Abstract
During the co-evolutionary arms race between plants and pathogens, plants evolved a sophisticated defense system to ward off their enemies. In this plant immune system , plant receptor proteins recognize non-self molecules of microbial origin, which leads to the activation of a basal level of disease resistance. The onset of these local plant immune reactions often triggers a systemic acquired resistance (SAR) in tissues distal from the site of infection. Beneficial microbes in the rhizosphere microbiome stimulate a phenotypically similar induced systemic resistance (ISR) that, like SAR, is effective against a broad spectrum of pathogens. There are differences and similarities in the SAR and ISR signaling pathways. The plant defense hormone salicylic acid is a major regulator of SAR, whereas jasmonic acid and ethylene play important roles in ISR. Priming of systemic tissue to express an accelerated defense response upon attack by a pathogen is a common phenomenon in both SAR and ISR. This chapter will outline the current concept of the plant immune system, with special emphasis on mechanisms of systemically induced disease resistance and priming for enhanced defense.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Induce Systemic Resistance
- Systemic Acquire Resistance
- Beneficial Microbe
- Induce Disease Resistance
- Plant Immune System
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 The Plant Immune System
In the past decade, ground-breaking conceptual advances have been made in the understanding of the evolutionary development and functioning of the plant immune system (Jones and Dangl 2006) . In the current concept of the plant immune system, pattern-recognition receptors (PRRs) have evolved to recognize pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs), such as bacterial flagellin or fungal chitin (Boller and Felix 2009) . MAMP recognition is translated into a basal defense called pattern-triggered immunity (PTI) (Dodds and Rathjen 2010) . Successful pathogens evolved virulence effector molecules to bypass this first line of defense, either by preventing detection by the host, or by suppressing PTI signaling (Dodds and Rathjen 2010; Pel and Pieterse 2013) . To fight these successful pathogens, plants developed a second line of defense in which resistance (R) proteins mediate recognition of attacker-specific effectors (formerly known as avirulence factors), resulting in highly powerful effector-triggered immunity (ETI) (Dodds and Rathjen 2010) . ETI is a manifestation of the classic gene-for-gene resistance that is accompanied by a hypersensitive response that prevents biotrophic pathogens from further entry (Chap. 10) .
Activation of PTI and ETI in locally infected tissues often triggers an induced resistance in tissues distal from the site of infection and involves one or more long-distance signals that propagate an enhanced defensive capacity in still undamaged plant parts. This pathogen-induced systemic resistance is known as systemic acquired resistance (SAR) (Fu and Dong 2013) . While PTI and ETI are activated rapidly and act locally to limit growth of the specific invader at the site of infection, SAR takes more time to develop but confers an enhanced defensive capacity that is typically effective against a broad spectrum of pathogens (Walters et al. 2013; Fu and Dong 2013) .
Besides pathogen infection, also colonization of plant roots by beneficial microbes has been shown to stimulate the plant immune system, resulting in a phenotypically similar type of broad-spectrum disease resistance , commonly referred to as induced systemic resistance (ISR) (Pieterse et al. 2014) . Moreover, insect herbivory and specific chemicals can also induce resistance (Howe and Jander 2008; Pastor et al. 2013) . After more than three decades of research, the picture is emerging that the different forms of induced resistance are regulated by a complex network of interconnecting signaling pathways in which plant hormones play an important regulatory role (Pieterse et al. 2012). Induced resistance signaling pathways that are triggered by pathogens, beneficial microbes, and insects partly overlap and share common signaling components (Pieterse et al. 2014). This provides plants with an enormous regulatory potential to rapidly adapt to their biotic environment and to utilize their limited resources for growth and survival in a cost-efficient manner. Intriguingly, successful pathogens evolved mechanisms to rewire the plant’s hormone signaling network to suppress or evade the host immune system (Robert-Seilaniantz et al. 2011; Pieterse et al. 2012) , highlighting the central role of plant hormones in the regulation of immunity .
The concepts of PTI and ETI that act locally in the plant immune system will be discussed in more depth elsewhere in this issue (see Chap. 10). In this chapter we will focus on the important principles and recent findings of induced disease resistance that acts systemically throughout the plant.
