodeling
- BAF:
-
BRG1/BRM associated factors
- C elegans :
-
Caenorhabditis.elegans
- EDC:
-
Epidermal differentiation complex
- EPB:
-
Epidermal permeability barrier
- HF:
-
Hair follicle;
- HFSCs:
-
Hair follicle stem cells
- ISWI:
-
Imitation SWI2
- KLF4:
-
Kruppel-like factor 4
- SNF2:
-
Sucrose Non Fermentation
- SWI2:
-
Mating type Switching 2
References
Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. https://doi.org/10.1146/annurev.biochem.77.062706.153223. Epub 2009/04/10. PubMed PMID: 19355820.
Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178(4):853–68. Epub 1984/10/05. PubMed PMID: 6436497.
Eberharter A, Becker PB. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci. 2004;117(Pt 17):3707–11. https://doi.org/10.1242/jcs.01175. Epub 2004/08/03. PubMed PMID: 15286171.
Wu JI, Lessard J, Crabtree GR. Understanding the words of chromatin regulation. Cell. 2009;136(2):200–6. https://doi.org/10.1016/j.cell.2009.01.009. Epub 2009/01/27. PubMed PMID: 19167321; PubMed Central PMCID: PMC2770578.
Kalinin AE, Kajava AV, Steinert PM. Epithelial barrier function: assembly and structural features of the cornified cell envelope. BioEssays. 2002;24(9):789–800. https://doi.org/10.1002/bies.10144. Epub 2002/09/05. PubMed PMID: 12210515.
Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–89. Epub 2003/03/07. PubMed PMID: 12620226.
Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 1998;17(23):6979–91. https://doi.org/10.1093/emboj/17.23.6979. Epub 1998/12/08. PubMed PMID: 9843504; PubMed Central PMCID: PMC1171046.
Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–95. Epub 2001/02/13. PubMed PMID: 11163203.
Li M, Chiba H, Warot X, Messaddeq N, Gerard C, Chambon P, Metzger D. RXR-alpha ablation in skin keratinocytes results in alopecia and epidermal alterations. Development. 2001;128(5):675–88. Epub 2001/02/15. PubMed PMID: 11171393.
Li M, Indra AK, Warot X, Brocard J, Messaddeq N, Kato S, Metzger D, Chambon P. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature. 2000;407(6804):633–6. https://doi.org/10.1038/35036595. Epub 2000/10/18. PubMed PMID: 11034212.
Vassar R, Rosenberg M, Ross S, Tyner A, Fuchs E. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc Natl Acad Sci U S A. 1989;86(5):1563–7. Epub 1989/03/01. PubMed PMID: 2466292; PubMed Central PMCID: PMC286738.
Indra AK, Li M, Brocard J, Warot X, Bornert JM, Gerard C, Messaddeq N, Chambon P, Metzger D. Targeted somatic mutagenesis in mouse epidermis. Horm Res. 2000;54(5–6):296–300. Epub 2001/10/12. doi: 53275. PubMed PMID: 11595821.
Indra AK, Dupe V, Bornert JM, Messaddeq N, Yaniv M, Mark M, Chambon P, Metzger D. Temporally controlled targeted somatic mutagenesis in embryonic surface ectoderm and fetal epidermal keratinocytes unveils two distinct developmental functions of BRG1 in limb morphogenesis and skin barrier formation. Development. 2005;132(20):4533–44. https://doi.org/10.1242/dev.02019. Epub 2005/09/30. PubMed PMID: 16192310.
Kaufman MH, BLB J. The anatomical basis of mouse development. San Diego: Academic; 1999. 291 p.
Byrne C, Hardman M, Nield K. Covering the limb – formation of the integument. J Anat. 2003;202(1):113–23. Epub 2003/02/18. PubMed PMID: 12587926; PubMed Central PMCID: PMC1571060.
Sumi-Ichinose C, Ichinose H, Metzger D, Chambon P. SNF2beta-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol Cell Biol. 1997;17(10):5976–86. Epub 1997/10/07. PubMed PMID: 9315656; PubMed Central PMCID: PMC232446.
Bao X, Tang J, Lopez-Pajares V, Tao S, Qu K, Crabtree GR, Khavari PA. ACTL6a enforces the epidermal progenitor state by suppressing SWI/SNF-dependent induction of KLF4. Cell Stem Cell. 2013;12(2):193–203. https://doi.org/10.1016/j.stem.2012.12.014. Epub 2013/02/12. PubMed PMID: 23395444; PubMed Central PMCID: PMC3661004.
