Abstract
Breast cancer is the leading cause of cancer death in women worldwide. Gene expression studies have been used over the last decades to define the signature of different breast cancer subtypes and to predict outcome and response to therapies. Recently, microRNAs (miRNAs) have been linked to several human diseases, including cancer. An aberrant miRNA expression in breast cancer was first reported in 2005. Now, an increasing body of experimental evidences supports the role of these small molecules in the tumorigenic process and their potential use as cancer specific biomarkers. Indeed, miRNAs are detectable as circulating molecules in the blood. In this chapter, we summarize our knowledge about the involvement of miRNAs in breast cancer and their potential as diagnostic, prognostic and therapeutic tools.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Breast cancer is one of the most frequent cancers worldwide and the most frequent affecting women. In 2012, 227,000 new cases of breast cancer were counted and 39,500 people died from this neoplasm in United States (Siegel et al. 2012). Breast cancer is a heterogeneous disease, that comprehends several histotypes, characterized by different biological and phenotypic features and that presents different prognostic and therapeutic procedures. The currently used factors for breast cancer diagnosis, classification and treatment include patient age, gender, menopausal status, lymph node status, tumor size, histological features (grade and type, peritumoral vascular invasion), hormone receptor status [Estrogen receptor (ER) and progesterone receptor (PR) expression], proliferation index (Mib1) and HER2 over-expression/amplification, as provided by St. Gallen criteria (Goldhirsch et al. 2009), Nottingham Prognostic Index (Galea et al. 1992) and Adjuvant Online (www.adjuvantonline.com). The definition of these guidelines for patient risk stratification, together with the improvement of chemotherapeutic regimens, enhanced breast cancer survival rate. However, these rules do not consider the individual molecular complexity of each neoplasm and for this reason some tumors belonging to a risk group do not behave in the expected way or do not respond to the chosen therapeutic strategy. The right prediction of aggressiveness and metastatic potential of a lesion are key factors in breast cancer patient management. The recent development of technologies (i.e. microarray, deep sequencing) helped in overcoming this problem. These methods are able to evaluate the global genomic and transcriptomic profile of tissues and tumors. Indeed, specific genome-wide profiles created for each kind of breast tumor subtype, improved histotype classification and prognosis definition (Perou et al. 2000; Koboldt et al. 2012; Curtis et al. 2012).
MicroRNAs (miRNAs) are a class of regulatory, non-coding small RNA that mainly post-transcriptionally regulate gene expression. The evaluation of cancer-specific miRNA profiles revealed to be useful in stratifying breast tumors. Breast cancer was among the first tumor types for which the evaluation of miRNA profile was performed. The miRNA microarray analysis has initially showed a panel of 29 miRNAs that were deregulated in breast cancer, if compared to healthy breast tissue (Iorio et al. 2005). Interestingly, further studies on other human cancers found that several of the 29 miRNAs were deregulated in other neoplasms, indicating that this first set of miRNAs could affect pathogenetic mechanisms potentially shared among tumors and suggesting the important role of miRNA in tumor development. Nowadays, the involvement of miRNAs in each step of cancerogenesis, from transformation to metastatic spreading, is well known and universally recognized.
MiRNA profiling has allowed for the identification of signatures associated with the diagnosis, staging, progression, prognosis, and response to treatment of human tumors (Dvinge et al. 2013). The miRNA-based classifier is much better than the mRNA classifier at establishing the correct diagnosis for metastatic cancer of unknown primary site and for metastases coming from or retrieved inside breast tissue (Ferracin et al. 2011). The maintenance of a strong nucleus of tissue-specific miRNAs after the spreading of cancer cells to distant sites is probably the reason why miRNA profiling is more effective in the diagnosis of cancers of unknown primary site. In this chapter we review the role of miRNAs in breast cancer transformation, progression and their possible application in breast cancer diagnosis, prognosis and therapy.
