Abstract
Selenoproteins play a crucial role in various complex biological processes, including cell growth, differentiation, apoptosis, and ferroptosis. In response to increased oxidative damage and altered nutrient metabolism known as the Warburg switch, cancer cells adapt via selenoprotein synthesis that are essential for their survival. Especially, selenoprotein thioredoxin reductase 1 (TrxR1) and glutathione peroxidase-4 (GPX4) have significant roles in controlling hydroperoxides and resisting ferroptosis. These selenoproteins offer protection to cancer cells from ferroptosis and oxidative damage by utilizing selenium-related mechanisms to counteract the toxic effects of chemotherapy and radiotherapy. The bioavailability of selenium is often overlooked as a determinant of oxidative defense status and has emerged as a critical factor in cancer cells. Hence, the impact of dietary selenium depletion in the context of anticancer treatment highlighted the need for further investigation into the potential therapeutic applications of selenium or selenoproteins in cancer treatment. This chapter explores the potential application of selenoproteins as therapeutic targets in cancer treatment, with a specific focus on ferroptosis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
17.1 Introduction
Ferroptosis, a form of regulated cell death, is controlled by a selenium-dependent antioxidant glutathione peroxidase 4 (GPX4) in the presence of iron and selenium (Seibt et al. 2019). A reduction in GPX4 activity due to decreased selenium levels can lead to oxidative stress and ferroptosis in cancer (Ingold et al. 2018). GPX4 and other 25 selenoproteins in humans are notably ferroptosis sensitive. Despite their potential role in preventing the progression of cancer, the precise role of other selenoproteins and the thioredoxin system in the ferroptosis pathway is poorly understood (Cai et al. 2020; Kalimuthu et al. 2022). However, due to the antioxidant properties of selenoproteins, they may play a significant role in the regulation of ferroptosis. A pan-cancer study on selenoprotein expression found that the expression of genes belonging to the GPX and TrxR families can vary between different cancer types and even within individual cancer cases (Wu et al. 2021). Few studies in mice models have shown that administering selenomethionine (SeMet) before irradiation can delay tumor formation. On the other hand, continued use of SeMet can lead to uncontrolled growth of tumors (Jeter et al. 2019). The complexity of selenoproteins classified is based on the type and stage of cancer. Hence, further investigation is needed to fully understand their role in carcinogenesis. In addition, selenoproteins play a role in various molecular signaling pathways and can influence the expression of transcription factors (Ghelichkhani et al. 2022).
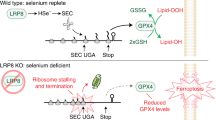
Ferroptosis may serve as a therapeutic target cancer cells with enhanced sensitivity, as evidenced by the altered regulation of selenoproteins in the cancer cells. Apolipoprotein E receptor 2 LRP8 (also known as ApoER2) can promote resistance to ferroptosis in cancer cells by regulating selenium and selenoprotein levels. In response to limited selenium availability, the levels of GPX4 rapidly decrease in cancer cells leading to ferroptosis (Li et al. 2022). Eagle and group highlight the potential of targeting selenophosphate synthetase 2 (SEPHS2) and selenium detoxification as a therapeutic strategy for acute myeloid leukemia (AML). The authors also observed other selenoproteins that help to suppress programmed cell death caused by iron-dependent lipid peroxidation, i.e., ferroptosis. Few other studies also suggested that selenium depletion may be used as a part of anticancer therapies (Eagle et al. 2022; Jankowski and Rabinowitz 2022). This book chapter points out the possibility to use selenium or selenoprotein levels as a therapeutic strategy against cancer and for regulating the ferroptosis pathway.
