Abstract
The aim of the study was to analyze the amino acid sequences of the SARS-CoV-2 coronavirus and homologous sequences with the use of bioinformatics tools. In result, phylogenetic trees were created for all selected sequences that illustrated the changes taking place in coronavirus proteins that could lead to the formation of SARS-CoV-2.
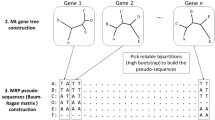
Four structural proteins of coronaviruses were selected for analysis: S - forming coronavirus spikes, M - building the membrane, N - occurring in the nucleocapsid and E - being part of the virus envelope.
In order to create phylogenetic trees, multiple sequence alignments were created using the progressive method and the PAM210 amino acid substitution matrix. Then, on the basis of the sequences, distance matrices were created and phylogenetic trees were built using two methods: UPGMA and neighbor joining. As a result, eight trees were obtained, two for each protein. The obtained trees were validated using the Bootstrap method for 100 replicates.
The analysis of the obtained proteins led to the discovery of a group among the studied viruses that contained the sequences most similar to the SARS-CoV-2 sequence, and thus the closest to it. This group includes the bat viruses RaTG13, Rp3, bat-SL-CoVZXC21 and bat-SL-CoVZC45, the pangolin virus, and the SARS-CoV virus. Regardless of the protein tested or the method used, the closest relationship was found for RaTG13, followed by the pangolin coronavirus.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
MathWorks. https://www.mathworks.com/help/bioinfo/ref/seqpdist.html
Dabravolski, S.A., Kavalionak, Y.K.: SARS-COV-2: structural diversity, phylogeny, and potential animal host identification of spike glycoprotein. J. Med. Virol. 92(9), 1690–1694 (2020)
Giovanetti, M., et al.: Evolution patterns of SARS-COV-2: snapshot on its genome variants. Biochem. Biophys. Res. Commun. 538, 88–91 (2020)
Jo, W.K., de Oliveira-Filho, E.F., Rasche, A., Greenwood, A.D., Osterrieder, K., Drexler, J.F.: Potential zoonotic sources of SARS-COV-2 infections. Transbound. Emerg. Dis. 68(4), 1824–1834 (2021)
Li, T., et al.: Phylogenetic supertree reveals detailed evolution of SARS-COV-2. Sci. Rep. 10(1), 22,366 (2020)
MATLAB: R2020a. The MathWorks Inc., Natick (2020)
Payne, S.: Family coronaviridae. Viruses 149–158 (2017)
Pevsner, J.: Bioinformatics and Functional Genomics. Wiley, Hoboken (2015)
Tabibzadeh, A., et al.: Evolutionary study of covid-19, severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) as an emerging coronavirus: phylogenetic analysis and literature review. Vet. Med. Sci. 7(2), 559–571 (2021)
Wiley, E.: Why trees are important. Evol.: Educ. Outreach 3, 499–505 (2010)
Xiong, J.: Essential Bioinformatics. Cambridge University Press, New York (2006)
Zhou, P., et al.: A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Tamulewicz, A., Uzdowska, J. (2024). Analysis of the SARS-COV-2 Molecular Sequences Using Bioinformatics Tools. In: Gzik, M., Paszenda, Z., Piętka, E., Tkacz, E., Milewski, K., Jurkojć, J. (eds) Innovations in Biomedical Engineering 2023. Lecture Notes in Networks and Systems, vol 875. Springer, Cham. https://doi.org/10.1007/978-3-031-52382-3_25
Download citation
DOI: https://doi.org/10.1007/978-3-031-52382-3_25
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-52381-6
Online ISBN: 978-3-031-52382-3
eBook Packages: Intelligent Technologies and RoboticsIntelligent Technologies and Robotics (R0)