Abstract
Max-tree (or component-tree) is a hierarchical representation which associates to a scalar image a descriptive data structure induced by the inclusion relation between the binary components obtained at successive level-sets. Various attributes related to these binary components can be computed and stored into the tree.
Max-trees have been involved in many applications, enabling to perform attribute filtering in an efficient algorithmic way. Since the resulting images do not contain any new contour, these kind of filters are called connected operators.
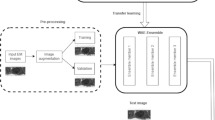
In this paper, we propose to rely on max-trees and attribute filters to enrich the input of a convolutional neural network (CNN) to improve a task of segmentation. More precisely, two approaches are considered: a first approach in which images are preprocessed using attribute filters and a second approach in which maps of attributes relying on max-trees are computed. Based on these two different approaches, the resulting maps are used as additional input in a standard CNN in a context of semantic segmentation.
We propose to compare different attributes and nodes selection strategies and to experiment their usage on a practical problem: the segmentation of the mitochondria and endoplasmic-reticulum in Focused Ion Beam milling combined with Scanning Electron Microscopy (FIB-SEM) images.
We provide original images, annotations, source code and a documentation to reproduce the experimentation results.
IdEx Doctoral contract, Université de Strasbourg.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Mathematical Morphology
- Connected Operators
- Max-tree
- Segmentation
- Deep Learning
- Convolutional Neural Network
- Electron Microscopy
- FIB-SEM
1 Introduction
The max-tree structure [16, 21] allows to perform efficiently attribute filtering [7] and has been involved in many image processing applications. The resulting operators are called connected [22] since they do not create new contours nor modify their position. In [3], Farfan et al. have suggested that max-tree attributes could be used to feed a deep convolutional neural network (CNN) in order to improve the results of detection and segmentation tasks. Following and generalizing this approach, the aim of this paper is to provide a reproducible framework enabling to perform various experiments involving max-trees and CNN in a context of semantic segmentation of cellular FIB-SEM images.
Our contributions are twofold: in a first approach, input images are preprocessed using various attribute filters [12] and then concatenated as additional inputs of a CNN. In a second approach, maps of attributes are computed from the max-tree and then added in a CNN, following Farfan et al. approach.
Finally, our work aims to be handy. For this purpose, all the methods we propose can be used on a high-end workstation and do not require large GPU/TPU clusters. Also, our source code, datasets (original images, annotations) and documentation are publicly available, allowing everybody to reproduce the results, but also to reuse the code for their own needs.
2 State of the Art
To address the segmentation of cellular electron microscopy images, the state-of-the-art methods are currently based on CNN [2, 5, 9, 12, 18, 19, 24] and the U-Net architecture remains mainly used. However, despite the good accuracy that can be obtained using these methods, the resulting segmentations can still suffer from various imperfections. In particular, thin and elongated objects such as endoplasmic reticulum can be disconnected and some parts may be distorted [12]. These effects may be the result of the context window that is fixed and too narrow in the first layer of the CNN, preventing to capture sufficient global information.
To overcome this, Farfan et al. [3] have proposed to enrich a CNN with attributes computed from the max-tree, enabling to capture at a pixel level, an information that may be non-local.
In the sequel of this paper, we will explore various strategies in order to incorporate max-tree attributes into CNN with the aim of potentially improving segmentation results.
3 Methods
3.1 Max-Tree
Let \(I:E \rightarrow V\) be a discrete, scalar (i.e. grayscale) image, with \(E\subseteq \mathbb Z^n\) and \(V\subseteq \mathbb Z\). A cut of I at level v is defined as: \(X_v(I) = \{p \in E | I(p) \ge v\}\). Let C[X] be the set of connected components of X. Let \(\varPsi \) be the set of all the connected components of the cuts of I:
The relation \(\subseteq \) is a partial order on \(\varPsi \). The transitive reduction of the relation \(\subseteq \) on \(\varPsi \) induces a graph called the Hasse diagram of \((\varPsi ,\subseteq )\). This graph is a tree, the root of which is E. The rooted tree \(\mathcal T=(\varPsi ,L, E)\) is called the max-tree of I, with \(\varPsi ,L,E\) being respectively the set of nodes, the set of edges and the root of \(\mathcal T\). The parent node of N, denoted Par(N), is the unique node such that: \((Par(N),N)\in L\) and \(N \subseteq Par(N)\) for \(N\ne E\). The branch associated to a node is the set of its ancestors and is defined for a node \(N\in \varPsi \) by: \(Br(N)=\{X\in \varPsi \mid X\supseteq N\}\).