2 Pathogen-Induced Systemic Acquired Resistance (SAR)
Hallmarks of SAR
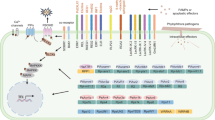
The term SAR was first coined by Ross for the phenomenon that uninfected systemic plant parts become more resistant in response to a prior infection elsewhere in the plant (Ross 1961) . SAR is typically triggered upon local activation of a PTI or ETI response (Shah and Zeier 2013) . In systemic tissues, SAR is characterized by increased levels of the hormone salicylic acid (SA), one of the hallmarks of SAR (Vlot et al. 2009) (Fig. 14.1). Early genetic studies in tobacco showed that SA accumulation and signaling is essential for the establishment of SAR (Vernooij et al. 1994) . Another hallmark of SAR is the coordinate activation of PATHOGENESIS-RELATED (PR) genes, several of which encode PR proteins with antimicrobial activity (Van Loon et al. 2006) . PR −1 is amongst the best characterized PR genes and is in many plant species used as a marker for SAR (Van Loon et al. 2006; Fu and Dong 2013) .
a Schematic representation of biologically induced disease resistance triggered by pathogen infection (SAR; red arrow) and colonization of the roots by beneficial microbes (ISR; purple arrow). Induced resistance involves long-distance signals that are transported through the vasculature or as airborne signals, and systemically propagate an enhanced defensive capacity against a broad spectrum of attackers in still healthy plant parts. b Schematic representation of molecular components and mechanisms involved in pathogen-induced SAR and rhizobacteria-mediated ISR. Solid black lines indicate established interactions; dashed black lines indicate hypothetical interactions. Colored arrows indicate systemic translocation of long-distance signals (indicated in the same color at the base of the arrows). Ac acetylation, ET ethylene, ETI effector-triggered immunity, Fe iron, ISR induced systemic resistance, JA jasmonic acid, MAMP microbe-associated molecular pattern, Me methylation, PAMP pathogen-associated molecular pattern, PRR pattern-recognition receptor, PTI PAMP-triggered immunity, R protein Resistance protein, SA salicylic acid, SAR systemic acquired resistance, TF transcription factor
Long-Distance Signals
Because the expression of SAR occurs in plant parts that are distant from the site of induction, a long-distance mobile signal is required that is produced locally and is responsible for the systemic onset SAR in still healthy tissues. The identity of the mobile SAR signal(s) has been a subject of controversy for many years. The lipid-transfer protein DEFECTIVE IN INDUCED RESISTANCE1 (DIR1) was shown to act as a chaperone for an unknown mobile SAR signal in the vascular tissue (Maldonado et al. 2002; Champigny et al. 2011) . Despite the fact that SA accumulates in the phloem sap of SAR-expressing plants, grafting experiments with tobacco showed that SA itself is not the mobile SAR signal (Vernooij et al. 1994). Recent genetic and biochemical studies uncovered several plant metabolites involved in long-distance SAR signaling. These include the methyl ester of SA (MeSA), the diterpenoid (DA), a glycerol -3-phosphate (G3P)-dependent factor, azelaic acid (AzA), and pipecolic acid (Pip) (Fig. 14.1). From these findings a more comprehensive view on the identity and functioning of the long-distance SAR signals started to emerge in which different signals may be operative under different environmental conditions (Shah and Zeier 2013; Dempsey and Klessig 2012; Kachroo and Robin 2013) . In systemic tissues, the onset of SAR requires the function of FLAVIN-DEPENDENT MONOOXYGENASE 1 (FMO1) (Mishina and Zeier 2006) , possibly to transduce or amplify long-distance signals originating from primary leaves, which then results in enhanced SA biosynthesis in still healthy tissues.
SAR Signaling
Upon activation of SAR, the SA signal is transduced by the redox-regulated protein NONEXPRESSOR OF PR GENES1 (NPR1), which functions as a transcriptional co-activator of a large set of PR genes (Fu and Dong 2013) . In non-stimulated cells, NPR1 is sequestered in the cytoplasm as an oligomer through intermolecular disulfide bonds. Upon SA accumulation, changes in the cellular redox state mediate monomerization of NPR1, which allows translocation of NPR1 into the nucleus. In the nucleus, NPR1 interacts with TGA transcription factors that together with WRKY transcription factors activate SA-responsive PR genes. Proper functioning of NPR1 requires that the protein is broken down by the proteasome, possibly to allow new NPR1 proteins to reinitiate the PR transcription cycle (Spoel et al. 2009) . Recently, NPR1 and its paralogues NPR3 and NPR4 were identified as SA receptors that bind to SA with different affinity thereby influencing the stability of NPR1 (Fu et al. 2012; Wu et al. 2012) .
