Abstract
Individual genetic composition has a fundamental role in the variations observed in drug response and tolerance. Pharmacogenomics aims to delineate the individual genetic profiles and drug response/toxicity. Nowadays, there are several medical disciplines where pharmacogenomics is readily applicable, while in others its usefulness is yet to be shown. Recent experimental evidence suggest that single nucleotide polymorphisms (SNPs) in modifier genes residing outside the human β-globin cluster are significantly associated with response to hydroxyurea (HU) treatment in β-type haemoglobinopathies patients, deducted from the increase in foetal haemoglobin levels. This chapter aims to provide an update and to discuss future challenges on the application of pharmacogenomics for β-type haemoglobinopathies therapeutics in relation to the current pharmacological treatment modalities for those disorders and the complexity of their pathophysiology.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- β-thalassaemia
- Sickle cell disease
- Pharmacogenomics
- Biomarkers
- Single nucleotide polymorphisms
- Hydroxyurea treatment
1 Haemoglobinopathies
Being easily isolated from blood, the study of haemoglobin has shed light on the understanding of the fundamental principles of gene regulation, gene function and the molecular basis of human genetic disorders. Haemoglobin is the key tetramer oxygen transport protein of red blood cells, consisting of two α-like and two β-like globin polypeptide chains [1–4]. Notably, various types of haemoglobins are found at different developmental stages; just before birth, foetal haemoglobin (HbF) represents the bulk of haemoglobin production, while ten months after birth it gradually declines to reach almost 1 % of the total haemoglobin production, being restricted to a distinct erythrocyte population, also known as F-cells [4]. In adults, both HbF and the number of F-cells share a genetic determination, and the latter varies among populations. The primary adult and children haemoglobin is the HbA (α2β2), while HbA2 (α2δ2) and foetal haemoglobin HbF (α2γ2) are found in amounts of less than 3 %, respectively.
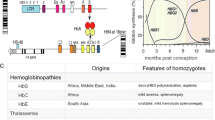
Haemoglobinopathies are divided into two main categories: thalassaemia syndromes and structural haemoglobin variants, both resulting from genomic variations that can be found in cis and/or in trans of the globin gene cluster. Thalassaemia syndromes (namely, α-thalassaemia and β-thalassaemia) are inherited autosomal recessive disorders with defects in globin synthesis and not in haemoglobin structure. The structural haemoglobin variants (or abnormal haemoglobins) are disorders characterised by defects in haemoglobin structure. The majority of haemoglobin variants are extremely rare, with the exception of HbS, HbC and HbE, that are found in certain populations, presumably due to positive natural selection. Abnormal haemoglobins can cause: (a) sickle cell disease (SCD) that results from red cell membrane deformation, (b) haemolytic anaemia, as a result of unstable haemoglobins, (c) methaemoglobinemia owing to rapid oxidation of haemoglobin, and (d) erythrocytosis due to unnatural oxygen affinity [3–7]. Particularly, SCD and β-thalassaemia are one of the commonest single gene disorders and at the same time one of the most serious health problems worldwide. These disorders are most prevalent in Asian and African populations, as well as those residing in the Mediterranean basin. Nowadays, however, due to the international migration, haemoglobinopathies extend worldwide. These genetic disorders are caused by genetic quantitative and qualitative defects in haemoglobin production [4].
Nowadays, more than 1000 Hb variants have been discovered and characterised [8], being a milestone in the history of haemoglobin research, where T.H.J. Huisman could not be omitted. A registry of these Hb variants and related information has been available online, at HbVar database (http://globin.bx.psu.edu/hbvar). During the last century, major developments in Hb research have been made using physical, chemical, physiological and genetic methods, impacting our understanding and management of the thalassaemias and sickle cell disease.
2 Therapeutic Approaches
The β-thalassaemias are considered as one of a few clinical conditions in which a mutant gene that is normally expressed later in development can be functionally replaced by a gene, which is transiently expressed during foetal life [9]. Foetal γ globin expression can be re-activated, being an appealing therapeutic approach as the foetal globin genes are universally present and noteworthy, appropriately contextually integrated in the β-globin locus in human haematopoietic stem cells. However, anaemia in β-thalassaemia syndromes can be also due to the rapid cellular apoptosis (α-globin chain precipitation) and/or the relatively low levels of endogenous erythropoietin (EPO) [10]. Thus, the ultimate goal of transfusion independence for thalassaemia patients should be approached via the stimulation of both foetal globin gene expression and erythropoiesis. In this context, chemotherapeutic agents, erythropoietin (EPO) preparations and short chain fatty acid derivatives (SCFADs) have demonstrated proof-of-principle in animal models and clinical trials .