2 Deregulation of MicroRNAs in Breast Cancer
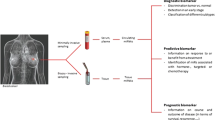
Several miRNAs were demonstrated to play important roles in breast cancer development (Le Quesne and Caldas 2010; Yu et al. 2010a). Like protein-coding genes, miRNAs could be classified as oncogenes (oncomiRs) or tumor suppressor genes, according to their expression levels in cancer and the cellular functions of miRNAs and their targets. In breast cancer, miR-145, miR-125, let-7 family and miR-200 family are the most known tumor suppressor miRNAs, while among the oncomiRs we can remind miR-21 and miR-155 (Fig. 14.1).
MicroRNAs involved in specific processes linked to breast cancer, such as cancer predisposition (genomic alterations), cancer phenotype (transcriptional profile), cancer progression (metastatic miRNAs) and their release as circulating miRNAs in the blood of cancer patients (circulating miRNAs) (Figure modified from Elsevier (Ferracin and Calin 2011))
MiR-125 is considered as a tumor suppressor in breast cancer, since it was found down-regulated in cancer, compared to normal mammary tissue (Iorio et al. 2005). Its tumor suppressor function was confirmed by the finding that many miR-125 targets, among which ERBB2, ERBB3 and MUC1, are frequently over-expressed in breast cancers, contributing to aggressiveness of the pathology (Scott et al. 2007).
Let-7 is a tumor suppressor miRNA and it was found to be down-regulated in several human neoplasms including breast cancer. It was demonstrated that let-7 is a negative regulator of several stemness properties and is reduced in breast tumor-initiating cells (BT-ICs). Indeed, its forced up-regulation decreased proliferation, self-renewal and metastasizing capability of cells (Yu et al. 2007). Known targets of let-7 are ESR1 (Bhat-Nakshatri et al. 2009) and RAS (Johnson et al. 2005), two known oncogenes, supporting its tumor suppressive function.
The well-known oncogenic miR-17-92 cluster plays a controversial role in breast cancer. Li and colleagues demonstrated that the over-expression of miR-17-5p promotes invasion of breast cancer cell lines (Li et al. 2011). In contrast, Yu and colleagues found a reduced expression of this cluster in breast tumors, if compared to normal tissue, and they proved that the cluster was able to inhibit cell proliferation and metastasizing capability by targeting cyclin D1 and IL-8 (Yu et al. 2008, 2010b), suggesting a potential tumor suppressive role for these miRNAs. Further studies will be necessary to better elucidate the role of this cluster in breast cancer. However, these results indicate that the classification of a miRNA, as tumor suppressor miRNA or oncomiR, is sometime difficult, since miRNAs are complex players in the molecular pathways of the cell and it is possible that they cooperate both in inducing and inhibiting oncogenic pathways.
3 MicroRNA Profile of Breast Cancer Subtypes
Some miRNA expression studies have been performed in order to find molecular profiles able to distinguish among different breast cancer subtypes and clinical-pathological features (Blenkiron et al. 2007). The expression of specific miRNAs has been associated to the activity of ER, PR and epidermal growth factor 2 receptor (HER2). These three receptors are key elements for breast cancer patient management. Indeed, their expression is routinely evaluated and contributes to define breast cancer diagnosis, prognosis and therapeutic regimen. In particular, triple-negative breast cancers (ER-/PR-/HER2-) are usually more aggressive and associated with poor prognosis, while neoplasms over-expressing at least one of the three receptors are characterized by better prognosis (because drugs targeting these molecules are available and effective in treating positive cancers).
It has been demonstrated that specific miRNAs can regulate the expression of estrogen, progesterone and epidermal growth factor 2 receptors. Several groups performed microarray miRNA profiling of breast cancers characterized by different ER, PR and HER2 status, in order to identify miRNAs associated with these features (Iorio et al. 2005; Mattie et al. 2006; Lowery et al. 2009). We discuss here some results that emerged from these and other studies, in order to highlight the miRNAs that influence receptors expression in breast cancer.