3 Induced Systemic Resistance (ISR) by Beneficial Microbes
Besides microbial pathogens, also large communities of commensal and mutualistic microbes interact with plants providing them with essential services, such as enhanced mineral uptake, nitrogen fixation , growth promotion, and protection from pathogens (Chap. 20; Lugtenberg and Kamilova 2009; Zamioudis and Pieterse 2012; Pieterse et al. 2014) . This community of microbes is predominantly hosted by the root system and is also referred to as the rhizosphere microbiome (Chap. 28; Berendsen et al. 2012; Mendes et al. 2011) . In 1991, it was demonstrated that colonization of plant roots by selected strains of plant growth-promoting rhizobacteria (PGPR) can stimulate the plant immune system in above-ground plant parts, resulting in a broad-spectrum disease resistance called rhizobacteria-ISR (Fig. 14.1) (Van Peer et al. 1991; Wei et al. 1991; Alström 1991) . Since then, hundreds of studies in dicots and monocots have reported on the ability of PGPR to promote plant health via ISR. These studies mainly involved Bacillus , Pseudomonas, and Serratia PGPR strains. In addition, non-pathogenic plant growth-promoting fungi (PGPF) strains from species like Fusarium oxysporum, Trichoderma spp., and Piriformospora indica strains, but also symbiotic arbuscular mycorrhizal fungi have been shown to trigger ISR (Pieterse et al. 2014).
Microbial Elicitors of ISR
In order to stimulate ISR, beneficial microbes must produce elicitors that are responsible for the onset of systemic immunity. Early research on MAMPs and other elicitors of ISR-inducing Pseudomonas and Bacillus PGPR focused on the involvement of lipopolysaccharides (LPS) and the iron-regulated metabolites pyoverdin and SA (De Vleesschauwer and Höfte 2009) . Other microbial ISR elicitors include antibiotics , like 2,4-diacetylphloroglucinol (DAPG) and pyocyanin, flagella, N-acyl homoserine lactones , siderophores, and biosurfactants (De Vleesschauwer and Höfte 2009). Also specific volatile organic compounds produced by beneficial microbes were demonstrated to elicit ISR (Ryu et al. 2004; Lee et al. 2012) . Several of these ISR elicitors were shown to act redundantly, indicating that multiple microbial elicitors can trigger common signaling pathway leading to systemic immunity (Bakker et al. 2003) . This resembles PTI in plant-pathogen interactions, where recognition of multiple PAMPs is channeled into the same PTI signaling pathway (Boller and Felix 2009) .
Rhizobacteria-ISR Signaling Pathways
Because of its broad spectrum effectiveness, rhizobacteria-ISR was initially thought to be mechanistically similar to pathogen-induced SAR. However, in radish it was shown that Pseudomonas fluorescens WCS417r (hereafter called WCS417r) triggered ISR without stimulating the accumulation of the PR proteins that are characteristic for SAR (Hoffland et al. 1995) . Also in Arabidopsis thaliana (Arabidopsis) WCS417r-ISR developed without the activation of PR genes (Pieterse et al. 1996). Transgenic SA-nonaccumulating Arabidopsis NahG plants mounted wild-type levels of ISR upon colonization of the roots by WCS417r, providing genetic evidence that ISR can be mediated via an SA-independent signaling pathway (Pieterse et al. 1996) . Hence, rhizobacteria-mediated ISR and pathogen-induced SAR are regulated by distinct signaling pathways (Fig. 14.1). Analysis of a large number of ISR-triggering plant-beneficial microbe interactions in which a role for SA had been functionally tested, revealed that the ability to activate an SA-independent ISR pathway is common for beneficial microbes and occurs in a broad range of plant species (Van Loon and Bakker 2005; Van Wees et al. 2008) .
Although ISR by beneficial microbes is often regulated through SA-independent mechanisms, certain strains of beneficial microbes have been reported to trigger ISR in an SA-dependent fashion, which resembles pathogen-induced SAR (De Vleesschauwer and Höfte 2009; Van de Mortel et al. 2012) . In these cases, reactive oxygen species that accumulate at the site of tissue colonization seem to act as important elicitors (De Vleesschauwer and Höfte 2009). Since SA-dependent signaling triggered by beneficial microbes is likely to follow the SAR signaling pathway, we refer to the above section on pathogen-induced SAR for information on mechanisms underlying this phenomenon.