Focusing on the pharmacological reactivation of HbF to compensate for the loss of HbA, the only pharmacological agent recognised from FDA used to increase the HbF in adults is hydroxycarbamine (or hydroxyurea, HU). Even though the mechanism underlying the HU action still remains elusive, it has been shown to inhibit cellular ribonucleotide reductase, whose role is critical for the DNA synthesis in the dividing late progenitor cells. HU is broadly used for ameliorating SCD symptoms and to a lesser extend in β-thalassaemia patients, while it also serves as a chemotherapeutic agent for many myeloproliferative conditions [11]. Nevertheless, HU is cytotoxic and can lead to cytopenia, hyperpigmentation, weight gain, hypomagnesemia or it may have a teratogenic effect [5]. The patient response to HU as well as the HU toxicity incidents vary [4, 12–14]. Thus, the discrimination between responders and non-responders is of fundamental importance towards patient stratification.
Collins and colleagues have observed that haematologic responses to the foetal globin inducer sodium phenylbutyrate occurred only in those subjects who had high endogenous EPO levels, being unrelated to any particular pattern of globin gene mutation [15]. Hence, a red cell survival advantage of increased endogenous EPO in β-thalassaemia has been suggested that may facilitate an effective γ-globin induction. In agreement to the above, a subset of subjects with inappropriately low levels of endogenous EPO has responded to combined therapy with butyrate plus EPO, whereas each agent alone had a lesser or minimal effect in the same time frame [10]. It seems, therefore, that the exogenously administered EPO acts both as a survival factor and an erythropoietic stimulant. Prolonging erythroid precursor cell survival could be beneficial, allowing a foetal globin inducer to act towards the correction of the pro-apoptotic chain imbalance and hence, improving the anaemia found in β + - thalassaemia patients. In thalassaemia intermedia and major, EPO has been combined with HU or arginine butyrate. Combinations of EPO with stem cell factor (SCF) to stimulate proliferation and transforming growth factor-β (TGF-β) to induce premature differentiation (like HU) could be also considered for more severe phenotypes, although pricing could be an obstacle for long-term use.
In other studies, SCFADs and hydroxamic acids have been shown to induce foetal globin gene expression, stimulate erythroid proliferation and prolong erythroblast survival [10, 16, 17]. In particular, they activate the Aγ globin gene promoter in cells cultured from β-thalassaemia patients , without inhibiting the erythroid cell growth. Sodium 2,2-dimethylbutyrate and α-methyl-hydrocinnamate stimulate erythroid colony formation more than the optimal haematopoietic growth factors alone and at the same time, signal through STAT-5 phosphorylation that is common to EPO and IL-3 signalling pathways. Most importantly, some of these agents hold the promise of oral administration, being more tolerable for long-term treatments, providing two therapeutic effects via one tolerable agent [10, 17].
Although the development of drugs to increase foetal haemoglobin has been the major therapeutic strategy in the treatment of both disorders, SCD and thalassaemias, and new foetal haemoglobin–modulating agents have been studied, only HU has shown long-term benefit. To this end, the gradual elucidation of the pathophysiology of the disease(s) has led to alternative strategies, treating the associated complications (decreasing the iron overload and reducing the oxidative stress). More recently, novel agents have been developed targeting the multiple pathways causing vascular injury in haemoglobinopathies; the increased adhesion of cells to the vascular endothelium, the NO dysregulation, inflammation, oxidative injury and the altered iron metabolism. Such agents (propranolol, statins, niacin, curcuminoids, hepcidin agonists/antagonists) have reached phase 1 and 2 clinical trials [18].
3 Pathophysiological Features and Obstacles
The reduction of the globin chain imbalance has been well accepted as the way to improve red cell survival and blood counts in β-thalassaemia. Nevertheless, a number of factors seem to collectively contribute to anaemia, such as ineffective erythropoiesis and erythroid precursor apoptosis. Thus, combination therapy is required with more than one agent acting at various molecular levels to achieve a tolerable and long-term therapeutic response. Moreover, different magnitudes of therapeutic effect for different thalassaemia patients are required to achieve functional clinical endpoints that could result in abolishing or decreasing needs for regular red blood cell transfusion. In thalassaemia intermedia patients with basal total haemoglobin levels of 6–8 g/dL, a 1–2 g/dL increase would be quite adequate to prevent the need for a regular transfusion program, being highly beneficial, whereas thalassaemia major patients having baseline haemoglobin levels below 5 g/dL would require higher levels of foetal globin induction.
The paediatric pathophysiology of both SCD and thalassaemia should also be considered when haemoglobinopathies’ therapeutics are in question. In SCD, acute chest syndrome, resulting from pulmonary microvascular occlusion and being a common cause of death, occurs in all age groups, but is most common in childhood. In children, acute sequestration of sickled cells in the spleen may also occur, exacerbating anaemia. Chronic spleen damage increases susceptibility to pneumococcal and Salmonella infections (including Salmonella osteomyelitis) that are especially common in early childhood and can be rapidly fatal [19]. Children with thalassaemia intermedia (mild to severe anaemia) or thalassaemia major need blood transfusions coupled to chelation therapy. Children with β-thalassaemia have elevated plasma levels of conjugated dienes and thiobarbituric acid-reactive substances (markers of lipid oxidation), while the RBC glutathione levels are much reduced [20]. There is evident oxidant injury to RBC haemoglobin and lipids.