Among the miRNAs targeting ER, we remind miR-22 (Pandey and Picard 2009; Xiong et al. 2010) and miR-145 (Spizzo et al. 2010). On the other hand, some miRNAs are regulated by ERα itself (miR-21, miR-181, miR-26) (Bhat-Nakshatri et al. 2009; Maillot et al. 2009; Wickramasinghe et al. 2009), establishing an interesting regulatory loop. Finally, there are some miRNAs including let-7 (Zhao et al. 2011; Bhat-Nakshatri et al. 2009), miR-17-92 cluster, miR-106a/363 (Castellano et al. 2009), miR-221/222 (Zhao et al. 2008; Di Leva et al. 2010) and miR-206 that are both regulated and regulating, suggesting the presence of feedback loops acting on ER pathway. Just to give an example, miR-206 (a miRNA found up-regulated in ER- tumors) targets ERα mRNA and its expression is impaired by ER agonists (Adams et al. 2007; Kondo et al. 2008). Moreover, this miRNA seems to play a role in the repression of estrogenic response mediated by epidermal growth factor (EGF) in MCF-7 that is responsible for the switch from luminal-A to basal-like phenotype (Adams et al. 2009).
To what concern the other two receptors, miR-26 and miR-181, two miRNAs down-regulated by estrogen treatment in MCF-7 cells, are able to modulate PR (Maillot et al. 2009). HER2 is a direct target of miR-125 (Scott et al. 2007) and its expression is indirectly reduced by miR-205, a miRNA that targets HER3, belonging to the same family of HER2 (Iorio et al. 2009). All these findings highlight the important contribution of miRNAs in defining the molecular subtypes and the clinical-pathological features of breast neoplasms and could represent important molecular targets in breast cancer management.
4 Role of MicroRNAs in Epithelial-Mesenchymal Transition and Metastasis
Important features of tumor aggressiveness are epithelial-mesenchymal transition (EMT) and metastatic capability. EMT is a process where epithelial cells lose their epithelial features, cell polarity and cell adhesion, and become mesenchymal cells, gaining migratory capability, invasive potential and, finally, metastasize. Breast cancers presenting an invasive phenotype or metastasis at diagnosis are characterized by poor prognosis. Several miRNAs play important role in regulating EMT and metastasis in breast cancer (Fig. 14.1).
MiR-10b was found highly expressed in breast cancer metastases and it was demonstrated that its expression is under the control of Twist, a transcription factor implicated in EMT process. MiR-10b up-regulation inhibits HOXD10 protein translation and induces RHOC expression, leading to an invasive and metastatic behavior of tumor cells (Ma et al. 2007). Furthermore, antagomiR-10b administration in a mouse-model of breast cancer induced a reduction in tumor proliferation and metastasis formation, introducing the promising role of miRNAs as therapeutic targets (Ma et al. 2010).
Other metastasis-promoting miRNAs are miR-373 and miR-520c (Huang et al. 2008). It was demonstrated that they are able to promote migration and invasion capability of MCF-7 breast cancer cell lines, by directly suppressing of CD44, a cell-surface glycoprotein involved in cell-cell interactions. Supporting this thesis, increased levels of miR-373 and CD44 reduction were found together in human metastatic breast cancers.
The well-known oncomiR-21 was extensively studied for its involvement in breast cancer development, because it is a master regulator of cell proliferation and survival. In addition, it is mightily implicated in metastasis, since it targets TPM1, a component of cytoskeleton (Zhu et al. 2008), maspin and PDCD4 (Lu et al. 2008; Qi et al. 2009) that are inhibitors of the pro-metastatic factor UPAR, and PTEN and TIMP3 (Qi et al. 2009), inhibitors of matrix metalloproteinases.