Role of Jasmonic Acid and Ethylene in ISR
After the discovery of SA as an important defense hormone, also the plant hormones jasmonic acid (JA) and ethylene (ET) emerged as important regulators of plant immunity (Pieterse et al. 2012) . By using JA or ET signaling mutants of Arabidopsis, it was shown that not SA, but JA and ET are central regulators of WCS417r-ISR (Pieterse et al. 1998). For many other PGPR and PGPF genetic evidence pointed to a role for JA and/or ET in the regulation of ISR (Pieterse et al. 2014), supporting the notion that JA and ET are dominant players in the regulation of SA-independent systemic immunity conferred by beneficial soil-borne microbes.
Master Regulators of ISR
The first regulatory protein identified as being essential for rhizobacteria-ISR was NPR1 (Pieterse et al. 1998). While in SAR, NPR1 functions as a transcriptional co-activator of SA-responsive PR genes, JA/ET-dependent ISR typically functions without PR gene activation. Hence, the role of NPR1 in ISR seems to be different from that in SAR. In SA signaling, NPR1 is clearly connected to a nuclear function (Fu and Dong 2013) , while in JA/ET signaling and ISR evidence is accumulating for a cytosolic function of NPR1 (Spoel et al. 2003; Stein et al. 2008; Pieterse et al. 2012) . Interestingly, simultaneous activation of SAR and ISR leads to an additively enhanced defensive capacity (Van Wees et al. 2000) . Whether this is based on the notion that SAR and ISR do not seem to compete for the same subcellular pool of NPR1 is unknown, as the exact molecular mechanism by which NPR1 functions in JA/ET-dependent ISR remains to be investigated.
Although ISR involves long-distance root-to-shoot signaling, only few studies have investigated the signaling components of the plant root that are involved in the onset of ISR. Analysis of the transcriptome of WCS417-colonized Arabidopsis roots revealed the R2R3 type MYB transcription factor gene MYB72 as one of the significantly induced genes (Verhagen et al. 2004) . In non-stimulated plants, MYB72 is lowly expressed in the root vascular bundle, but becomes highly expressed in root epidermis and cortical cells upon colonization by ISR-inducing PGPR or their volatiles (Zamioudis et al. 2014b) . Knockout myb72 mutants are impaired in their ability to express ISR, indicating that this root-specific transcription factor is essential for the onset of ISR (Van der Ent et al. 2008) . MYB72 is also induced in Trichoderma-colonized Arabidopsis roots and shown to be crucial for Trichoderma-ISR (Segarra et al. 2009) , suggesting that MYB72 is a node of convergence in the ISR signaling pathway triggered by different beneficial microbes. Being a transcriptional regulator, it was postulated that MYB72 plays an important role in the generation and/or translocation of a long-distance ISR signal. Besides its crucial role in the onset of ISR, MYB72 is also implicated in the iron-deficiency response of plant roots (Zamioudis et al. 2014a; Zamioudis et al. 2014b) . How ISR and the iron-deficiency response are interconnected is currently unknown.
4 Induced Disease Resistance: Priming for Enhanced Defense
While SA accumulation and PR gene expression are hallmarks of SAR, ISR triggered by beneficial microbes is lacking such universal characteristics associated with the onset of systemic immunity. In many cases, colonization of plant roots by beneficial microbes does not lead to major changes in defense-related gene expression in the above-ground plant parts. Instead, pathogen infection or insect herbivory on ISR-expressing plants often leads to an accelerated expression of defense-related gene expression in comparison to similarly attacked control plants (Van Wees et al. 1999; Van Oosten et al. 2008) . Large-scale analysis of the WCS417r-ISR transcriptome of Arabidopsis before and after pathogen challenge showed that ISR is associated with potentiated expression of a large set of JA/ET-regulated defense genes that are induced upon pathogen challenge (Fig. 14.1) (Verhagen et al. 2004) . This preparation of the whole plant to better combat pathogen or insect attack is called ‘priming’ and is characterized by a faster and/or stronger activation of cellular defenses upon invasion, resulting in an enhanced level of resistance (Conrath 2011) . To date, a large number of studies with PGPR and PGPF have supported the notion that ISR by beneficial microbes is commonly based on defense priming (Pieterse et al. 2014) .