In this work, the computation of the max-tree is based on the recursive implementation of Salembier [21], and node attributes are computed during the construction of the tree. In the rest of this paper, we will focus on the following attributes which have been proposed in the literature:
-
The height H is the minimum gray level of the connected component [21].
-
The area A is the number of pixels in the connected component [21].
-
The contour length CT represents the number of pixels that have both a neighbor inside and outside the component [20].
-
The contrast C is the difference between the maximum and minimum grey-level in the connected component [21].
-
The complexity CPL represents the contour length CT divided by the area A [20].
-
The compacity (sometimes named compactness or circularity) CPA is the area A divided by the square of the contour length \(CT^2\) [22].
-
The volume V is the sum of the difference between the pixels values in the node and the node height [16].
-
The mean gradient border MGB represents the mean value of the gradient magnitude for the contour pixels of the component [3].
The tree attributes can be merged in order to compute an image, by associating to each pixel an attribute value computed from its corresponding nodes [3]. Each pixel p belongs to several nodes: the connected component N including p in the level-set \(X_{I(p)}(I)\) and all the nodes belonging to its branch Br(N). To associate a unique value to each pixel, different policies can be implemented: for example, by keeping the maximum, the minimum or the mean value of the attributes of the branch nodes [3].
In this work, we propose the following strategy. For each pixel p, the set of nodes belonging to the branch of p is retrieved, and only a subset of nodes having an attribute value in a certain range (given as a parameter) is kept. From this set, the node \(N_{best}\) optimizing a certain stability criterion is kept. Finally, the value of p in the resulting image is set to the attribute value of \(N_{best}\). The resulting image is normalized in the range \(V=\llbracket 0, 255 \rrbracket \).
The criterion used to retrieve the optimal node is based on the concept of Maximally Stable Extremal Regions proposed by Matas et al. [10]. The idea is to retrieve the most stable regions based on the area variation between successive nodes since these regions represent salient objects of the image. For each node \(N\in \varPsi \), with \(N\ne E\) (i.e. different from the root), we define two stability attributes as follows:
where Par(N) defines the parent node of N.
3.2 Segmentation
We base our segmentation method on fully convolutional networks for semantic segmentation. We use attribute filtered images or max-tree attribute maps as additional input of our network. We feed the network with the original and the preprocessed images, concatenating them in the color (spectral) channels.
For model architectures, we use a 2D U-Net [19], which is a reference for biomedical image segmentation. A 3D U-Net [2] could also be used, but the results are not necessarily better [17, 23, 25] and the computational cost of training the model is much higher. In a preliminary experiment, we have compared 2D and 3D models, using an equal number of parameters and same input size and the 2D U-Net performs as well, if not better than the 3D one.
Each block of the network is composed of convolutions with ReLU activation, followed by batch normalization [6] and a residual connection [4], see Fig. 1. We use a 50% dropout entering the deepest block to avoid over fitting. We always use padded convolution to maintain the spatial dimension of the output. The model starts with 64 filters at the first level, for a total of 32.4 millions parameters.
We train our models by minimizing the Dice loss. For the binary segmentation task, we use classical Dice loss and for multi-class segmentation problems, we use a weighted mean of the loss for each class, with the same weight (\(W=0.5\)) for our two classes. We note X the ground truth, Y the prediction, W the weight list and C the class list. The \(\varepsilon \) term is used for stability when \(\sum {(X + Y)} = 0\) and is set to \(10^{-4}\).
The model is trained for 128 epochs, each epoch is composed of 512 batches, a batch is composed of 8 patches and a patch is a \(256 \times 256 \times C\) subpart of the image, with C the number of channels. We use random \(90^{\circ }\) rotation, horizontal and vertical flips for data augmentation on the patches. We trained our model using Adam [8] optimizer with the following parameters: \(\alpha =0.001\), \(\beta _1=0.9\), \(\beta _2=0.999\), \(\varepsilon =1\textrm{e}{-07}\).
3.3 Evaluation Metrics
To evaluate our models, we binarize the model prediction with a threshold at 0.5. To predict a slice, we use the whole slice to avoid the negative border effects of padding.