Park J, Wood MA, Cole MD. BAF53 forms distinct nuclear complexes and functions as a critical c-Myc-interacting nuclear cofactor for oncogenic transformation. Mol Cell Biol. 2002;22(5):1307–16. Epub 2002/02/13. PubMed PMID: 11839798; PubMed Central PMCID: PMC134713.
Tea JS, Luo L. The chromatin remodeling factor Bap55 functions through the TIP60 complex to regulate olfactory projection neuron dendrite targeting. Neural Dev. 2011;6:5. https://doi.org/10.1186/1749-8104-6-5. Epub 2011/02/03. PubMed PMID: 21284845; PubMed Central PMCID: PMC3038883.
Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95(5):625–36. Epub 1998/12/09. PubMed PMID: 9845365.
Mulder KW, Wang X, Escriu C, Ito Y, Schwarz RF, Gillis J, Sirokmany G, Donati G, Uribe-Lewis S, Pavlidis P, Murrell A, Markowetz F, Watt FM. Diverse epigenetic strategies interact to control epidermal differentiation. Nat Cell Biol. 2012;14(7):753–63. https://doi.org/10.1038/ncb2520. Epub 2012/06/26. PubMed PMID: 22729083.
Bao Y. Chromatin response to DNA double-strand break damage. Epigenomics. 2011;3(3):307–21. https://doi.org/10.2217/epi.11.14. Epub 2011/11/30. PubMed PMID: 22122340.
Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10(3):345–55. Epub 1999/04/16. PubMed PMID: 10204490.
Kashiwagi M, Morgan BA, Georgopoulos K. The chromatin remodeler Mi-2beta is required for establishment of the basal epidermis and normal differentiation of its progeny. Development. 2007;134(8):1571–82. https://doi.org/10.1242/dev.001750. Epub 2007/03/16. PubMed PMID: 17360773.
Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY. Epigenetic regulation of gene expression in keratinocytes. J Invest Dermatol. 2012;132(11):2505–21. https://doi.org/10.1038/jid.2012.182. Epub 2012/07/06. PubMed PMID: 22763788; PubMed Central PMCID: PMC3650472.
Cremer T, Cremer M. Chromosome territories. In: Misteli T, Spector D, editors. The Nucleus. New York: Cold Spring Harbor Laboratory Press; 2011. p. 93–114.
Sanyal A, Bau D, Marti-Renom MA, Dekker J. Chromatin globules: a common motif of higher order chromosome structure? Curr Opin Cell Biol. 2011;23(3):325–31. https://doi.org/10.1016/j.ceb.2011.03.009. Epub 2011/04/15. PubMed PMID: 21489772; PubMed Central PMCID: PMC3109114.
Koster MI, Roop DR. Mechanisms regulating epithelial stratification. Annu Rev Cell Dev Biol. 2007;23:93–113. https://doi.org/10.1146/annurev.cellbio.23.090506.123357. Epub 2007/05/11. PubMed PMID: 17489688.
Truong AB, Khavari PA. Control of keratinocyte proliferation and differentiation by p63. Cell Cycle. 2007;6(3):295–9. Epub 2007/02/01. PubMed PMID: 17264679.
Mardaryev AN, Gdula MR, Yarker JL, Emelianov VU, Poterlowicz K, Sharov AA, Sharova TY, Scarpa JA, Joffe B, Solovei I, Chambon P, Botchkarev VA, Fessing MY. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development. 2014;141(1):101–11. https://doi.org/10.1242/dev.103200. Epub 2013/12/19. PubMed PMID: 24346698; PubMed Central PMCID: PMC3865752.
Lopez-Pajares V, Qu K, Zhang J, Webster DE, Barajas BC, Siprashvili Z, Zarnegar BJ, Boxer LD, Rios EJ, Tao S, Kretz M, Khavari PA. A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell. 2015;32(6):693–706. https://doi.org/10.1016/j.devcel.2015.01.028. PubMed PMID: 25805135; PubMed Central PMCID: PMCPMC4456036.
Cavazza A, Miccio A, Romano O, Petiti L, Malagoli Tagliazucchi G, Peano C, Severgnini M, Rizzi E, De Bellis G, Bicciato S, Mavilio F. Dynamic transcriptional and epigenetic regulation of human epidermal keratinocyte differentiation. Stem Cell Reports. 2016;6(4):618–32. https://doi.org/10.1016/j.stemcr.2016.03.003. PubMed PMID: 27050947; PubMed Central PMCID: PMC4834057.