Considering the overall complexity in the pathophysiology of haemoglobinopathies as well as the still unresolved pathophysiological issues in thalassaemias [20], patient stratification is critical towards effective cure or mitigation of the disease. In this context, pharmacogenomics are expected to have a fundamental role.
4 Pharmacogenomics for Haemoglobinopathies
Pharmacogenomics aims to determine how the genetic background of a patient influences his response to a drug or the probability to develop adverse drug reactions, via the correlation of gene expression or Single Nucleotide Polymorphisms (SNPs) with drug efficacy and toxicity [21]. The application of pharmacogenomics in haemoglobinopathies is particularly appealing due to the limitation of the therapeutic approaches and the complexity of disease pathophysiology. So far, the role of HU treatment towards the cure or mitigation of the disease has been vital. HU increases the HbF levels mainly in SCD, but also in compound heterozygous SCD/β-thalassaemia patients, ameliorating their clinical manifestations. Additionally, β-thalassaemia intermedia patients have been also shown to respond to HU treatment. Herein, we summarise the current knowledge regarding the genetic factors that have been reported to influence HbF expression levels in relation to HU treatment, including a large number of genomic variations residing inside or outside the human β-globin gene cluster (Tables 1, 2).
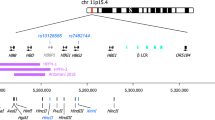
4.1 Genomic Biomarkers Linked to the Human β-Globin Gene Cluster
Patients’ response to HU varies in terms of amplitude and velocity, leading to the in depth investigation of the determinants of this differential response. This investigation began years ago and continues until today, revealing several genetic factors including SNPs in various genes that are linked (or not) to the β-globin gene cluster. These genes are believed to act by modulating HbF levels.
Steinberg et al. [23] studied 150 HbS homozygous patients treated with HU. A group of them showed almost a 40 % HU-induced HbF levels, compared to the remaining of the study group. In order to define the genetic factors that may have influenced these responses, they focused on genetic factors that are believed to influence the foetal globin (HBG1 and HBG2) gene expression, including the haplotypes of α- and β-globin gene clusters and the X-linked F-cell expression locus (FCP) [22]. As concluded, the FCP and the HbF levels before the treatment were not correlated with the HU-induced HbF response. Notably, the absence of a particular β-globin gene haplotype (namely, the Central African Republic-CAR) was related with higher HbF response [23].
Although HU is established to ameliorate the clinical manifestations of SCD, many patients die of this disease, mostly by acute chest syndrome (as it has been described earlier in the text, it occurs in all age groups, but it is most common in childhood), even upon HU treatment. Bakanay et al. [24] compared the β-globin gene cluster haplotype distributions (BAN, BEN, CAM, SEN) between the deceased and surviving patients treated with HU, concluding that homozygosity in the BAN haplotype or heterozygosity in the CAM haplotype were more likely to be observed in the deceased patient group [24].
In β-thalassaemia major , the increase of HbF expression by HU administration is not as effective as in the case of SCD. This may be due to the heterogeneity of the disease and also due to the complexity of the genetic elements involved in the HbF expression patterns of patients. Alebouyeh et al. [25] attempted to correlate the HU response in two different groups of β-thalassaemia major patients, from whom, 25 were blood transfusion-dependent and nine were non-dependent, in order to explore candidate genetic markers for the pharmacological HbF reactivation by HU. They have demonstrated that the XmnI polymorphism (HBG2: g.-158C > T) and the IVSII-1G > A mutation, both at the homozygous state, were found in the responders of both groups. On the contrary, these markers were either not present or in a heterozygous state in the non-responders groups tested. Nevertheless, the small number of patients included in the study as well as the fact that two siblings (responders group) were found to bear the common allele for both the XmnI polymorphism and the IVSII-1G > A mutation suggest that further research must be carried out in order to identify and elucidate the genetic modifiers for the HbF upregulation upon HU treatment [25].
Yavarian et al. [26] studied 133 Iranian transfusion-dependent β-thalassaemia patients, treated with HU in order to determine their response to the drug and the associated genetic background; 61 % of the patients became blood transfusion-independent, after 4 months of HU administration (good responders), 23 % of the patients, remained blood transfusion-dependent, albeit at a less frequent rate (moderate responders) than before HU treatment, whereas in the remaining 16 % of the patients HU, even after a year of administration, had no effect in their clinical manifestation and the frequency of blood transfusion. The authors examined the genetic aetiology of β-thalassaemia in these patients, the human β-gene cluster haplotype and their molecular background in the promoter region of the globin genes as well as at the HS2 hypersensitive site of the β-globin Locus Control Region (LCR). They concluded that the XmnI polymorphism was the most important genetic factor correlated with HU response and that its linkage with the human β-globin gene cluster haplotype I and with the HBB gene framework 2 is the “favourable genetic background” for good response to HU [26]. Also, Ansari et al. [27], studied 143 β-thalassaemia patients , treated with HU, confirming that XmnI polymorphism in homozygous or heterozygous state is a genomic marker to predict HU response. This finding was also demonstrated in two other studies including 54 Algerian β-thalassaemia patients [28] and 18 homozygous β-thalassaemia patients treated with HU for a period of 4 years [29].