17.2 Ferroptosis
Before the term “ferroptosis” was coined, Dolma and group discovered inducers of this form of cell death through a high-throughput screening method in which they evaluated 23,550 compounds for their cell-killing ability on tumorigenic cells induced by multiple oncogenes, including RAS (which is mutated in approximately 25% of cancers, as described by Dixon et al (Dixon et al. 2012)). Human fibroblast cells were transformed into tumor cells through the introduction of human telomerase reverse transcriptase (hTERT), the catalytic component of telomerase, and the large T (LT) and small T (ST) oncoproteins of SV40 and onco-RAS protein. The researchers found nine compounds that were four times more effective on these tumorigenic cells, with erastin being effective among the identified compounds and exhibiting specificity toward these cells (as an eradicator of RAS and ST cells). The morphological studies suggested that erastin did not result in any changes in nuclear morphology and neither caused DNA fragmentation nor activated caspases, indicating that the compound induces a type of non-apoptotic cell death (Dolma et al. 2003). In 2007, Yagoda and coworkers reported that this form of cell death is reliant on the RAS-RAF-MEK signaling pathway and can be suppressed by antioxidants, suggesting that reactive oxygen species (ROS), particularly lipids, might play a role in inducing cell death (Yagoda et al. 2007).
The researchers discovered that erastin binds to voltage-dependent anion channels (VDACs) and changes the permeability of the outer membrane of the mitochondria. Out of 47,725 compounds tested, two compounds, RSL3 and RSL5, were found to exhibit anticancer activity through a mode of action like erastin, which involves the activation of the RAS-RAF-MEK pathway, binding to VDACs, and the involvement of lipid-derived ROS (Yang and Stockwell 2008). The authors also observed that two iron chelators, including deferoxamine (DFX), can block the effects of RSLs and erastin, indicating a role for intracellular iron in this new form of cell death (Yang and Stockwell 2008). They characterized ferroptosis as being distinct from apoptosis, necrosis, and autophagy in terms of biochemistry, morphology, and genetics (Dixon et al. 2012).
17.3 Characteristics of Ferroptosis
Ferroptosis can be distinguished from other forms of cell death as it does not result in chromatin condensation as seen in apoptosis or cytoplasmic swelling and plasma membrane rupture as seen in necrosis. Additionally, ferroptosis does not involve the formation of double-membrane vacuoles, which is a hallmark of autophagy (Dixon et al. 2012). Biochemically, ferroptosis can be differentiated from apoptosis as it does not involve the activation of caspases, cytochrome c release, or cleavage of PARP-1 (Yagoda et al. 2007). Furthermore, ferroptosis does not result in ATP depletion, a key feature of necrosis (Dixon et al. 2012). To identify the unique genetic network underlying ferroptosis, Dixon and researchers used an shRNA library targeting 1087 genes involved in mitochondrial functions. The results of this study showed no correlation between shRNAs that protected cells from erastin-induced ferroptosis and staurosporine-induced apoptosis, indicating that the genetic networks involved in these two forms of cell death are distinct (Dixon et al. 2012). They also reported six genes that are essential for ferroptosis: RPL8 (ribosomal protein L8), IREB2 (iron response element-binding protein 2), ATP5G3 (ATP synthase F0 complex subunit C3), CS (citrate synthase), TTC35 (tetratricopeptide repeat domain 35), and ACSF2 (acyl-CoA synthetase family member 2). The knockout (KO) of these genes was found to protect cells against ferroptosis, but not against cell death induced by other agents such as staurosporine, rotenone, or rapamycin (Dixon et al. 2012). These findings support the notion that ferroptosis is regulated by a specific genetic network.