Other important pro-metastatic miRNAs belong to miR-103/107 family, whose high levels are associated with metastasis and poor outcome in breast cancer. Martello and colleagues proved that injection of miR-103/107 antagomiR in a breast cancer xenograft in mouse induced a reduction of number of metastatic foci in the animal. They also demonstrated that down-regulation of Dicer, a molecular target of miR-103/107, and the consequent global down-regulation of miRNAs biosynthesis, played an important role in metastatic effects induced by these miRNAs. Further, miR-103/107 promoted epithelial-to-mesenchymal transition by down-regulating miR-200 levels (Martello et al. 2010).
MiR-200 and miR-205 are frequently down-regulated in breast cancer, compared to healthy mammary tissue. It was demonstrated that they are able to protect cells from EMT by targeting ZEB1 and ZEB2, two strong activators of epithelial to mesenchymal transition (Gregory et al. 2008; Korpal et al. 2008; Park et al. 2008). Indeed, down-regulation of miR-200 family was associated with metastasis development in breast cancer, while miR-205 induction was responsible for a reduced invasion capability of breast cancer cell lines and in vivo metastasis formation (Gregory et al. 2008) and it has an important role in triple negative breast cancers (Piovan et al. 2012).
MiR-335 and miR-126 were found to be anti-metastatic miRNAs through the comparison of metastatic nodules versus primary tumors. Indeed, the two miRNAs showed reduced levels in metastasis and their normal expression in primary tumors correlated with increasing metastasis-free time. They were able to counteract metastasis development by blocking expression of SOX4 and TNC (Tavazoie et al. 2008). Similarly, miR-17-92 clusters seemed to play an anti-metastatic role in breast cancer, as previously discussed.
Altogether, these findings suggest that miRNAs play very crucial roles in invasion capability and metastatic development of cancer and for this reason they could be considered interesting targets for anti-cancer-tailored therapies.
5 Circulating MicroRNAs as Novel Diagnostic Markers
One of the major challenges in molecular oncology is the employment of miRNAs as biomarkers for early diagnosis of cancer. Until now, only few proteins have been used in the clinic as blood cancer biomarkers. MiRNAs are released in the blood from cancer cells, and they have a good potential of being employed for this purpose, since it was demonstrated that they are stable in human serum and plasma (Mitchell et al. 2008). Furthermore, it has been found that they are differentially represented in cancer patients compared to healthy controls. Concerning breast cancer, blood levels of several miRNAs were found differentially released in cancer patients compared to healthy controls (Chan et al. 2013; Cuk et al. 2013). Figure 14.1 summarizes the miRNAs that are deregulated in breast cancer and released into the circulation. As can be noticed, the circulating miRNA profile does not match the expression profile of solid tumors (Cookson et al. 2012).
Detection of miRNAs in serum or plasma for the early diagnosis of cancer is a very promising tool. However many issues should be solved before this tool could enter into the clinic. First of all, different technologies (RT-qPCR, digital PCR, microarray, next-generation sequencing) are now available for the evaluation of miRNA expression in the blood, but it was recently demonstrated that results of the same pathological condition achieved with different technologies and different preparation methods are very different. This is due to the fact that technical issues can dramatically influence the results of analysis. For this reason, the optimization of many preanalytical and analytical variables is necessary. As a consequence of the lack of optimized protocols, the majority of published studies do not reciprocally confirm their findings (Chan et al. 2013; Madhavan et al. 2013).
6 MicroRNAs as Prognostic Tools in Cancer
Several studies have demonstrated the potential of using miRNA to predict prognosis of human cancers. Some miRNAs have been negatively or positively associated with prognostic endpoints. MiR-21 is one of the most studied oncomiRs and its expression was linked to poor prognosis in several human cancers (Krichevsky and Gabriely 2009). In breast cancer, miR-21 over-expression correlates with negative hormone receptor status, advanced tumor stage, high grade, lymph node metastasis and poor survival (Yan et al. 2008; Qian et al. 2009). These findings clearly suggest that miR-21 over-expression is a negative prognostic factor. A similar function is suspected for miR-210, a miRNA whose transcription is induced by HIF-1 in hypoxic conditions (Kulshreshtha et al. 2007). A higher expression of this miRNA was associated with a shorter disease-free time, overall-survival (Camps et al. 2008) and time-to-metastasis in lymph node negative patients (Foekens et al. 2008). An opposite role has been described for miR-30c, whose expression was found to be an independent predictor of progression-free survival in a large cohort of advanced ER+ breast cancer patients treated with tamoxifen (Rodriguez-Gonzalez et al. 2011). Despite these promising findings, the use of miRNA expression as prognostic tool is not yet in clinical practice.