From a number of studies, it is evident that HU is more promising treating β-thalassaemia intermedia due to the lesser imbalance of α/β-globin chain. Dixit et al. [30] studied the response of 37 β-thalassaemia intermedia patients to HU, from whom almost 70 % were categorised as responders. The response to HU was not associated with the β-thalassaemia mutation. On the other hand, a statistically insignificant correlation of HU response and the XmnI polymorphism was observed, suggesting that the combination of other genetic elements can possibly influence the final response to HU treatment [30]. In a similar survey of 18 homozygous β-thalassaemia patients, 11 of who were transfusion-dependent were treated with HU for 50 months in order to correlate their response to HU and their genetic background. The results showed that 82 % of transfusion-dependent patients who were treated with HU turned into transfusion-independent, while 78 % of them were found homozygous or heterozygous for the XmnI polymorphism. Interestingly, this genomic variation was not present in the HU non-responders. Nevertheless, there were two responding patients, who were negative for the XmnI polymorphism. In total, these data suggest that there may be other genetic elements, which could determine the HU response. In the same study, no correlation between response to HU and the nature of β-thalassaemia mutation or α-thalassaemia deletion was observed [29]. However, in a similar study of 16 transfusion-independent Iranian patients, treated with HU for 6 months, Ehsani et al. [31] could not establish any correlation between the XmnI polymorphism and the response to HU treatment. This may be due to the fact that in this study, as with the previous one, the number of patients was too small to reach any significant conclusion.
Similarly, Italia et al. [13] attempted to correlate the response to HU of 79 β-thalassaemia patients of western Indian origin, from which 38 were β-thalassaemia intermedia and 41 were β-thalassaemia major, treated with HU for almost a year. The correlation (if any) of the HU response to the genetic factors residing within the human β-globin gene cluster was investigated. As it was shown, in the presence of α-thalassaemia, β-thalassaemia patients showed a better HU response, and the presence of the XmnI polymorphism in homozygosity also resulted in a better clinical response to HU [13]. In a subsequent study of 13 Indian HbE/β-thalassaemia patients with severe clinical manifestations, from whom 36.3 % of the patients were good responders, 36.3 % were partial responders and 27.2 % showed no response, Italia et al. [32] failed to correlate the HU response with specific genetic factors, focusing mainly to the HBB genotype and the human β-globin cluster haplotype and XmnI polymorphism, most likely due to the small number of patients [32].
Karimi et al. [33] attempted to correlate the response to HU with HBB gene mutations and the XmnI polymorphism in a much larger patient sample, consisting of 232 β-thalassaemia patients of Iranian origin, upon HU treatment for a 13-year period. These authors showed that β-thalassaemia patients with homozygous or heterozygous for a β0 mutation were better HU responders compared to patients who were homozygous for a β+ mutation. Interestingly, though, these authors could not establish any correlation between the XmnI polymorphic site and HU response [33].
Finally, Rigano et al. [34] studied the HU efficiency in a long and short term treatment of 24 β-thalassaemia intermedia patients and concluded that the presence of Hb Lepore and δβ-thalassaemia genotypes were indicators of a better HU response [34].
All these studies are summarised in Table 1.
4.2 Genomic Biomarkers Non-Linked to the Human β-Globin Gene Cluster
Apart from the numerous studies presented above with the aim to delineate the response to HU and the genomic markers present in the human β-globin gene cluster, a number of studies have been recently conducted, attempting to implicate the genomic loci residing on other chromosomes with HU response. Some of these genes, particularly BCLl1A, have been shown to be directly related with increasing HbF levels and as such, these genes might constitute excellent candidates for pharmacogenomic biomarkers to predict HU response (Table 2).
In the most comprehensive study so far, Ma et al. [35] studied 137 SCD patients in an effort to correlate the HU response to several genomic biomarkers that are linked trait loci (QTLs), located on chromosomes 6 and 8 as well as the X-chromosome (these genes have been previously linked to HU metabolism and to erythroid progenitor proliferation). In particular, the authors investigated the association of 327 tagSNPs within these loci to the HU response of patients, using HapMap data. It was concluded that the rs2182008 variation in the FLT1 gene, either in homozygosity or heterozygosity, was correlated with an almost 6-fold increase in HbF expression levels, following HU treatment. Moreover, two other FLT1 gene variants, namely the rs9319428 and rs8002446, were found to be associated with the HU response. Overall, there were various genomic variations residing in the MAP3K5, PDE7B, ASS, TOX, ARG1, ARG2, NOS2A and NOS1 genes, found to be correlated with the HU influence to the HbF expression. Notably, the majority of the above mentioned SNPs were located in intronic or in untranslated regions of the candidate genes [35].