17.4 Role of Selenium in Ferroptosis Pathway
The critical role of selenium in the ferroptosis pathway can be understood from the results of various biochemical assays such as western blot and promoter assay. However, the previous studies on ferroptosis suggest that these assays were often not reproducible even when the same cell lines were used in different laboratories. Why do the results differ? The findings of Karlenius and coworkers from gene promoter reporter assay indicate that the results were altered when the laboratory switched to a different vendor of FBS supplier. To investigate this further, they determined the serum selenium concentrations of FBS from various country vendors and their corresponding activity for gene promoter, cell proliferation, and antioxidant enzyme assays. The chemical composition of different batches of sera used for cell culture may vary including selenium concentration, which is not routinely tested before shipping. The significance of redox control systems in regulating a range of biological processes such as gene expression and cell signaling is now widely accepted. Different growth mediums may lead to the activation of different key redox systems in the cells grown which may have various downstream effects. For example, many transcription factors are redox-regulated which in turn regulates the activity of other gene promoters (Shahdadfar et al. 2005; Turanov et al. 2010). The variation in the serum-containing cell culture medium is a key factor in the output of conflicting results for many tests such as gene reporter assays, mRNA quantification, and western blot assay. A recent report suggests that the metabolic environment of tumors in vivo cannot be recreated using currently available cell culture media (Karlenius 2011). To test this, the effect of a new physiological medium called Plasmax was compared with commercial media for animal cell culture. It was found that cancer cells undergo metabolic disturbances because of the unbalanced nutrient composition of commercial media (Gardner et al. 2022; Kalimuthu et al. 2022). The high levels of pyruvate and arginine in this context have specific effects on the regulation of gene expression and the urea cycle. Pyruvate stabilizes HIF-1α in normal oxygen conditions, leading to a false hypoxia response. Arginine interferes with the normal functioning of the urea cycle enzyme argininosuccinate lyase; however, this effect can be prevented in vitro by Plasmax. Ferroptosis influenced by the varying levels of selenium in serum inhibited the ability of cancer cells to form colonies on commercial media, whereas predetermined selenium in Plasmax was able to restore this ability (Voorde Vande et al. 2019). Recent study on the effects of selenium on cancer stem cells is consistent with the previous reports which indicate that selenium offer resistance to cancer drug (Eagle et al. 2022). Hence, we propose that optimal levels of selenium in the medium could serve as a supplement and can be employed in drug discovery experiments for drug-resistant cancer.
17.5 Ferroptosis-Induced ER Stress
Endoplasmic reticulum (ER) stress is one of the attractive biological phenomena that ensue as part of ferroptosis (Lee et al. 2020). The unfolded protein response (UPR) pathway is triggered by three transmembrane proteins, IRE1 (inositol-requiring enzyme 1), ATF6 (activated transcription factor 6), and PERK (protein kinase RNA (PKR)-like endoplasmic reticulum kinase) (Moncan et al. 2021). In addition, the presence of seven selenoproteins in the endoplasmic reticulum membrane demonstrates the diversity within this subgroup and their impact on cancer processes such as proliferation, survival, and apoptosis. Recent studies have highlighted the role of selenoproteins in endoplasmic reticulum stress in promoting tumor growth and metastasis in various solid tumors. Although most selenoproteins, including SELENO F, SELENO K, SELENO M, SELENO N, SELENO S, and SELENO T are involved in protein degradation and calcium homeostasis (Fig. 17.1), the exact mechanism through which they protect cancer cells from apoptosis and ferroptosis cell death is still unclear (Varlamova et al. 2022). There is mounting evidence suggesting that the endoplasmic reticulum membrane represents a crucial location where lipid peroxidation occurs, leading to various cellular dysfunctions and pathologies (Varlamova et al. 2022; von Krusenstiern et al. 2023). In fact, GPX4 act as a protector against lipid peroxidation in ER and other subcellular membranes.