To evaluate our results, we use the F1-Score which is a region-based metric, and the average symmetric surface distance (ASSD) which is a boundary-based metrics. In fact, the F1-Score is equivalent to the Dice Score. We note respectively TP, FP and FN the cardinal of the sets of true positives, false positives and false negatives. X is the ground truth, Y the binary prediction and \(\partial X\) the boundary of X.
with \(d(x, A) = \min _{y \in A}{{\Vert x - y \Vert }_2}\).
4 Experiments
In this section, we test the improvement of the segmentation thanks to the addition of filtered images in the input. We compare the original image input with the enriched version. We also compare in the same time a multi-class segmentation model and two binary segmentations models. Finally, we repeat each of these configurations 11 times. In total, 363 models have been trained for this experiment, each training lasts about 5 h with an NVIDIA GeForce RTX 2080 Ti.
First, we define the filters we use as our experiment variable. Attributes are selected for their potential help for the segmentation, we list the selected filters in Table 1 (attribute maps strategy) and 2 (connected operators strategy), and renamed them for sake of simplicity.
Before processing the image with a max-tree, we apply a low pass filter (\(9 \times 9\) mean filter).
4.1 Data
We perform our experimentation on a stack of 80 slices from a 3D FIB-SEM image. Each slice has a size of \(1536 \times 1408\). The image represents a HeLa cell and has an \((x\times y\times z\)) resolution of 5 nm \(\times \) 5 nm \(\times \) 20 nm. A ground truth is available on the stack for two kinds of organelles (i.e. cell subunits) : mitochondria and endoplasmic reticulum. A default background class is affected to non-assigned pixel. An example slice with label is available in Figs. 3 and 4 in Sect. A.2. Figures 5 to 14 depict the slice for each applied filter.
We divide the stack into 3 sets: training (first 40 slices), validation (next 20 slices) and test (last 20 slices). The training set is used to train the network, the validation set to select the best model during the training and the test set to provide evaluation metrics.
4.2 Results
Figs. 2a and 2b depict the F1-score on the two classes of segmentation as box plots. Detailed mean and variation score are available in Table 3 and 4 in Sect. A.1.
Baseline Segmentation Results. Mitochondria are well segmented with a median F1-score up to 95% in multi-class segmentation and 94% in binary segmentation, which let a little possible improvement. Median F1-scores for reticulum up to 72% and 71% in multi-class and binary segmentation respectively. This thin organelle is indeed more difficult to segment.
Additional-input segmentation results On the mitochondria, the additional inputs improve the results for 11 cases out of 20 and 17 out of 20 for the reticulum. The gain on the reticulum is very interesting since it is the more difficult to segment for the baseline setup. The following additional inputs improve the result in all the four tests (binary and multi-class segmentations on mitochondria and reticulum): Contrast\(_{\varDelta _A}\), Complexity\(_{\varDelta _A}\), Contrast \(\beta \). Moreover, the whiskers show that only Contrast\(_{\varDelta _A}\)and Contrast \(\beta \) have a good stability in this experiment. These two inputs are therefore good candidates to be additional inputs to improve segmentation results. On the contrary, Compacity\(_{\varDelta _A}\)and MGB do not yield to any improvement.
4.3 Reproducibility
In this section, we present the required steps to reproduce the results we presented.
We follow the ACM definition of reproducibility: “The measurement can be obtained with stated precision by a different team using the same measurement procedure, the same measuring system, under the same operating conditions, in the same or a different location on multiple trials. For computational experiments, this means that an independent group can obtain the same result using the author’s own artifacts.” [1]
For this purpose, our codes and datasets are publicly available. The project is split between three subprojects. First, the experimentation detailed documentation, training scripts, evaluation scripts, preprocessing script, results logs [11]. Second, the max-tree related functions and preprocessing script [14]. Finally, the dataset with images and annotations [13]
The following information are also available on the documentation repository with more details.
Requirements. A system with Ubuntu 18.04.6 (or compatible) with g++ and git installed. Python 3.6.9 with an environment including TensorFlow 2.6.2, NumPy, SciPy, scikit-image and MedPy.
Image preprocessing
-
Prepare data for extraction with low pass filter. python 01_mean_filter.py
-
Extract attribute image from pre-processed images. ./build_bin_requirements.sh ./02_attribute_image.sh
-
Crop the image to the annotated area and construct a tiff stack. python 03_crop_roi.py
Network Training and Evaluation. For the following commands, $ID is a unique identifier for the train, $INPUT is the folder containing the dataset, $OUTPUT is the folder containing the trained models and evaluation metrics, $DATASET select the dataset to use (in our case, binary or multi-class dataset), $SETUP select the experiment to run. An automation bash script is available on the repository to run all the 33 setups once.
