Abstract
To obtain a better insight in tissue microstructures using diffusion MRI, a high resolution and dense sampling of q-space is required. In clinical settings, however, this can often not be achieved due to limited acquisition time. Reduced field-of-view (FOV) approaches counteract this limitation but may pose a challenge for the post-processing steps such as motion and artifact correction. We present an evaluation of the potential problems that arise with reduced FOV data during the standard post-processing. The acquisition with reduced FOV is extracted from a full FOV dataset. We select three different registration tools to perform the standard data post-processing pipeline. We first evaluate the spatial error and then measure its impact on the tensor reconstruction as well as on the derived fractional anisotropy (FA). With reduced FOV images, the multi-scale registration methods showed high sensitivity to parameter selection and produced up to 30 % outliers. With an optimized parameter set, all registration methods yielded spatial errors of 1 mm (±0.572). The spatial error resulted in a mean error of 0.03 (±0.013) in the estimated FA values, and was thus of the same magnitude as group differences as they are typically reported in DTI studies. Regions with large FA differences were located especially in the corpus callosum. The evaluation indicates that diffusion-weighted MR acquisitions with reduced FOV require careful selection of registration parameters and also cautious interpretation when quantifying derived indices.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Since the inception of diffusion MRI more and more complex models have been developed to provide better insights into tissue microstructures. To achieve these improvements, acquisitions sequences with higher resolution and denser sampling of the q-space are required. In consequence, acquisition time has risen to the point where they are the limiting factor in clinical scenarios. One possible solution to address this limitation is the acquisition of reduced field of views (FOV) in order to enclose an object of interest. There are several approaches described in the literature for limited FOV diffusion-weighted MR acquisition, i.e. the usage of localized volumes [5] and zoom imaging [14].
For all subject acquisitions, it is typically the head movement of the patient during acquisition together with distortions by eddy currents that corrupt the acquired diffusion weighted (DW) images and thus make correction steps mandatory [2, 11]. In practice, both movement and distortions are commonly corrected by means of affine registration [10]. After resampling each of the DW images according to the estimated transformation, the rotational component of the transformation is then used for correcting the respective gradient direction [8].
The precision of the registration approach and its influence on the DW data post-processing have been subject of several studies. Ling et al. [9] reported a positive bias in fractional anisotropy (FA) in their study on retrospective subject head-motion correction. Kim et al. [7] evaluated the sensitivity of the FA and the mean diffusivity (MD) to the degrees-of-freedom of the transformation estimated for motion correction and showed significant differences in FA, especially in comparison between corrected and non-corrected datasets.
In this paper, we evaluate potential problems that can occur when applying the standard DW image post-processing to datasets with limited FOV. We evaluate alignment errors that are introduced by the head motion and eddy currents correction step and follow them through the pipeline to estimate their impact on the tensor and their derived scalar images. We also analyze the parameter settings for the registration methods and derive practical suggestions for reduced FOV data correction.
2 Methods
The first part of the evaluation focused on the initial alignment error introduced by the specific properties of the partial datasets. Typically, the performance of registration procedures is evaluated by quantifying a fiducial or target registration error, e.g. the mean spatial distance of anatomical structures labeled by an expert in both fixed and moving image.
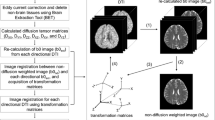
To allow such an assessment without having to label the structures in all images involved, a partial data acquisition was simulated by region extraction from the whole brain images. The transformations, the tensor data and the FA indices estimated upon the whole brain data were considered as a reference for the respective results obtained on partial data . For each subject, the partial data was computed by leaving out the lower and upper 10 slices from the original DW acquisition as illustrated in Fig. 1. The cropping parameters were selected so that the white matter structures of the limbic system were kept in the reduced field of view.
2.1 MRI Acquisition
The group of participants represents a typical collective used in diffusion tensor imaging studies on disease quantification. The collective consisted of 15 healthy controls, 15 AD patients and 17 MCI patients with mean age of 66 ( ± 7), 72 ( ± 7) and 70 ( ± 5) years respectively.