The AAA+ ATPase p97 (also known as VCP in metazoans and Cdc48 in lower eukaryotes) regulates the entire autophagy process, from initiation to maturation and autophagy-mediated degradation. This highly abundant protein constitutes about 1% of total cellular protein and is essential for endoplasmic reticulum-associated protein degradation (ERAD). During the ERAD process, the recruitment of VCP/p97 to the ER membrane is necessary for substrate degradation and is facilitated by the selenoproteins SELENO S and SELENO K. The binding of SELENO K to p97 is essential for the recruitment of SELENO S. The binding between SELENO K and SELENO S is not influenced by the redox state of SELENO S and does not involve the Cys173 and Sec188 residues of SELENO S. (Lee et al. 2015). Notably, both SELENO S and SELENO K bind to derlins that are involved in the ERAD complex with an association of p97 (VCP). It is well known that selenoprotein S functions in cell survival by adaptive regulation of ER stress. Selenoprotein K is also one of the ER stress-regulated proteins and plays an important role in resistance against ER stress-mediated apoptosis. This extensive list corresponds with the reports that at least some of SELENO S and SELENO K cellular roles are tied to the protein degradation process and lipid storage (Shchedrina et al. 2011).
According to some reports, SELENO K promotes melanoma tumor progression and metastasis. It binds to the DHHC6 (each letter represents amino acids Asp-His-His-Cys) acyltransferase enzyme to catalyze the palmitoylation of the IP3R which is required for ER membrane stability. Although, it has been seen that SELENO K disturbs the IP3R palmitoylation and Ca2+ in the ER membrane, however, increasing SELENO K expression may not be in proportionality for IP3R palmitoylation as well as the Ca2+ in the ER membrane (Marciel and Hoffmann 2019). Genetic manipulation of the SELENO K gene results in significantly lower levels of store-operated calcium (Ca2+) entry protein in melanoma cancer cells in vitro and in vivo. According to recent studies, selenium deficiency may be used in cancer treatment (Eagle et al. 2022; Jankowski and Rabinowitz 2022). Recently, few studies have revealed that targeting SELENO K can effectively reduce tumor size by modulating ferroptosis, thereby contributing a favorable establishment for the development of potential therapies for cervical cancer (Abdurahman et al. 2023). Previously, it was believed that proteins that trigger ferroptosis such as RSL3 and ML162 were inhibited by GPX4, TNXRD1 and SELENO K. Recent discovery suggests that GPX4 does not directly inhibit RSL3 and ML162 (Cheff et al. 2023). However, SELENO K and TNXRD1 may directly inhibit those ferroptosis triggerers.
17.6 p62/Keap1/NRF2 Axis Protects against Ferroptosis
Brigelius-Flohe proposed that selenium metabolites like selenols and selenenic acids can modify protein thiols to form selenylsulfides (Se-S), selenotrisulfides (S-Se-S), selenocysteine (SeSec), and selenomethionine (SeMet) (Brigelius-Flohé 2008). Recently, some reports revealed that selenium-containing natural compounds have opened new avenues for selenometabolomics in eukaryotic cells (Hou and Xu 2022; Kayrouz et al. 2022; Abdalla and Mühling 2023). The changes in the levels of these selenometabolites may play an important role in the survival of cancer cells. Selenium metabolites mainly regulate the nuclear factor erythroid 2-related factor 2 (NRF2) pathway (Fig. 17.2). First, NRF2 protects cells against oncogenic attacks such as ROS and electrophilic carcinogenic species. Conversely, once malignant transformation occurs within a cell, the NRF2-KEAP1 signaling pathway can protect the tumor from oxidative stress and chemo- or radiotherapy-induced cytotoxicity through the cooperation of various cells populations and functions.