-
Train the networks python train.py $ID $INPUT $OUTPUT $DATASET $SETUP
-
Evaluate the networks python eval.py $ID $INPUT $OUTPUT $DATASET $SETUP BEST
Result Analysis and Figures Reproduction. Since the output of each model evaluation is a comma separated values (CSV) file, the analysis of the results can be done using various tools. We propose to use a Jupyter notebook with Pandas and Seaborn, merging the CSV files into a single dataframe. An example analysis.ipynb notebook is provided on the GitHub, which we use to produce our result figures and tables.
5 Conclusion
In this paper, we have presented a detailed experimental setup to evaluate the use of additional input in a CNN based segmentation task. The additional inputs are attribute maps obtained from a max-tree representation of the image. The evaluation is made on segmentation tasks in the context of 3D electronic microscopy. If most of the additional inputs improve one segmentation task, two of them – namely Contrast\(_{\varDelta _A}\)and Contrast \(\beta \)– improve all the tested segmentation tasks in terms of median F1-score and stability. Further than the segmentation results, the setup inspired from [3] has been entirely implemented in C++ and Python and is proposed in open access to make it reproducible.
As a perspective of this work, the feature extraction method based on max-tree attributes presented in this paper could be used for other applications. For example, it would be interesting to compute the max-tree directly inside the model and to use the attributes images as a nonlinear filter. Also, the attribute maps could be used as feature maps for more simple and explainable classifier as random forests or even decision tree. Besides, the \(\varDelta _A\) attributes we defined could be used in an interactive segmentation setup, where a single pixel will allow to select an interesting node, selecting an object connected component over a background. Finally, we proposed here a max-tree based method, but an extension to the tree of shapes [15] could be interesting and add more information to the image.
References
ACM: Artifact Review and Badging - vol 1.1. https://www.acm.org/publications/policies/artifact-review-and-badging-current
Çiçek, Ö., Abdulkadir, A., Lienkamp, S.S., Brox, T., Ronneberger, O.: 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016, pp. 424–432. Lecture Notes in Computer Science, Springer International Publishing, Cham (2016). https://doi.org/10.1007/978-3-319-46723-8_49
Farfan Cabrera, D.L., Gogin, N., Morland, D., Naegel, B., Papathanassiou, D., Passat, N.: Segmentation of Axillary and Supraclavicular Tumoral Lymph Nodes in PET/CT: A Hybrid CNN/Component-Tree Approach. In: 2020 25th International Conference on Pattern Recognition (ICPR), pp. 6672–6679 (Jan 2021). https://doi.org/10.1109/ICPR48806.2021.9412343
He, K., Zhang, X., Ren, S., Sun, J.: Deep Residual Learning for Image Recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778. IEEE, Las Vegas, NV, USA (2016). https://doi.org/10.1109/CVPR.2016.90
Heinrich, L., et al.: Whole-cell organelle segmentation in volume electron microscopy. Nature pp. 1–6 (2021). https://doi.org/10.1038/s41586-021-03977-3
Ioffe, S., Szegedy, C.: Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. In: Proceedings of the 32nd International Conference on Machine Learning, pp. 448–456. PMLR (2015)
Jones, R.: Connected filtering and segmentation using component trees. Comput. Vis. Image Underst. 75(3), 215–228 (1999). https://doi.org/10.1006/cviu.1999.0777
Kingma, D.P., Ba, J.: Adam: A Method for Stochastic Optimization. In: Bengio, Y., LeCun, Y. (eds.) 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7–9, 2015, Conference Track Proceedings (2015)
Liu, J., et al.: Automatic reconstruction of mitochondria and endoplasmic reticulum in electron microscopy volumes by deep learning. Front. Neurosci. 14 (2020). https://doi.org/10.3389/fnins.2020.00599
Matas, J., Chum, O., Urban, M., Pajdla, T.: Robust wide-baseline stereo from maximally stable extremal regions. Image Vis. Comput. 22(10), 761–767 (2004). https://doi.org/10.1016/j.imavis.2004.02.006
Meyer, C.: CTAISegmentationCNN. GitHub repository (2022). https://github.com/Cyril-Meyer/DGMM2022-RRPR-CTAISegmentationCNN