The datasets used throughout the paper were acquired on a 1.5 T whole body clinical scanner equipped with a quadrature head coil (Magnetom Symphony, Siemens Medical Solutions, Erlangen, Germany) with a gradient strength of 40 mT/m. A single shot echo planar imaging technique with a twice refocused spin echo diffusion preparation was employed using the following parameters: repetition time (TR)/echo time (TE) 4,700/78 ms, field of view 240 mm, data matrix of 96 × 96 yielding an in plane resolution of 2.5 mm, 50 axial slices with a thickness of 2.5 mm and no gap. A b-value of 1,000 s/mm2 was used for a DTI protocol with N = 6 gradient directions and one non-DW (b = 0) acquisition. The DTI protocol was repeated 10 times resulting in 70 acquired volumes per dataset.
2.2 Eddy Current and Head Motion Correction Schemes
Three correction tools were selected to perform the DW image alignment . The widely used FLIRT method [4] from the FSL library applies a multi-scale global search combined with local parameter optimization. As second method, BRAINSFit [6], which performs a single scale image registration, was selected and finally an in-house implementation, which extends the BRAINSFit to a multi-scale pyramid registration scheme, was included. Both BRAINSFit and our in-house implementation are based on ITK [3] and use the regular step gradient descent optimizer.
For the alignment of weighted images onto the unweighted reference, the mutual information (MI) metric was chosen, since it is known to perform well in this multi-modal setting [13]. The 10 reference images with b = 0 were aligned to the first reference image by minimizing the normalized correlation (NC) metric.
2.3 Registration Parameters
The ability of an image registration method to find the optimal transformation depends strongly on the chosen parameter values. Each of the correction tools provides default parameter settings, that are often used for whole data registration. But applying the default parameter settings to the multi-scale methods for partial data often resulted in registration outlier. These are characterized by small or even complete lack of overlap between the transformed moving and the fixed reference image. In order to improve the registration, suitable parameter values for partial data registration were determined as described in following paragraph. The evaluation was then performed with the optimized parameter settings.
Since the patient movement is limited in the MR, the search space for rotation was restricted to [−15, 15] degrees for all axes. The image information available for optimization is reduced by the downsampling applied for each scale. Because of the reduced size of the partial datasets, only one downsampling step was allowed in the optimized settings for FSL. For the same reason, the image pyramid was limited to two levels when registering partial data for the in-house ITK method. The optimizer settings of the lowest pyramid level correspond to the settings in BRAINSFit. The translation scales were set to 10−3, all other to 1. The optimizer was set to a maximal iteration count of 100 and a minimal step size of 10−4.
2.4 Performance Metrics
2.4.1 Precision of Registration
The spatial error of the image registration step was evaluated by the target registration error (TRE) . The fiducials, represented by a point set P, were uniformly distributed over the spatial bounding box of the partial image. With A i denoting the estimated transformation for the whole image and A i (p) for the corresponding partial image, the mean TRE for a transformation pair \((A_{i},A_{i}^{(p)})\) is defined by
The TRE was evaluated separately for all transformation pairs of the 45 × (10 × 6) gradient images and 45 × 9 unweighted images.
2.4.2 Tensor Fit Quality
Assessment of the influence of the registration error on the tensor fit quality was performed by means of a residual error. In order to ensure comparison only of voxels within the original FOV and within the brain, the Brain Extraction Tool (BET) [12] was used to determine brain masks Ω i in all whole images, which were then cropped to fit the partial data (Ω i (p)). The mask of valid voxels was then defined as
Over all valid voxels x ∈ Ω, the normalized squared residual error between the observed signal S i (x) and the signal modelled by the estimated tensor \(\tilde{S}_{i}(x)\) as introduced by Kim et al. [7]:
was measured.
2.4.3 Deviation in Fractional Anisotropy
In order to evaluate the deviation of FA values between the whole and partial datasets, the mean absolute error (MAE)
over the valid voxels in the partial FA image was considered.
3 Results
According to the mean TRE, the individual registration tasks could be clearly separated in two groups. Either the registration fails completely, i.e. transforms the moving image almost to the outside of the reference image FOV, or the transformation on the partial data differs only slightly. Computing the statistics only over the different subjects would assign an disproportionally large role to each outlier resulting into an increased TRE for the respective dataset. The presence of outliers underlines the necessity of considering the results separately.
3.1 Registration Parameters
With default settings for registering weighted data on the unweighted reference, the FSL method resulted into an outlier ratio of 32 % of the cases and the in-house ITK pyramid method with a three-level pyramid produced 15 % outliers. With optimized settings, the outlier ratio decreased to 8.6 % for FSL and 0.48 % for the ITK pyramid method. For the unweighted data, only the ITK method produced 1.15 % outliers. The BRAINSFit remained outlier-free on both weighted and unweighted data.