Similarly, Tafrali et al. [36] attempted to elucidate a probable association between the genetic variations in the MAP3K5 and PDE7B gene with the β-thalassaemia disease severity and response to HU in two groups of 38 β-thalassaemia homozygous and SCD/β-thalassaemia compound heterozygous patients of western Greek origin. The authors showed that there is a significant correlation between two single nucleotide polymorphisms residing in the region of MAP3K5 intron 1 (rs9483947, rs9376230) and improved HU response. Also, by comparative whole-transcriptome analysis in erythroid progenitor cell cultures from normal Maltese adults and Maltese HPFH haploinsufficient cases, bearing the KLF1:p.K288X nonsense mutation [37], before and after HU treatment, MAP3K5 gene expression was increased upon HU treatment [36].
A retrospective association study was conducted with the purpose of detecting genetic determinants of the HU response in 81 transfusion-dependent β-thalassaemia patients from Iran. Genomic variants, located in three QTLs that have been previously shown to have an effect on HbF and F-cell levels, namely the XmnI polymorphism (rs7482144), two SNPs in the intron 2 of the BCL11A gene (rs766432, rs4671393) and two SNPs in the intergenic region of HBS1L-MYB gene (rs9399137, rs4895441) were investigated. The authors failed to obtain a correlation between the HBSL1L-MYB SNPs and HU response. On the contrary, the presence of the XmnI polymorphism, as well as the minor alleles of the BCL11A SNPs, namely rs766432C and rs4671393A, were significantly associated with good response to HU treatment [38]. Similarly, Flanagan et al. [39] attempted to elucidate the effect of HU on the erythroid gene expression in 93 children suffering from SCD, in order to explore how HU can influence both the red cell development and the HbF reactivation. Although the existence of the rs1186868 or rs1427407 SNPs in the BCL11A gene, in either homo- or heterozygosity, led to the down-regulation of the BCL11A expression and higher HbF levels, the authors failed to observe any difference in HbF levels, subject to HU treatment, between the patients who were homozygous or heterozygous for the above SNPs and the wild type ones [39].
Borg et al. [40] conducted a pharmacogenomic study on the HU effect on HbF levels of Hellenic SCD/β-thalassaemia compound heterozygotes, the first to be carried out using a whole transcriptome analysis approach. The authors have comparatively analysed, using whole transcriptome analysis, human erythroid progenitor cells, treated with HU, derived from SCD/β-thalassaemia patients that responded or not to HU. They also studied the effect of the HU on erythroid progenitor cells of healthy and KLF1-haploinsufficient Maltese adult patients ex vivo, expressing low and high HbF levels, respectively, aiming to reveal differential expression profiles in genes implicated in augmenting HbF levels. KLF10 was shown to be the strongest candidate, among 43 identified genes, in both analyses [40]. Subsequently, the authors used an independent cohort of SCD/β-thalassaemia compound heterozygotes so as to corroborate their results. Their genotyping analysis demonstrated that the presence of the rs3191333 SNP in the 3′ UTR of the KLF10 gene can be correlated with the severity of β-thalassaemia, as well as with efficacy to the HU therapy. In conclusion, KLF10 has not only been shown to be a pharmacogenomic marker to predict β-thalassaemia patient response to HU, but has also been implicated for the first time in erythropoiesis [40].
Finally, Kumkhaek et al. [41] examined 386 SCD patients in an effort to correlate polymorphisms in the SAR1A gene promoter region with differential response to HU and differences in HbF levels among different patients. It was concluded that 5 SNPs in the SAR1A regulatory region were correlated with patients’ response to HU and with different HbF levels, after a 2 year treatment with HU [41].
5 Concluding Remarks
In this chapter, we (i) provided an succinct overview of haemoglobinopathies, (ii) presented the challenges of their pathophysiology and the limitations of their therapeutics and (iii) summarised our current knowledge regarding the genetic factors that have been reported to influence HbF expression levels in relation to HU treatment, including a large number of genomic variations residing inside or outside the human β-globin gene cluster (Tables 1, 2).
Contrary to other medical specialties, such as oncology, and treatments, such as anticoagulation therapies, experimental data supporting the use of pharmacogenomics for haemoglobinopathies therapeutics using HU are currently very limited, and clearly, more pharmacogenomic studies are needed, not only in larger, but also in ethnically diverse β-thalassaemia and SCD patients groups. In this way, a better picture will be obtained as to whether it is possible to stratify those patients who are likely to benefit from HU therapy. In addition, similar studies may be also conducted for more pharmacological agents and different treatment modalities, such as decitabine and/or butyrate, although presently at experimental stage. However, although drug-induced augmentation therapies towards HbF levels have been demonstrated as a therapeutic modality for β-type haemoglobinopathies patients, it should be clarified that these cannot correct per se the numerous events that underlie the pathophysiology of this group of disorders. In addition, one should bear in mind that no straightforward correlation between HbF increase and clinical improvements in β-type haemoglobinopathies patients has been demonstrated. Therefore, all possible phenotype and clinical indicators should be determined to categorise the “responder” and the “non-responder” patient groups for pharmacogenomic studies, which do not necessarily have to be correlated with HbF increment alone, particularly in the case of SCD. Similar complexities also exist for other thalassaemia-related treatments, such as the use of iron chelators.