The p62-KEAP1-NRF2 pathway is involved in the protection of cancer cells against ferroptosis through the up-regulation of several genes such as quinone oxidoreductase 1 [NQO1], heme oxygenase-1 [HO1], glutathione peroxidase (GPx), thioredoxin reductase (Trx), and ferritin heavy chain 1 [FTH1] that are intricately involved in iron metabolism (Sun et al. 2016). Upon treatment with an electrophilic precursor of methylselenol, methylseleninic acid first activates the transcription factor NRF2, leading to the induction of the gene encoding NQO-1 through direct modification of selenium-KEAP1 (Park et al. 2018). These findings provide a novel mechanism by which methylseleninic acid exhibits a significant ability to induce oxidative/stress-responsive gene expression associated with selenium metabolites. There is documented evidence that inhibition of NRF2 significantly enhances the anticancer effects of ferroptosis inducers in several cancers (Lei et al. 2019). Selenium is an essential micronutrient that is required for the proper functioning of various cellular pathways, including the antioxidant defense system. Recently, the researcher has discovered an unexpected outcome associated with the plant-derived compound known as artesunate. While the mechanism of how artesunate alters KEAP1 protein expression is not yet fully understood, it has been suggested that it may be related to changes in selenium metabolism. However, further research is needed to fully elucidate the relationship between artesunate, KEAP1, and selenium metabolism (Hill et al. 2021). Lee et al. 2003 demonstrated that selenium can maintain the growth of specific human hepatocellular carcinoma cell lines in the serum-free medium containing 0.1% of serum (Lee et al. 2003). We propose for the first time that selenium-dependent cell growth and proliferation can be defined as selenium-dependent selenoplasia which is one of several types of metalloplasia. It is in line with other metalloplasia such as cuproplasia which is copper-dependent cell proliferation resulting from the modulation of signaling pathways (Ge et al. 2022). However, the precise signaling mechanism of selenoplasia that underlies cancer formation has not yet been identified and it is the survival, particularly via acquiring resistance to ferroptosis.
17.7 BRF2 Selenoprotein: Cancer Cell
The UGA stop codon is responsible for encoding selenocysteines in the protein molecule. During translation, cellular processes depend on specific machinery to ensure the correct insertion of specific residues. Selenocysteine insertion sequence-binding protein 2 (SBP2) can recognize a specific stem-loop in the mRNA known as the selenocysteine insertion sequence (SECIS) element which allows the recruitment of eEFsec (selenocysteine-specific elongation factor. In addition, eEFsec is responsible for binding SeCystRNA which is an essential step in the process of ensuring that selenoproteins contain SeCys (Howard et al. 2007). It is important to note that the reduced level of GPx4 is the result of the depletion of SBP2 or eEFSec. Some reports demonstrate that SBP2 knockout increases ROS production and leads to ferroptosis and apoptosis (Arimbasseri and Maraia 2016; Jehan et al. 2022). Ultimately, the researcher suggests that the SeCystRNA transcription is a response to control redox reaction. More specifically, RNA polymerase III is a core transcription factor composed of three subunits (TATA-binding protein) TBP, Bdp1, and Brf2). Researchers highlighted a direct link between Brf2 and the overexpression of many cancers including breast and lung cancer. SeCystRNA contributes to the regulation of the selenoprotein level which is the main transcription factor of Brf2 (Jenkins and Gouge 2021). The researchers believe that Brf2 plays the role of a master regulator in the oxidative stress response which is accomplished by regulating SeCystRNA levels in a redox-dependent manner (Fig. 17.3). Cancer cells suppress this regulation and prevent apoptosis by overexpressing Brf2. Notably, this phenomenon occurs even in the presence of activated Nrf2 (Gouge and Vannini 2018; Rashidieh et al. 2021).