Meyer, C., Mallouh, V., Spehner, D., Baudrier, É., Schultz, P., Naegel, B.: Automatic Multi Class Organelle Segmentation For Cellular Fib-Sem Images. In: 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), pp. 668–672 (2021). https://doi.org/10.1109/ISBI48211.2021.9434075
Meyer, C., Mallouh, V., Spehner, D., Schultz, P.: DGMM2022-RRPR-MEYER-DATA. GitHub repository (2022). https://github.com/Cyril-Meyer/DGMM2022-RRPR-MEYER-DATA
Meyer, C., Naegel, B.: ComponentTreeAttributeImage. GitHub repository (2022). https://github.com/Cyril-Meyer/DGMM2022-RRPR-ComponentTreeAttributeImage
Monasse, P., Guichard, F.: Fast computation of a contrast-invariant image representation. IEEE Trans. Image Process. 9(5), 860–872 (2000). https://doi.org/10.1109/83.841532
Najman, L., Couprie, M.: Building the component tree in quasi-linear time. IEEE Trans. Image Process. 15(11), 3531–3539 (2006). https://doi.org/10.1109/TIP.2006.877518
Nemoto, T., et al.: Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J. Radiat. Res. 61(2), 257–264 (2020). https://doi.org/10.1093/jrr/rrz086
Oztel, I., Yolcu, G., Ersoy, I., White, T., Bunyak, F.: Mitochondria segmentation in electron microscopy volumes using deep convolutional neural network. In: 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 1195–1200 (2017). https://doi.org/10.1109/BIBM.2017.8217827
Ronneberger, O., Fischer, P., Brox, T.: U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, pp. 234–241. Lecture Notes in Computer Science, Springer International Publishing, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Salembier, P., Brigger, P., Casas, J., Pardas, M.: Morphological operators for image and video compression. IEEE Trans. Image Process. 5(6), 881–898 (1996). https://doi.org/10.1109/83.503906
Salembier, P., Oliveras, A., Garrido, L.: Antiextensive connected operators for image and sequence processing. IEEE Trans. Image Process. 7(4), 555–570 (1998). https://doi.org/10.1109/83.663500
Salembier, P., Wilkinson, M.H.: Connected operators. IEEE Signal Process. Mag. 26(6), 136–157 (2009). https://doi.org/10.1109/MSP.2009.934154
Srikrishna, M., et al.: Comparison of two-dimensional- and three-dimensional-based u-net architectures for brain tissue classification in one-dimensional brain CT. front. Comput. Neurosci. 15, 785244 (2022)
Xiao, C., et al.: Automatic mitochondria segmentation for EM data using a 3D supervised convolutional network. Front. Neuroanat. 12 (2018). https://doi.org/10.3389/fnana.2018.00092
Zettler, N., Mastmeyer, A.: Comparison of 2D vs. 3D Unet Organ Segmentation in abdominal 3D CT images. In: WSCG’2021 - 29. International Conference in Central Europe on Computer Graphics, Visualization and Computer Vision’2021 (2021). https://doi.org/10.24132/CSRN.2021.3101.5
Acknowledgements
We acknowledge the High Performance Computing Center of the University of Strasbourg for supporting this work by providing scientific support and access to computing resources. Part of the computing resources were funded by the Equipex Equip@Meso project (Programme Investissements d’Avenir) and the CPER Alsacalcul/Big Data. We thank D. Spehner from the Institut de Génétique et de Biologie Moléculaire et Cellulaire for providing the images and V. Mallouh for providing the annotations. We acknowledge the use of resources of the French Infrastructure for Integrated Structural Biology FRISBI ANR-10-INBS-05 and of Instruct-ERIC. We acknowledge that this work is supported by an IdEx doctoral contract, Université de Strasbourg.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
A Appendix
A Appendix
1.1 A.1 Results
In the following tables, the mean and deviation are computed over the 8 best model out of 11, selected using the F1-score on the validation set.
1.2 A.2 Example Preprocessing Visualization
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Meyer, C., Baudrier, É., Schultz, P., Naegel, B. (2023). Combining Max-Tree and CNN for Segmentation of Cellular FIB-SEM Images. In: Kerautret, B., Colom, M., Krähenbühl, A., Lopresti, D., Monasse, P., Perret, B. (eds) Reproducible Research in Pattern Recognition. RRPR 2022. Lecture Notes in Computer Science, vol 14068. Springer, Cham. https://doi.org/10.1007/978-3-031-40773-4_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-40773-4_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-40772-7
Online ISBN: 978-3-031-40773-4
eBook Packages: Computer ScienceComputer Science (R0)