Rishikaysh P, Dev K, Diaz D, Qureshi WM, Filip S, Mokry J. Signaling involved in hair follicle morphogenesis and development. Int J Mol Sci. 2014;15(1):1647–70. https://doi.org/10.3390/ijms15011647. Epub 2014/01/24. PubMed PMID: 24451143; PubMed Central PMCID: PMC3907891.
Bhattacharya S, Wheeler H, Leid M, Ganguli-Indra G, Indra AK. Transcription factor CTIP2 maintains hair follicle stem cell pool and contributes to altered expression of LHX2 and NFATC1. J Invest Dermatol. 2015. https://doi.org/10.1038/jid.2015.281. Epub 2015/07/16. PubMed PMID: 26176759.
Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, Cotsarelis G. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11(12):1351–4. https://doi.org/10.1038/nm1328. Epub 2005/11/17. PubMed PMID: 16288281.
Liang X, Bhattacharya S, Bajaj G, Guha G, Wang Z, Jang HS, Leid M, Indra AK, Ganguli-Indra G. Delayed cutaneous wound healing and aberrant expression of hair follicle stem cell markers in mice selectively lacking Ctip2 in epidermis. PLoS One. 2012;7(2):e29999. https://doi.org/10.1371/journal.pone.0029999. Epub 2012/03/03. PubMed PMID: 22383956; PubMed Central PMCID: PMC3283611.
Botchkarev VA, Paus R. Molecular biology of hair morphogenesis: development and cycling. J Exp Zool B Mol Dev Evol. 2003;298(1):164–80. https://doi.org/10.1002/jez.b.33. Epub 2003/09/02. PubMed PMID: 12949776.
Millar SE. Molecular mechanisms regulating hair follicle development. J Invest Dermatol. 2002;118(2):216–25. https://doi.org/10.1046/j.0022-202x.2001.01670.x. Epub 2002/02/14. PubMed PMID: 11841536.
Suda T, Arai F. Wnt signaling in the niche. Cell. 2008;132(5):729–30. https://doi.org/10.1016/j.cell.2008.02.017. Epub 2008/03/11. PubMed PMID: 18329358.
Jaks V, Kasper M, Toftgard R. The hair follicle-a stem cell zoo. Exp Cell Res. 2010;316(8):1422–8. https://doi.org/10.1016/j.yexcr.2010.03.014. Epub 2010/03/27. PubMed PMID: 20338163.
Mardaryev AN, Meier N, Poterlowicz K, Sharov AA, Sharova TY, Ahmed MI, Rapisarda V, Lewis C, Fessing MY, Ruenger TM, Bhawan J, Werner S, Paus R, Botchkarev VA. Lhx2 differentially regulates Sox9, Tcf4 and Lgr5 in hair follicle stem cells to promote epidermal regeneration after injury. Development. 2011;138(22):4843–52. https://doi.org/10.1242/dev.070284. Epub 2011/10/27. PubMed PMID: 22028024; PubMed Central PMCID: PMC4067271.
Plikus MV, Gay DL, Treffeisen E, Wang A, Supapannachart RJ, Cotsarelis G. Epithelial stem cells and implications for wound repair. Semin Cell Dev Biol. 2012;23(9):946–53. https://doi.org/10.1016/j.semcdb.2012.10.001. Epub 2012/10/23. PubMed PMID: 23085626; PubMed Central PMCID: PMC3518754.
Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132(2):299–310. https://doi.org/10.1016/j.cell.2007.11.047. Epub 2008/02/05. PubMed PMID: 18243104; PubMed Central PMCID: PMC2546702.
Xiong Y, Li W, Shang C, Chen RM, Han P, Yang J, Stankunas K, Wu B, Pan M, Zhou B, Longaker MT, Chang CP. Brg1 governs a positive feedback circuit in the hair follicle for tissue regeneration and repair. Dev Cell. 2013;25(2):169–81. https://doi.org/10.1016/j.devcel.2013.03.015. Epub 2013/04/23. PubMed PMID: 23602386.
Alonso L, Fuchs E. The hair cycle. J Cell Sci. 2006;119(Pt 3):391–3. https://doi.org/10.1242/jcs02793. Epub 2006/01/31. PubMed PMID: 16443746.
Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118(5):635–48. https://doi.org/10.1016/j.cell.2004.08.012. Epub 2004/09/02. PubMed PMID: 15339667.
Levy V, Lindon C, Zheng Y, Harfe BD, Morgan BA. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 2007;21(7):1358–66. https://doi.org/10.1096/fj.06-6926com. Epub 2007/01/27. PubMed PMID: 17255473.
Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3(1):33–43. https://doi.org/10.1016/j.stem.2008.05.009. Epub 2008/07/03. PubMed PMID: 18593557; PubMed Central PMCID: PMC2877596.
Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, Stange DE, Toftgard R, Clevers H. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327(5971):1385–9. https://doi.org/10.1126/science.1184733. Epub 2010/03/13. PubMed PMID: 20223988.
Na J, Lee K, Na W, Shin JY, Lee MJ, Yune TY, Lee HK, Jung HS, Kim WS, Ju BG. Histone H3K27 Demethylase JMJD3 in cooperation with NF-kappaB regulates keratinocyte wound healing. J Invest Dermatol. 2016;136(4):847–58. https://doi.org/10.1016/j.jid.2015.11.029. PubMed PMID: 26802933.
Odorisio T. Epigenetic control of skin Re-Epithelialization: the NF-kB/JMJD3 connection. J Invest Dermatol. 2016;136(4):738–40. https://doi.org/10.1016/j.jid.2016.01.010. PubMed PMID: 27012558.
Menchon C, Edel MJ, Izpisua Belmonte JC. The cell cycle inhibitor p27Kip(1) controls self-renewal and pluripotency of human embryonic stem cells by regulating the cell cycle, Brachyury and Twist. Cell Cycle. 2011;10(9):1435–47. Epub 2011/04/12. PubMed PMID: 21478681; PubMed Central PMCID: PMC3685623.
Moloney FJ, Lyons JG, Bock VL, Huang XX, Bugeja MJ, Halliday GM. Hotspot mutation of Brahma in non-melanoma skin cancer. J Invest Dermatol. 2009;129(4):1012–5. https://doi.org/10.1038/jid.2008.319. Epub 2008/10/17. PubMed PMID: 18923443.
Halliday GM, Zhou Y, Sou PW, Huang XX, Rana S, Bugeja MJ, Painter N, Scolyer RA, Muchardt C, Di Girolamo N, Lyons JG. The absence of Brm exacerbates photocarcinogenesis. Exp Dermatol. 2012;21(8):599–604. https://doi.org/10.1111/j.1600-0625.2012.01522.x. Epub 2012/07/11. PubMed PMID: 22775994.
Hassan NM, Painter N, Howlett CR, Farrell AW, Di Girolamo N, Lyons JG, Halliday GM. Brm inhibits the proliferative response of keratinocytes and corneal epithelial cells to ultraviolet radiation-induced damage. PLoS One. 2014;9(9):e107931. https://doi.org/10.1371/journal.pone.0107931. Epub 2014/09/26. PubMed PMID: 25254962; PubMed Central PMCID: PMC4177874.
Halliday GM, Cadet J. It’s all about position: the basal layer of human epidermis is particularly susceptible to different types of sunlight-induced DNA damage. J Invest Dermatol. 2012;132(2):265–7. https://doi.org/10.1038/jid.2011.281. Epub 2012/01/14. PubMed PMID: 22241442.
Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W. Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet. 2010;6(5):e1000941. https://doi.org/10.1371/journal.pgen.1000941. Epub 2010/05/14. PubMed PMID: 20463888; PubMed Central PMCID: PMC2865526.
Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27(22):4324–7. Epub 1999/10/28. PubMed PMID: 10536138; PubMed Central PMCID: PMC148712.
Metzger D, Indra AK, Li M, Chapellier B, Calleja C, Ghyselinck NB, Chambon P. Targeted conditional somatic mutagenesis in the mouse: temporally-controlled knock out of retinoid receptors in epidermal keratinocytes. Methods Enzymol. 2003;364:379–408. Epub 2003/11/25. PubMed PMID: 14631857.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer International Publishing AG, part of Springer Nature
About this chapter
Cite this chapter
Ganguli-Indra, G., Indra, A.K. (2018). The Role of ATP-dependent Chromatin Remodeling in the Control of Epidermal Differentiation and Skin Stem Cell Activity. In: Botchkarev, V., Millar, S. (eds) Epigenetic Regulation of Skin Development and Regeneration. Stem Cell Biology and Regenerative Medicine. Humana Press, Cham. https://doi.org/10.1007/978-3-319-16769-5_6
Download citation
DOI: https://doi.org/10.1007/978-3-319-16769-5_6
Published:
Publisher Name: Humana Press, Cham
Print ISBN: 978-3-319-16768-8
Online ISBN: 978-3-319-16769-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)