3.2 Precision of Registration
The TRE evaluation as depicted on Fig. 2 reveals multiple effects. First, all selected registration methods are comparable in their performance on both weighted and unweighted images. Second, for the unweighted datasets, there is no significant difference between whole and partial case, the TRE remains far below a 10th of the voxel size ( = 2.5 mm). Third, and most important, the TRE for the weighted images reaches almost 1 mm.
3.3 Tensor Fit Quality
In spite of the large, 1 mm TRE of the weighted images for partial data, the residual error of the tensor fit (see Fig. 3) showed only a minimal difference between partial and whole data for all methods except for FSL (FSL Partial in Fig. 3). This shows that the residual error is not capable of revealing the registration error.
The larger residual error of FSL turned out to be caused by the high number of outliers. When the outliers were removed, which is possible because the 10-fold repetition of the gradient scheme during acquisition guarantees a sufficient amount of gradient images for tensor reconstruction, the residual error is of about the same size as for the other methods (FSL* Partial in Fig. 3).
3.4 Deviation in Fractional Anisotropy
Contrary to the residual error of the tensor fit, the FA images computed on partial images differ significantly from the reference FA images. Consistent for outlier-free registration tools, the MAE yields values of almost 0. 03 as shown in Fig. 4. Again the presence of outliers also increases the MAE.
Fractional anisotropy evaluation. (a) MAE for all registration tools with numerical median values and the percentage of outliers in the upper plot part (outliers were removed before plotting and calculating the median). (b) FA difference map (FA −FA (p)) for a single slice, the arrows point to the high error areas in the corpus callosum. The color scale range is set to [−0. 2, 0. 2]
By visual inspection of the differences (Fig. 4b), the area of the corpus callosum (marked by arrows) proximate to the ventricle border show the highest FA error.
4 Discussion
Following recently published results, the increasing trend towards high-resolution DW in clinical practice inevitably leads to the application of partial data acquisitions. To quantify the effects introduced by the partial data into the standard DW image post-processing, we have presented an evaluation of the registration error and its propagation into the tensor and FA images.
We have demonstrated that the standard post-processing fails for partial data and the registration error affects the FA values. Compared to the recent findings in voxel-based FA evaluation studies, the MAE of almost 0.03 is of the same magnitude as the mean group differences [1, 15] and the variance for each of the groups [1]. Moreover, the corpus callosum, one of the structures frequently evaluated in disease quantification studies, shows the highest difference in the FA values. Our findings correspond with the sensitivity of FA indices to the subject motion and its correction reported by Ling et al. [9] and Kim et al. [7]. However, our result does not reproduce the FA differences to a full extent, which may be attributed to the smaller registration error introduced by the partial data.
A limitation to our work is the missing real ground truth for the TRE evaluation. But as the standard correction of whole datasets was approved in several quantification studies, it provides a reliable base for evaluating the error trend which we focused on. More importantly, using the whole images as reference made the DTI and FA image difference evaluation possible in the first place.
The downscaling is a factor limiting the stability of the weighted partial data registration as shown by the ratio of outliers for the standard settings. The FSL method optimizes in its first step only the translation parameters for fixed rotation values from the given range. The combination of the downsampling and maximal rotation around the in-plane-axes reduces the information available for the metric to an extent, that a stronger optimizer minimum is found for a minimal overlap of the moving and fixed images.
The residual tensor error seems to be insensitive to small registration errors, as it increased significantly only in the presence of outliers. Compared to Kim et al. [7], the error magnitude in the outlier-free case is about 50 %. However, this can result from the small number (6) of different directions we acquired, making it easier to explain the data with a six parameter model as compared to fitting the same model to many directions. In case of outliers, the missing image value caused by the misregistration is replaced by a default background value (0). The effect on the tensor fit is minimal due to the tenfold repetition of the acquisition, but the difference enumerated by the residual error increases significantly.
Interesting for further investigations are the different error magnitudes achieved on unweighted images and the gradient images respectively. In future work, we want to focus on improving the post-processing of partial data by using the precise registration results on the unweighted images to control the alignment of the gradient images.