Being better than individual miRNAs, miRNA profiles (i.e. a panel of miRNAs) could constitute more effective prognostic factors. Gene expression profiling has been used since 2000 to study breast cancer biology and to find gene signatures able to distinguish between different subtypes. Perou et al. (2000) performed the first microarray analysis of 65 breast cancer samples allowing the identification of five different “intrinsic” subtypes, characterized by different gene expression profiles: luminal A, luminal B, basal-like, HER2-enriched and normal-like. These profiles, confirmed also by subsequent studies, were able to depict different kinds of breast cancer, associated with different prognosis and clinical course. Later, also Blenkiron and colleagues performed gene-expression and miRNA analysis using microarray technology on 51 breast cancer samples. Starting from gene-expression results, he classified the samples according to Perou’s profile and compared miRNA expression analysis in samples belonging to different subtypes, allowing the identification of several subtypes-related miRNAs (Blenkiron et al. 2007). This discovery suggested that Perou’s classification could be improved adding miRNA expression results to gene expression profiles.
Besides predicting prognosis of breast cancer, miRNA profiling was demonstrated to be a good tool also to predict therapeutic response. The identification of neoplasms that will respond to a given chemotherapeutic drug is extremely important to reduce breast cancer morbidity and mortality. Using the gene-expression data from NCI-60 panel of cell lines tested for sensitivity to several drugs, Salter and colleagues developed mRNA and miRNA profiles associated with resistance to chemotherapeutic agents and validated their results in a cohort of 133 breast cancer patients treated with TFAC (paclitaxel, 5-fluorouracil, adriamycin, and cyclophosphamide) regimen. Profiles-derived predictions were compared with predictions based on traditional criteria (ER, PR, HER2 and Topoisomerase IIA expression levels), suggesting the importance of integrating classical prediction methods with new molecular tests and concluding that molecular profiling could represent a rational strategy to identify alternative therapeutic opportunities (Salter et al. 2008). In another study, miRNA expression profile of doxorubicine-resistant versus sensitive MCF-7 breast cancer cell line was performed and it was found that resistant cells over-express miR-106, miR-21, miR-206 and miR-28, while they down-regulate miR-127, miR-200c, miR-34a, miR-27b and let-7. Moreover, they demonstrated that miR-451, whose expression was lost in resistant cells, targets multidrug resistance 1 gene (mdr1/ABCB1) (Kovalchuk et al. 2008).
With similar approaches, two studies found miRNAs involved in tamoxifen and cisplatin resistance: Miller and colleagues studied miRNAs related to tamoxifen sensitivity, finding miR-221, miR-222 and miR-181 among the up-regulated miRNAs in resistant cells and miR-21, miR-342 and miR-489 among the down-regulated ones (Miller et al. 2008). Pogribny et al. (2010) performed miRNA profiling of cisplatin-resistant and sensitive MCF-7 cell lines and identified a large panel of miRNAs between the two phenotypes. In another study, aimed at identifying miRNAs associated with chemotherapy response in ovarian cancers, let-7i down-regulation emerged to be associated to cisplatin resistance both in ovarian and breast tumors (Yang et al. 2008). Other miRNAs related to chemoresistance are miR-125b, that was able to reduce taxol sensitivity (Zhou et al. 2010), and miR-155 that targets the proapoptotic gene FOXO3 and was able to induce doxorubicine resistance (Kong et al. 2010).