It should be also noted that not all genomic loci that have been shown to increase HbF levels can be also considered as pharmacogenomic markers for HU response. KLF1, one of the key players participating in HBB gene activation that is recently shown to be also indirectly involved in human foetal globin gene silencing [36] is not correlated with increased HbF levels upon HU treatment (Kaimakis and Patrinos, unpublished). The same is true for genomic variations in the HBBP1 pseudogene and PDE7B gene that although recently shown to be related with β-thalassaemia disease severity [37, 42], genomic alterations in these genes cannot be correlated with response to HU treatment.
Whole genome association and whole transcriptome pharmacogenomic studies are only beginning and there are only few reports in the field [35, 37, 40]. Such studies may identify novel gene candidates that participate in different pathways related to HU treatment, such as stage-specific transcription factors, novel erythroid genes and/or genes involved in HbF-inducing HU metabolism. Also, the scarcity of β-thalassaemia intermedia patients and the need to stratify these patients not only according to their response status, but also, and most importantly, according to their HBB genotype, makes the formation of large multi-center consortia more than ever urging to better orientate pharmacogenomic marker identification in good and poor responders to HbF-inducing therapy. This will in turn facilitate the design of customised high throughput pharmacogenomic tests for β-type haemoglobinopathies.
Pharmacogenomic studies may be also extended to other therapeutic modalities for β-thalassaemia, such as iron chelation therapy. In particular, a fraction of β-thalassaemia patients present a number of adverse effects to iron chelators, which result in early death [43]. The correlation of genomic variations located in genes that influence, e.g. iron homeostasis with tolerance or response to iron chelation treatment would potentially better stratify patients for iron chelation therapies and enable the emergence of new and improved iron chelators. Similarly, as with β-thalassaemia and SCD patients, whole genome pharmacogenomic studies in these patient groups can also establish genes involved in iron chelators’ metabolism pathways, hence allowing identifying putatively useful pharmacogenomic markers for iron chelation therapies, leading to the individual tailoring of chelation therapy to maximise iron excretion.
Pharmacogenomics in children bring on additional challenges. It is well established that there are differences in drug response among children and adults [44], especially in drug metabolism and gene expression, as the latter is a highly dynamic process functioning from the neonatal period over childhood and the adult life later on. Thus, the data quality and its analysis/ interpretation is challenging per se. Ethical and legal aspects also accompany this, since the child in question is incapable of giving informed consent himself [44]. Data interpretation difficulties and ethical considerations are clearly needed to be addressed.
In essence, although pharmacogenomics for β-type haemoglobinopathies is currently in its infancy, there is definitely a big potential to determine whether genomic biomarkers can be exploited in the clinic to stratify β-thalassaemia and SCD patients that are likely to benefit from therapy.
References
Pace BS, Zein S (2006) Understanding mechanisms of gamma-globin gene regulation to develop strategies for pharmacological foetal hemoglobin induction. Dev Dyn 235(7):1727–1737
Schechter AN (2008) Hemoglobin research and the origins of molecular medicine. Blood 112(10):3927–3938
Weatherall DJ (2001) Towards molecular medicine; reminiscences of the haemoglobin field, 1960–2000. Br J Haematol 115(4):729–738
Patrinos GP, Antonarakis SE (2010) Human hemoglobin, vol 11. Vogel and Motulsky’s human genetics: problems and approaches. Springer, Berlin
Kohne E (2011) Hemoglobinopathies: clinical manifestations, diagnosis, and treatment. Dtsch Arztebl Int 108(31–32):532–540
Higgs DR (2013) The molecular basis of α-thalassaemia. Cold Spring Harb Perspect Med 3(1):a011718