References
Alves, G.S., O’Dwyer, L., Jurcoane, A., Oertel-Knöchel, V., Knöchel, C., Prvulovic, D., Sudo, F., Alves, C.E., Valente, L., Moreira, D., Fuer, F., Karakaya, T., Pantel, J., Engelhardt, E., Laks, J.: Different patterns of white matter degeneration using multiple diffusion indices and volumetric data in mild cognitive impairment and alzheimer patients. PLoS ONE 7(12), e52,859 (2012). doi:10.1371/journal.pone.0052859
Andersson, J.L., Skare, S.: A model-based method for retrospective correction of geometric distortions in diffusion-weighted epi. NeuroImage 16(1), 177–199 (2002). doi:10.1006/ nimg.2001.1039
Ibanez, L., Schroeder, W., Ng, L., Cates, J.: The ITK Software Guide, 2nd edn. Kitware, Clifton Park (2005). ISBN 1-930934-15-7, http://www.itk.org/ItkSoftwareGuide.pdf
Jenkinson, M., Smith, S.: A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5, 143–156 (2001)
Jeong, E.K., Kim, S.E., Kholmovski, E.G., Parker, D.L.: High-resolution DTI of a localized volume using 3d single-shot diffusion-weighted STimulated echo-planar imaging (3D ss-DWSTEPI). Magn. Reson. Med. 56(6), 1173–1181 (2006). doi:10.1002/mrm.21088
Johnson, H., Harris, G., Williams, K.: BRAINSFit: mutual information registrations of whole-brain 3D images, using the Insight Toolkit (2007). http://hdl.handle.net/1926/1291
Kim, D.J., Park, H.J., Kang, K.W., Shin, Y.W., Kim, J.J., Moon, W.J., Chung, E.C., Kim, I.Y., Kwon, J.S., Kim, S.I.: How does distortion correction correlate with anisotropic indices? A diffusion tensor imaging study. Magn. Reson. Imaging 24(10), 1369–1376 (2006). doi:10.1016/j.mri.2006.07.014
Leemans, A., Jones, D.K.: The B-matrix must be rotated when correcting for subject motion in DTI data. Magn. Reson. Med. 61(6), 1336–1349 (2009). doi:10.1002/mrm.21890
Ling, J., Merideth, F., Caprihan, A., Pena, A., Teshiba, T., Mayer, A.R.: Head injury or head motion? Assessment and quantification of motion artifacts in diffusion tensor imaging studies. Hum. Brain Mapp. 33(1), 50–62 (2012). doi:10.1002/hbm.21192
Mohammadi, S., Möller, H.E., Kugel, H., Müller, D.K., Deppe, M.: Correcting eddy current and motion effects by affine whole-brain registrations: evaluation of three-dimensional distortions and comparison with slicewise correction. Magn. Reson. Med. 64(4), 1047–1056 (2010). doi:10.1002/mrm.22501
Rohde, G., Barnett, A., Basser, P., Marenco, S., Pierpaoli, C.: Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magn. Reson. Med. 51(1), 103–114 (2004). doi:10.1002/mrm.10677
Smith, S.M.: Fast robust automated brain extraction. Hum. Brain Mapp. 17(3), 143–155 (2002)
Thevenaz, P., Unser, M.: Optimization of mutual information for multiresolution image registration. IEEE Trans. Image Process. 9, 2083–2099 (2000)
Wargo, C.J., Gore, J.C.: Localized high-resolution DTI of the human midbrain using single-shot EPI, parallel imaging, and outer-volume suppression at 7 T. Magn. Reson. Imaging (2013, in press). doi:10.1016/j.mri.2013.01.013
Zhuang, L., Wen, W., Trollor, J.N., Kochan, N.A., Reppermund, S., Brodaty, H., Sachdev, P.: Abnormalities of the fornix in mild cognitive impairment are related to episodic memory loss. J. Alzheimers Dis. 29(3), 629–639 (2012). doi:10.3233/JAD-2012-111766
Acknowledgements
Dr. Maier-Hein (né Fritzsche) and Jan Hering received support from the German Research Foundation (DFG), grant# ME 833/15-1 and WO 1218/3-1 respectively.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this paper
Cite this paper
Hering, J., Wolf, I., Meinzer, HP., Stieltjes, B., Maier-Hein, K.H. (2014). A Quantitative Evaluation of Errors Induced by Reduced Field-of-View in Diffusion Tensor Imaging. In: Schultz, T., Nedjati-Gilani, G., Venkataraman, A., O'Donnell, L., Panagiotaki, E. (eds) Computational Diffusion MRI and Brain Connectivity. Mathematics and Visualization. Springer, Cham. https://doi.org/10.1007/978-3-319-02475-2_4
Download citation
DOI: https://doi.org/10.1007/978-3-319-02475-2_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-02474-5
Online ISBN: 978-3-319-02475-2
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)