17.8 Protein Synthesis
Ferroptosis is related to deregulated redox metabolism. Lee and coworkers were initially interested in examining the potential role of glucose starvation in promoting ferroptosis since it reduces ATP generation and increases the production of reactive oxygen species (ROS). However, they made an unexpected discovery that glucose starvation along with other energy stress-inducing conditions prevents cells from ferroptosis (Lee et al. 2020). AMP-activated protein kinase (AMPK) response to energy stress depletes ATP and temporarily slows protein synthesis. Also, AMPK along with one of its major downstream targets, rapamycin (mTOR) regulates the rate of cell proliferation. mTOR is a serine/threonine protein kinase that regulates two distinct protein complexes called mTOR complex 1 (mTORC1) and 2 (mTORC2) by acting as a catalytic component. mTORC1 plays an important role in cancer cell growth and cell division. In addition, activation of p70S6 kinase (p70S6K) and 4E-binding protein (4E-BP) increases protein synthesis and effectively involves de novo lipid synthesis. It is well known that human eukaryotic initiation factor 4G (eIF4G) is a binding site on eIF4E which is essential for translation initiation (Rehman et al. 2014). However, when 4E-BP is phosphorylated by mTOR, it can no longer bind with eIF4E. Multiple types of stress inhibit mTORC1 activity resulting in phosphorylated 4E-BPs that block the translation initiation factor eIF4E. In contrast, mTORC1 inhibition results in only a 60% decrease in the total translation of proteins. The remaining 40% of protein translation may not necessarily be occurring via the cap-independent mechanism. Over the past decade, researchers have believed that cap-dependent translation is normally mediated by the protein eIF4F. In recent years, researchers have postulated that cap-dependent translation is typically mediated by the eIF4F protein. Surprisingly, the discovery of eIF3d as a new player in cap-dependent mRNA translation has expanded our understanding of the regulation of protein synthesis. The requirement of conserved stem-loop structures in the 5’ UTR of mRNAs for eIF3d-mediated translation raises questions about the mechanism of recognition and how these structures block canonical eIF4F binding. The specific role of eIF3d in glucose deprivation-mediated translation raises questions about the presence of other types of switches for stimulus-specific translation reprogramming (Lamper et al. 2020; Jia and Qian 2021). These questions highlight the need for further research to fully understand the complex regulation of protein synthesis and the role of eIF3d in this process. Similarly, the interactions of eIF3 with selenoprotein mRNAs may provide a means to steer mRNAs into specific translation pathways. Further studies will be necessary to determine if eIF3 regulates the translation of selenoprotein mRNAs and how it contributes to the establishment of the selenoprotein expression hierarchy in stressful conditions (Hayek et al. 2022). If potent and selective AMPK inhibitors are developed and used in clinical settings, combining them with other specific inhibitory eIF3d chemotherapy agents may help to kill cancer through the ferroptosis pathway.
The production of ribosomes is a complex process that involves the assembly of ribosomal subunits, the transcription of ribosomal RNA, and the folding and modification of ribosomal proteins. These events are tightly regulated to ensure proper ribosome biogenesis and optimal protein synthesis, making ribosomes a critical component of cell biology (Harish and Caetano-Anollés 2012). Malinouski et al. suggest that deficiency in levels of certain ribosomal proteins may affect the ribosome structure and result in an increased insertion of Selenocysteine(SeSec). This could also happen if the knockdown of these proteins decreases the rate of protein synthesis, which supports the slow SeSec insertion process. The control of selenoprotein expression appears to be regulated globally through ribosome structure and function, but more research is needed to understand the specific mechanism. The same group suggests that the knockdown of ribosomal structural protein ribosomal protein L14 (RPL14) has been shown to increase selenium levels and increase the expression of the genes TrxR1 and selenoprotein S (Seleno S). In addition to that some other proteins were deficient in the expression of a potassium voltage-gated channel subfamily A member 1 (KCNA1). KCNA1 deficiency has been found to also decrease the expression of several selenoproteins, with molecular weights under 25 kilodaltons (kDa). This study found that several genes have an impact on selenium levels in HeLa cells by altering selenoprotein expression. These findings suggest that RPL14 may play a role in regulating selenium metabolism and the expression of related genes (Malinouski et al. 2014).
The recent study suggests that regulating the metabolism of selenoaminoacids in pancreatic cancer cell lines can be achieved through the interaction of diaphanous-related formin 3(DIAPH3) with key proteins involved in selenoaminoacid metabolism. It has been previously reported that ribosomal proteins, including RPL6, play a crucial role in maintaining selenium and selenoprotein levels, and a deficiency in these proteins can affect the ribosome structure, leading to increased Sec insertion and selenoprotein expression. The interaction between DIAPH3 and RPL6 may alter the ribosome structure, leading to increased expression of the selenoprotein TrxR1. It is also possible that this interaction decreases the rate of protein synthesis, indirectly supporting slow Sec insertion and increased TrxR1 expression. TrxR1 may be a potential target for inhibiting the progression of pancreatic cancer (Rong et al. 2021).