Many of the above mentioned studies were performed only in cellular models of breast cancer. A further validation in clinical samples is therefore necessary to identify miRNAs that could really influence therapeutic decisions. Indeed, these findings clearly highlight the complexity of the scenario where several miRNA concur in determining sensitivity or resistance to chemotherapeutics. However, the potential of miRNAs expression profile as a tool to predict prognosis seems to be achievable very soon and constitutes a real tool to improve clinical patient management.
7 MicroRNAs as Therapeutic Targets or Therapeutic Agents
Since their first discovery, miRNAs were considered of special interest as cancer therapeutics. Indeed, the modulation (induction or inhibition) of a single miRNA is able to change the expression of hundreds of target genes in the cell. Moreover, their small size, high stability and endogenous nature, indicate they have suitable features to be used as therapeutic molecules. MiRNAs represent interesting targets in cancer treatment with two possible approaches: silencing of an over-expressed miRNA or introduction of a down-regulated miRNA.
To inhibit miRNA activity, different molecules have been tested and demonstrated their efficacy in in vitro and/or in in vivo models: anti-miRs (AMO), antagomiRs, locked-nucleic acid (LNA), sponge vectors; they have different molecular structures, but they are all able to block their target miRNA by directly linking mature miRNA sequences via complementarity. For example, the administration of antagomiR-10b in breast tumor-bearing mice was demonstrated to reduce metastasis formation (Ma et al. 2010).
The restoration of a down-regulated miRNA in tumor cells could be achieved in different ways. In cellular models, the simple transfection of pre-miR precursors is effective, while for in vivo delivery different kind of miRNA-expressing vectors have been studied. It was demonstrated that miR-145 transfection in different kind of breast cancer cell lines induced a significant block of cancer cell proliferation and enhanced apoptosis (Spizzo et al. 2010). In another study adeno-associated virus (AAV) expressing miR-26a was proved to be effective in reducing liver-tumor growth in mice, without relevant toxicity (Kota et al. 2009). Another interesting possibility for cancer therapy is the use of miRNA-dependent oncolytic vectors, that take advantage of our knowledge on cancer-specific miRNAs to control the viral infection (Callegari et al. 2013).
Even if some issues have yet to be extensively considered, like target delivery and systemic toxicity, several studies involving miRNAs as cancer therapeutics have been performed (Tong and Nemunaitis 2008) and some clinical trials have been approved (www.clinicaltrials.gov), highlighting the promising role of miRNAs in tumor treatment in the future.
8 Conclusions
MiRNA global expression profiling could be used for the classification of breast cancers, establishing specific diagnoses and offering prognostic values. Recent studies have demonstrated that circulating miRNAs could serve as biomarkers for the early diagnosis of tumors. Moreover, they represent interesting targets for a new-generation of targeted therapies. For all these reasons, miRNAs appear to be a very promising tool in each step of breast cancer management.