Nienhuis AW, Nathan DG (2012) Pathophysiology and clinical manifestations of the β-thalassaemias. Cold Spring Harb Perspect Med 2(12):a011726
Thein SL (2011) Milestones in the history of hemoglobin research (in memory of professor Titus H.J. Huisman). Hemoglobin 35(5–6):450–462
Steinberg MH, Rodgers GP (2001) Pharmacologic modulation of foetal hemoglobin. Medicine (Baltimore) 80(5):328–344
Perrine SP (2005) Foetal globin induction-can it cure beta thalassaemia? Hematol Am Soc Hematol Educ Program 38–44
Karimi M, Darzi H, Yavarian M (2005) Haematologic and clinical responses of thalassaemia intermedia patients to hydroxyurea during 6 years of therapy in Iran. J Pediatr Hematol Oncol 27(7):380–385
Patrinos GP, Grosveld FG (2008) Pharmacogenomics and therapeutics of hemoglobinopathies. Hemoglobin 32(1–2):229–236
Italia KY, Jijina FJ, Merchant R, Panjwani S, Nadkarni AH, Sawant PM, Nair SB, Ghosh K, Colah RB (2009) Response to hydroxyurea in beta thalassaemia major and intermedia: experience in western India. Clin Chim Acta 407(1–2):10–15
Karimi M, Cohan N, Mousavizadeh K, Moosavizadeh K, Falahi MJ, Haghpanah S (2010) Adverse effects of hydroxyurea in beta-thalassaemia intermedia patients: 10 years’ experience. Pediatr Hematol Oncol 27(3):205–211
Collins AF, Pearson HA, Giardina P, McDonagh KT, Brusilow SW, Dover GJ (1995) Oral sodium phenylbutyrate therapy in homozygous beta thalassaemia: a clinical trial. Blood 85(1):43–49
Boosalis MS, Bandyopadhyay R, Bresnick EH, Pace BS, Van DeMark K, Zhang B, Faller DV, Perrine SP (2001) Short-chain fatty acid derivatives stimulate cell proliferation and induce STAT-5 activation. Blood 97(10):3259–3267
Pace BS, White GL, Dover GJ, Boosalis MS, Faller DV, Perrine SP (2002) Short-chain fatty acid derivatives induce foetal globin expression and erythropoiesis in vivo. Blood 100(13):4640–4648
Vichinsky E (2012) Emerging ‘A’ therapies in hemoglobinopathies: agonists, antagonists, antioxidants, and arginine. Hematol Am Soc Hematol Educ Program 2012:271–275
Di Nuzzo DV, Fonseca SF (2004) [Sickle cell disease and infection]. J Pediatr (Rio J) 80(5):347–354
Schrier SL (2002) Pathophysiology of thalassaemia. Curr Opin Hematol 9(2):123–126
Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, Krauss RM, McLeod HL, Ratain MJ, Relling MV, Ring HZ, Shuldiner AR, Weinshilboum RM, Weiss ST, Network PR (2006) Pharmacogenomics: challenges and opportunities. Ann Intern Med 145(10):749–757
Chang YC, Smith KD, Moore RD, Serjeant GR, Dover GJ (1995) An analysis of foetal hemoglobin variation in sickle cell disease: the relative contributions of the X-linked factor, beta-globin haplotypes, alpha-globin gene number, gender, and age. Blood 85(4):1111–1117
Steinberg MH, Lu ZH, Barton FB, Terrin ML, Charache S, Dover GJ (1997) Foetal hemoglobin in sickle cell anaemia: determinants of response to hydroxyurea. Multicenter Study of Hydroxyurea. Blood 89(3):1078–1088
Bakanay SM, Dainer E, Clair B, Adekile A, Daitch L, Wells L, Holley L, Smith D, Kutlar A (2005) Mortality in sickle cell patients on hydroxyurea therapy. Blood 105(2):545–547
Alebouyeh M, Moussavi F, Haddad-Deylami H, Vossough P (2004) Hydroxyurea in the treatment of major beta-thalassaemia and importance of genetic screening. Ann Hematol 83(7):430–433
Yavarian M, Karimi M, Bakker E, Harteveld CL, Giordano PC (2004) Response to hydroxyurea treatment in Iranian transfusion-dependent beta-thalassaemia patients. Haematologica 89(10):1172–1178
Ansari SH, Shamsi TS, Munzir S, Khan MT, Erum S, Perveen K, Farzana T, Ashraf M, Mehboob T, Moinuddin M (2013) Gγ-Xmn I polymorphism: a significant determinant of β-thalassaemia treatment without blood transfusion. J Pediatr Hematol Oncol 35(4):e153–e156
Bradai M, Pissard S, Abad MT, Dechartres A, Ribeil JA, Landais P, de Montalembert M (2007) Decreased transfusion needs associated with hydroxyurea therapy in Algerian patients with thalassaemia major or intermedia. Transfusion 47(10):1830–1836
Koren A, Levin C, Dgany O, Kransnov T, Elhasid R, Zalman L, Palmor H, Tamary H (2008) Response to hydroxyurea therapy in beta-thalassaemia. Am J Hematol 83(5):366–370.