Priming for enhanced defense emerged as an important cellular process in many types of biologically and chemically induced systemic immunity, including SAR, ISR, and herbivore-induced resistance (Frost et al. 2008; Luna et al. 2014; Pastor et al. 2013; Conrath 2011) . For instance, low doses of SAR-inducing agents do not directly activate PR gene expression, but prime systemic tissues for enhanced PR gene expression after pathogen challenge, indicating that priming is also an important component of this type of induced resistance (Conrath 2011). By studying the costs and benefits of defense priming, it was shown that the fitness costs of priming are lower than those of constitutively activated defenses (Van Hulten et al. 2006; Walters et al. 2008; Vos et al. 2013) . The fitness benefit of priming was shown to outweigh its cost when under pathogen pressure, suggesting that priming functions as an ecological adaptation of the plant to respond faster to its hostile environment.
Priming: A Molecular Memory of Immunization
Because defense priming is clearly expressed at the transcriptional level, research on the mechanisms underlying the primed state has focused on the expression of signaling intermediates in transcriptional networks. These factors are thought to remain inactive in the absence of an attacker, but their accumulation can provide the plant with the capacity to react with an accelerated defense response upon perception of a pathogen- or insect-derived stress signal. In Arabidopsis, the ISR-primed state was shown to be associated with elevated transcript levels of genes that encode transcription factors of the AP2/ERF family and MYC2, both of which have been implicated in the regulation of JA- and/or ET-dependent defenses (Van der Ent et al. 2009; Pozo et al. 2008) . This is in agreement with the observation that in particular JA/ET-regulated genes show a primed expression pattern in challenged ISR-expressing plants (Verhagen et al. 2004) .
Mitogen-activated protein kinases (MAPKs) have also been implicated in defense priming. Inactive forms of the MPK3 and MPK6 were shown to accumulate after treatment of plants with low concentrations of the SAR-inducing SA-analogue benzothiadiazole (BTH) (Beckers et al. 2009) . After pathogen challenge, these latent signaling molecules were activated, resulting in accelerated PR -1 gene expression and the development of enhanced disease resistance . Priming is also associated with chromatin modifications in the promoters of WRKY transcription factor genes that regulate SA-dependent defenses, thereby facilitating potentiated expression of these regulatory genes upon pathogen attack (Jaskiewicz et al. 2011) . Recently, epigenetic regulation of pathogen- and chemically-induced priming for SA-dependent defenses and herbivore-induced priming for JA-dependent defenses was shown to be inherited to the offspring via chromatin remodeling (Slaughter et al. 2012; Rasmann et al. 2012; Luna et al. 2012) . Hence, plants seem to have the capacity to “memorize” a stressful situation and subsequently immunize not only themselves, but also their next generation against future attacks (Pastor et al. 2013) .
5 Induced Resistance: Shaping the Plant’s Social Network
Exciting developments in induced disease resistance research provided a wealth of information on the molecular details of how this adaptive defense system functions. In nature, plants are attacked by a multitude of pathogens and pests. However, beneficial associations between plants and mutualistic microbes are abundant in nature as well, improving plant growth and health . Hormone-regulated plant defense signaling networks finely balance plant responses to beneficial microbes, pathogens, and insects to maximize both profitable and protective functions. Defense signaling pathways that are recruited in response to parasitic and beneficial organisms can overlap, indicating that the regulation of the plant’s adaptive response to its biotic environment is finely balanced between protection against aggressors and acquisition of benefits. Plant hormones play pivotal roles in the regulation of the defense signaling network. Their signaling pathways interact in a synergistic or antagonistic manner, providing the plant with the capacity to tailor its immune response to the attacker encountered (Pieterse et al. 2012) . In agricultural and ecological settings, plants often interact with a whole suite of other organisms that range from beneficial microbes on their root system to foliar pathogens and insect herbivores. Detailed mechanistic knowledge on how the plant immune signaling network functions during multi-organisms interactions is fundamental to develop novel strategies for sustainable protection of our future crops that need to produce more with less input of pesticides and fertilizers.