References
Adams BD, Furneaux H, White BA (2007) The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol 21(5):1132–1147
Adams BD, Cowee DM, White BA (2009) The role of miR-206 in the epidermal growth factor (EGF) induced repression of estrogen receptor-alpha (ERalpha) signaling and a luminal phenotype in MCF-7 breast cancer cells. Mol Endocrinol 23(8):1215–1230
Bhat-Nakshatri P, Wang G, Collins NR, Thomson MJ, Geistlinger TR, Carroll JS et al (2009) Estradiol-regulated microRNAs control estradiol response in breast cancer cells. Nucleic Acids Res 37(14):4850–4861
Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ et al (2007) MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol 8(10):R214
Callegari E, Elamin BK, D’Abundo L, Falzoni S, Donvito G, Moshiri F et al (2013) Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. PloS One 8(9):e73964
Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H et al (2008) hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res 14(5):1340–1348
Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P et al (2009) The estrogen receptor-alpha-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A 106(37):15732–15737
Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA et al (2013) Identification of circulating MicroRNA signatures for breast cancer detection. Clin Cancer Res 19(16):4477–4487
Cookson VJ, Bentley MA, Hogan BV, Horgan K, Hayward BE, Hazelwood LD et al (2012) Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell Oncol 35(4):301–308
Cuk K, Zucknick M, Heil J, Madhavan D, Schott S, Turchinovich A et al (2013) Circulating microRNAs in plasma as early detection markers for breast cancer. Int J Cancer 132(7):1602–1612
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ et al (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403):346–352
Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C et al (2010) MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst 102(10):706–721
Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A et al (2013) The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497(7449):378–382
Ferracin M, Calin GA (2011) Principles of MicroRNA involvement in breast cancer. Breast Dis Year Book Q 22(3):238–243
Ferracin M, Pedriali M, Veronese A, Zagatti B, Gafa R, Magri E et al (2011) MicroRNA profiling for the identification of cancers with unknown primary tissue-of-origin. J Pathol 225(1):43–53
Foekens JA, Sieuwerts AM, Smid M, Look MP, de Weerd V, Boersma AW et al (2008) Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc Natl Acad Sci U S A 105(35):13021–13026
Galea MH, Blamey RW, Elston CE, Ellis IO (1992) The Nottingham Prognostic Index in primary breast cancer. Breast Cancer Res Treat 22(3):207–219
Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ (2009) Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol 20(8):1319–1329
Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G et al (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10(5):593–601
Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S et al (2008) The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol 10(2):202–210
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16):7065–7070
Iorio MV, Casalini P, Piovan C, Di Leva G, Merlo A, Triulzi T et al (2009) microRNA-205 regulates HER3 in human breast cancer. Cancer Res 69(6):2195–2200
Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A et al (2005) RAS is regulated by the let-7 microRNA family. Cell 120(5):635–647
Koboldt DC, Fulton RF, McLellan MD, Schmidt M, Kalicki-Veizer J, TCGA N (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70
Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H (2008) miR-206 expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res 68(13):5004–5008
Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D et al (2010) MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem 285(23):17869–17879
Korpal M, Lee ES, Hu G, Kang Y (2008) The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem 283(22):14910–14914
Kota J, Chivukula RR, O’Donnell KA, Wentzel EA, Montgomery CL, Hwang HW et al (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137(6):1005–1017
Kovalchuk O, Filkowski J, Meservy J, Ilnytskyy Y, Tryndyak VP, Chekhun VF et al (2008) Involvement of microRNA-451 in resistance of the MCF-7 breast cancer cells to chemotherapeutic drug doxorubicin. Mol Cancer Ther 7(7):2152–2159
Krichevsky AM, Gabriely G (2009) miR-21: a small multi-faceted RNA. J Cell Mol Med 13(1):39–53
Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ et al (2007) A microRNA signature of hypoxia. Mol Cell Biol 27(5):1859–1867
Le Quesne J, Caldas C (2010) Micro-RNAs and breast cancer. Mol Oncol 4(3):230–241
Li H, Bian C, Liao L, Li J, Zhao RC (2011) miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat 126(3):565–575
Lowery AJ, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C et al (2009) MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res 11(3):R27
Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH et al (2008) MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene 27(31):4373–4379
Ma L, Teruya-Feldstein J, Weinberg RA (2007) Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 449(7163):682–688
Ma L, Reinhardt F, Pan E, Soutschek J, Bhat B, Marcusson EG et al (2010) Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nat Biotechnol 28(4):341–347
Madhavan D, Cuk K, Burwinkel B, Yang R (2013) Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front Genet 4:116
Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, Benes V et al (2009) Widespread estrogen-dependent repression of micrornas involved in breast tumor cell growth. Cancer Res 69(21):8332–8340
Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S et al (2010) A MicroRNA targeting dicer for metastasis control. Cell 141(7):1195–1207
Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK et al (2006) Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer 5:24
Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL et al (2008) MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem 283(44):29897–29903
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105(30):10513–10518
Pandey DP, Picard D (2009) miR-22 inhibits estrogen signaling by directly targeting the estrogen receptor alpha mRNA. Mol Cell Biol 29(13):3783–3790
Park SM, Gaur AB, Lengyel E, Peter ME (2008) The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22(7):894–907
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Piovan C, Palmieri D, Di Leva G, Braccioli L, Casalini P, Nuovo G et al (2012) Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol 6(4):458–472
Pogribny IP, Filkowski JN, Tryndyak VP, Golubov A, Shpyleva SI, Kovalchuk O (2010) Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 127(8):1785–1794
Qi L, Bart J, Tan LP, Platteel I, Sluis T, Huitema S et al (2009) Expression of miR-21 and its targets (PTEN, PDCD4, TM1) in flat epithelial atypia of the breast in relation to ductal carcinoma in situ and invasive carcinoma. BMC Cancer 9:163
Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R et al (2009) High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res Treat 117(1):131–140
Rodriguez-Gonzalez FG, Sieuwerts AM, Smid M, Look MP, Meijer-van Gelder ME, de Weerd V et al (2011) MicroRNA-30c expression level is an independent predictor of clinical benefit of endocrine therapy in advanced estrogen receptor positive breast cancer. Breast Cancer Res Treat 127(1):43–51
Salter KH, Acharya CR, Walters KS, Redman R, Anguiano A, Garman KS et al (2008) An integrated approach to the prediction of chemotherapeutic response in patients with breast cancer. PloS One 3(4):e1908
Scott GK, Goga A, Bhaumik D, Berger CE, Sullivan CS, Benz CC (2007) Coordinate suppression of ERBB2 and ERBB3 by enforced expression of micro-RNA miR-125a or miR-125b. J Biol Chem 282(2):1479–1486
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62(1):10–29
Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S et al (2010) miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ 17(2):246–254
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD et al (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451(7175):147–152
Tong AW, Nemunaitis J (2008) Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther 15(6):341–355
Wickramasinghe NS, Manavalan TT, Dougherty SM, Riggs KA, Li Y, Klinge CM (2009) Estradiol down regulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res 37(8):2584–2595
Xiong J, Yu D, Wei N, Fu H, Cai T, Huang Y et al (2010) An estrogen receptor alpha suppressor, microRNA-22, is down regulated in estrogen receptor alpha-positive human breast cancer cell lines and clinical samples. FEBS J 277(7):1684–1694
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL et al (2008) MicroRNA miR-21 over expression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14(11):2348–2360
Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K et al (2008) MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res 68(24):10307–10314
Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C et al (2007) let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell 131(6):1109–1123
Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M et al (2008) A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol 182(3):509–517
Yu Z, Baserga R, Chen L, Wang C, Lisanti MP, Pestell RG (2010a) microRNA, cell cycle, and human breast cancer. Am J Pathol 176(3):1058–1064
Yu Z, Willmarth NE, Zhou J, Katiyar S, Wang M, Liu Y et al (2010b) microRNA 17/20 inhibits cellular invasion and tumor metastasis in breast cancer by heterotypic signaling. Proc Natl Acad Sci U S A 107(18):8231–8236
Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X et al (2008) MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 283(45):31079–31086
Zhao Y, Deng C, Wang J, Xiao J, Gatalica Z, Recker RR et al (2011) Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer. Breast Cancer Res Treat 127(1):69–80
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi Y et al (2010) MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J Biol Chem 285(28):21496–21507
Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY (2008) MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res 18(3):350–359
Acknowledgements
Our work was supported by the Italian Association for Cancer Research (AIRC MFAG11676). We thank Miriam Ferracin for her assistance in generating the graphical illustration.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Ferracin, M., Lupini, L. (2014). MicroRNA Expression Profiling and Its Clinical Impact in Breast Cancer. In: Babashah, S. (eds) MicroRNAs: Key Regulators of Oncogenesis. Springer, Cham. https://doi.org/10.1007/978-3-319-03725-7_14
Download citation
DOI: https://doi.org/10.1007/978-3-319-03725-7_14
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-03724-0
Online ISBN: 978-3-319-03725-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)