Dixit A, Chatterjee TC, Mishra P, Choudhry DR, Mahapatra M, Tyagi S, Kabra M, Saxena R, Choudhry VP (2005) Hydroxyurea in thalassaemia intermedia-a promising therapy. Ann Hematol 84(7):441–446
Ehsani MA, Hedayati-Asl AA, Bagheri A, Zeinali S, Rashidi A (2009) Hydroxyurea-induced haematological response in transfusion-independent beta-thalassaemia intermedia: case series and review of literature. Pediatr Hematol Oncol 26(8):560–565
Italia KY, Jijina FF, Merchant R, Panjwani S, Nadkarni AH, Sawant PM, Nair SB, Ghosh K, Colah RB (2010) Effect of hydroxyurea on the transfusion requirements in patients with severe HbE-beta-thalassaemia: a genotypic and phenotypic study. J Clin Pathol 63(2):147–150
Karimi M, Haghpanah S, Farhadi A, Yavarian M (2012) Genotype-phenotype relationship of patients with β-thalassaemia taking hydroxyurea: a 13-year experience in Iran. Int J Hematol 95(1):51–56
Rigano P, Pecoraro A, Calzolari R, Troia A, Acuto S, Renda D, Pantalone GR, Maggio A, Di Marzo R (2010) Desensitization to hydroxycarbamide following long-term treatment of thalassaemia intermedia as observed in vivo and in primary erythroid cultures from treated patients. Br J Haematol 151(5):509–515
Ma Q, Wyszynski DF, Farrell JJ, Kutlar A, Farrer LA, Baldwin CT, Steinberg MH (2007) Foetal hemoglobin in sickle cell anaemia: genetic determinants of response to hydroxyurea. Pharmacogenomics J 7(6):386–394
Borg J, Papadopoulos P, Georgitsi M, Gutiérrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, van Ijcken W, Ozgür Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, von Lindern M, Felice AE, Patrinos GP, Philipsen S (2010) Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of foetal hemoglobin. Nat Genet 42(9):801–805
Tafrali C, Paizi A, Borg J, Radmilovic M, Bartsakoulia M, Giannopoulou E, Giannakopoulou O, Stojiljkovic-Petrovic M, Zukic B, Poulas K, Stavrou EF, Lambropoulou P, Kourakli A, Felice AE, Papachatzopoulou A, Philipsen S, Pavlovic S, Georgitsi M, Patrinos GP (2013) Genomic variation in the MAP3K5 gene is associated with β-thalassaemia disease severity and hydroxyurea treatment efficacy. Pharmacogenomics 14(5):469–483
Banan M, Bayat H, Azarkeivan A, Mohammadparast S, Kamali K, Farashi S, Bayat N, Khani MH, Neishabury M, Najmabadi H (2012) The XmnI and BCL11A single nucleotide polymorphisms may help predict hydroxyurea response in Iranian β-thalassaemia patients. Hemoglobin 36(4):371–380
Flanagan JM, Steward S, Howard TA, Mortier NA, Kimble AC, Aygun B, Hankins JS, Neale GA, Ware RE (2012) Hydroxycarbamide alters erythroid gene expression in children with sickle cell anaemia. Br J Haematol 157(2):240–248
Borg J, Phylactides M, Bartsakoulia M, Tafrali C, Lederer C, Felice AE, Papachatzopoulou A, Kourakli A, Stavrou EF, Christou S, Hou J, Karkabouna S, Lappa-Manakou C, Ozgur Z, van Ijcken W, von Lindern M, Grosveld FG, Georgitsi M, Kleanthous M, Philipsen S, Patrinos GP (2012) KLF10 gene expression is associated with high foetal hemoglobin levels and with response to hydroxyurea treatment in β-hemoglobinopathy patients. Pharmacogenomics 13(13):1487–1500
Kumkhaek C, Taylor JG, Zhu J, Hoppe C, Kato GJ, Rodgers GP (2008) Foetal haemoglobin response to hydroxycarbamide treatment and sar1a promoter polymorphisms in sickle cell anaemia. Br J Haematol 141(2):254–259
Giannopoulou E, Bartsakoulia M, Tafrali C, Kourakli A, Poulas K, Stavrou EF, Papachatzopoulou A, Georgitsi M, Patrinos GP (2012) A single nucleotide polymorphism in the HBBP1 gene in the human β-globin locus is associated with a mild β-thalassaemia disease phenotype. Hemoglobin 36(5):433–445
Hershko C, Link G, Konijn AM, Cabantchik ZI (2005) Objectives and mechanism of iron chelation therapy. Ann N Y Acad Sci 1054:124–135
Vanakker OM, De Paepe A (2013) Pharmacogenomics in children: advantages and challenges of next generation sequencing applications. Int J Pediatr 2013:136524
Acknowledgements
We wish to thank Professors Frank Grosveld, Alex E. Felice, Sjaak Philipsen, Drs. Adamantia Papacatzopoulou, Joseph Borg, Sonja Pavlovic, Marios Phylactides, Marina Kleanthous, Alexandra Kourakli, Marianthi Georgitsi and Mrs. Christina Tafrali, Marina Bartsakou-lia, Arsinoi Paizi, Emily Giannopoulou, and Olga Giannakopoulou for their contribution at the various stages of our projects related to pharmacogenomics for haemoglobinopathies. Our work is supported by a RDF (Cyprus, ΠΔΕ046_02) and European Commission (ITHANET Coordination action 026539) grants to GPP.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Gravia, A., Chondrou, V., Katsila, T., Patrinos, G. (2015). Pharmacogenomics for Haemoglobinopathies Therapeutics. In: Grech, G., Grossman, I. (eds) Preventive and Predictive Genetics: Towards Personalised Medicine. Advances in Predictive, Preventive and Personalised Medicine, vol 9. Springer, Cham. https://doi.org/10.1007/978-3-319-15344-5_7
Download citation
DOI: https://doi.org/10.1007/978-3-319-15344-5_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-15343-8
Online ISBN: 978-3-319-15344-5
eBook Packages: MedicineMedicine (R0)