References
Alström S (1991) Induction of disease resistance in common bean susceptible to halo blight bacterial pathogen after seed bacterization with rhizosphere pseudomonads. J Gen Appl Microbiol 37:495–501
Bakker PAHM, Ran LX, Pieterse CMJ et al (2003) Understanding the involvement of rhizobacteria-mediated induction of systemic resistance in biocontrol of plant diseases. Can J Plant Pathol 25:5–9
Beckers GJM, Jaskiewicz M, Liu Y et al (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell 21:944–953
Berendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486
Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60:379–406
Champigny M, Shearer H, Mohammad A et al (2011) Localization of DIR1 at the tissue, cellular and subcellular levels during systemic acquired resistance in Arabidopsis using DIR1:GUS and DIR1:EGFP reporters. BMC Plant Biol 11:125
Conrath U (2011) Molecular aspects of defence priming. Trends Plant Sci 16:524–531
De Vleesschauwer D, Höfte M (2009) Rhizobacteria-induced systemic resistance. In: Van Loon LC (ed) Plant Innate Immunity, vol 51. Advances in botanical research. Academic Press Ltd-Elsevier Science Ltd, London, pp 223–281
Dempsey DA, Klessig DF (2012) SOS—too many signals for systemic acquired resistance? Trends Plant Sci 17:538–545
Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11:539–548
Frost CJ, Mescher MC, Carlson JE et al (2008) Plant defense priming against herbivores: getting ready for a different battle. Plant Physiol 146:818–824
Fu ZQ, Dong X (2013) Systemic acquired resistance: turning local infection into global defense. Annu Rev Plant Biol 64:839–863
Fu ZQ, Yan S, Saleh A et al (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486:228–232
Hoffland E, Pieterse CMJ, Bik L et al (1995) Induced systemic resistance in radish is not associated with accumulation of pathogenesis-related proteins. Physiol Mol Plant Pathol 46:309–320
Howe GA, Jander G (2008) Plant immunity to insect herbivores. Annu Rev Plant Biol 59:41–66
Jaskiewicz M, Conrath U, Peterhansel C (2011) Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep 12:50–55
Jones JDG, Dangl JL (2006) The plant immune system. Nature 444:323–329
Kachroo A, Robin GP (2013) Systemic signaling during plant defense. Curr Opin Plant Biol 16:527–533
Lee B, Farag MA, Park HB et al (2012) Induced resistance by a long-chain bacterial volatile: elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE 7:e48744
Lugtenberg B, Kamilova F (2009) Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556
Luna E, Bruce TJA, Roberts MR et al (2012) Next-generation systemic acquired resistance. Plant Physiol 158:844–853
Luna E, Van Hulten M, Zhang Y et al (2014) Plant perception of β-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat Chem Biol 10:450–456
Maldonado AM, Doerner P, Dixon RA et al (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419:399–403
Mendes R, Kruijt M, De Bruijn I et al (2011) Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332:1097–1100
Mishina TE, Zeier J (2006) The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol 141:1666–1675
Pastor V, Luna E, Mauch-Mani B et al (2013) Primed plants do not forget. Environ Exp Bot 94:46–56
Pel MJC, Pieterse CMJ (2013) Microbial recognition and evasion of host immunity. J Exp Bot 64:1237–1248
Pieterse CMJ, Van Wees SCM, Hoffland E et al (1996) Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8:1225–1237
Pieterse CMJ, Van Wees SCM, Van Pelt JA et al (1998) A novel signaling pathway controlling induced systemic resistance in Arabidopsis. Plant Cell 10:1571–1580
Pieterse CMJ, Van der Does D, Zamioudis C et al (2012) Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol 28:489–521
Pieterse CMJ, Zamioudis C, Berendsen RL et al (2014) Induced systemic resistance by beneficial microbes. Annu Rev Phytopathol 52:347–375
Pozo MJ, Van der Ent S, Van Loon LC et al (2008) Transcription factor MYC2 is involved in priming for enhanced defense during rhizobacteria-induced systemic resistance in Arabidopsis thaliana. New Phytol 180:511–523
Rasmann S, De Vos M, Casteel CL et al (2012) Herbivory in the previous generation primes plants for enhanced insect resistance. Plant Physiol 158:854–863
Robert-Seilaniantz A, Grant M, Jones JDG (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49:317–343
Ross AF (1961) Systemic acquired resistance induced by localized virus infections in plants. Virology 14:340–358
Ryu C-M, Farag MA, Hu CH et al (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026
Segarra G, Van der Ent S, Trillas I et al (2009) MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol 11:90–96
Shah J, Zeier J (2013) Long-distance communication and signal amplification in systemic acquired resistance. Front Plant Sci 4:30
Slaughter A, Daniel X, Flors V et al (2012) Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol 158:835–843
Spoel SH, Koornneef A, Claessens SMC et al (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15:760–770
Spoel SH, Mou ZL, Tada Y et al (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137:860–872
Stein E, Molitor A, Kogel KH et al (2008) Systemic resistance in Arabidopsis conferred by the mycorrhizal fungus Piriformospora indica requires jasmonic acid signaling and the cytoplasmic function of NPR1. Plant Cell Physiol 49:1747–1751
Van de Mortel JE, De Vos RCH, Dekkers E et al (2012) Metabolic and transcriptomic changes induced in Arabidopsis by the rhizobacterium Pseudomonas fluorescens SS101. Plant Physiol 160:2173–2188
Van der Ent S, Verhagen BWM, Van Doorn R et al (2008) MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol 146:1293–1304
Van der Ent S, Van Hulten MHA, Pozo MJ et al (2009) Priming of plant innate immunity by rhizobacteria and ß-aminobutyric acid: differences and similarities in regulation. New Phytol 183:419–431
Van Hulten M, Pelser M, Van Loon LC et al (2006) Costs and benefits of priming for defense in Arabidopsis. Proc Natl Acad Sci U S A 103:5602–5607
Van Loon LC, Bakker PAHM (2005) Induced systemic resistance as a mechanism of disease suppression by rhizobacteria. In: Siddiqui ZA (ed) PGPR: biocontrol and biofertilization. Springer, Dordrecht, pp 39–66
Van Loon LC, Rep M, Pieterse CMJ (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Van Oosten VR, Bodenhausen N, Reymond P et al (2008) Differential effectiveness of microbially induced resistance against herbivorous insects in Arabidopsis. Mol Plant-Microbe Interact 21:919–930
Van Peer R, Niemann GJ, Schippers B (1991) Induced resistance and phytoalexin accumulation in biological control of fusarium wilt of carnation by Pseudomonas sp. strain WCS417r. Phytopathology 81:728–734
Van Wees SCM, Luijendijk M, Smoorenburg I et al (1999) Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol Biol 41:537–549
Van Wees SCM, De Swart EAM, Van Pelt JA et al (2000) Enhancement of induced disease resistance by simultaneous activation of salicylate- and jasmonate-dependent defense pathways in Arabidopsis thaliana. Proc Natl Acad Sci U S A 97:8711–8716
Van Wees SCM, Van der Ent S, Pieterse CMJ (2008) Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol 11:443–448
Verhagen BWM, Glazebrook J, Zhu T et al (2004) The transcriptome of rhizobacteria-induced systemic resistance in Arabidopsis. Mol Plant-Microbe Interact 17:895–908
Vernooij B, Friedrich L, Morse A et al (1994) Salicylic acid is not the translocated signal responsible for inducing systemic acquired resistance but is required in signal transduction. Plant Cell 6:959–965
Vlot AC, Dempsey DA, Klessig DF (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47:177–206
Vos IA, Pieterse CMJ, Van Wees SCM (2013) Costs and benefits of hormone-regulated plant defences. Plant Pathol 62:43–55
Walters DR, Paterson L, Walsh DJ et al (2008) Priming for plant defense in barley provides benefits only under high disease pressure. Physiol Mol Plant Pathol 73:95–100
Walters DR, Ratsep J, Havis ND (2013) Controlling crop diseases using induced resistance: challenges for the future. J Exp Bot 64:1263–1280
Wei G, Kloepper JW, Tuzun S (1991) Induction of systemic resistance of cucumber to Colletrotichum orbiculare by select strains of plant-growth promoting rhizobacteria. Phytopathology 81:1508–1512
Wu Y, Zhang D, Chu JY et al (2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1:639–647
Zamioudis C, Pieterse CMJ (2012) Modulation of host immunity by beneficial microbes. Mol Plant-Microbe Interact 25:139–150
Zamioudis C, Hanson J, Pieterse CMJ (2014a) β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron uptake responses in Arabidopsis roots. New Phytol 204:368–379
Zamioudis C, Korteland J, Van Pelt JA et al (2014b) Root bacteria stimulate iron uptake in plants via a novel photosynthesis-dependent iron sensing system. Submitted
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Pieterse, C., Van Wees, S. (2015). Induced Disease Resistance. In: Lugtenberg, B. (eds) Principles of Plant-Microbe Interactions. Springer, Cham. https://doi.org/10.1007/978-3-319-08575-3_14
Download citation
DOI: https://doi.org/10.1007/978-3-319-08575-3_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-08574-6
Online ISBN: 978-3-319-08575-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)