17.9 Selenoprotein’s Regulation of Multiple Transcription Factors
Selenoproteins are involved in a variety of molecular signaling pathways and may be closely linked to several complex cellular network signaling pathways. The cellular role of selenoproteins in the regulation of many networks at the transcriptional level was to protect antioxidant pathways. Many transcription factors play a significant role in regulating selenoprotein mRNA transcription. According to the report by Stoytcheva and Berry, the common potential promoter of the selenoprotein family is 21 transcription factors (Stoytcheva and Berry 2009). Other transcription factor binding sites for selenoprotein gene subsets were discovered. Experimentally, most of the transcriptional factors are responsible for the different selenoprotein gene expressions. MITF is one of the most important transcriptional regulators of melanocytes and melanoma cancer. Recently, another group published research related to TrxR1 (selenoprotein) and its crucial regulatory role in the redox control and stability of the transcriptional factor MITF (Kline et al. 2022). Some recent studies suggest that the metabolism of seleno-aminoacids is through the interaction of ferroptosis. Indisputably, in the future, studies will reveal the important role of selenoprotein in the ferroptosis pathway as well as in drug-resistant cancer cells to adapt to oxidative stress conditions at the transcription level.
17.10 Conclusion
The incorporation of selenium into the body occurs through the selenocysteine (SeSec) biosynthesis pathway, which is crucial for the production of several selenoproteins, including glutathione peroxidases and thioredoxin reductases. Selenoproteins regulate several intracellular biological processes including ferroptosis, drug resistance, protein quality control, and lipid production. In addition, selenium uptake during protein synthesis also guides many epigenetic processes. Selenoproteins act as a strategic moderator of cancer redox homeostasis in ferroptosis. Cancer cells are highly susceptible to chemodrugs and another therapeutic regimen which induce the production of free radicals. Interestingly, this susceptibility of cancer cells to free radicals is associated with selenoproteins. This suggests that deficiency of selenium through diet would aid in improving the efficacy of various anticancer therapies.
References
Abdalla MA, Mühling KH (2023) Selenium exerts an intriguing alteration of primary and secondary plant metabolites: advances, challenges, and prospects. CRC Crit Rev Plant Sci 42:34. https://doi.org/10.1080/07352689.2022.2158270
Abdurahman A, Li Y, Jia S-Z et al (2023) Knockdown of the SELENOK gene induces ferroptosis in cervical cancer cells. Metallomics 15:mfad019. https://doi.org/10.1093/mtomcs/mfad019
Arimbasseri AG, Maraia RJ (2016) RNA polymerase III advances: structural and tRNA functional views. Trends Biochem Sci 41:546
Brigelius-Flohé R (2008) Review) Selenium Compounds and Selenoproteins in Cancer. Chemistry and Biodiversity 5(3):389–395. https://doi.org/10.1002/cbdv.200890039
Cai LL, Ruberto RA, Ryan MJ et al (2020) Modulation of ferroptosis sensitivity by TXNRD1 in pancreatic cancer cells. bioRxiv. https://doi.org/10.1101/2020.06.25.165647
Cheff DM, Huang C, Scholzen KC, Gencheva R, Ronzetti MH, Cheng Q, Hall MD, Arnér ES (2023) The ferroptosis inducing compounds RSL3 and ML162 are not direct inhibitors of GPX4 but of TXNRD1. Redox Biol 62:102703
Dixon SJ, Lemberg KM, Lamprecht MR et al (2012) Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149:1060. https://doi.org/10.1016/j.cell.2012.03.042
Dolma S, Lessnick SL, Hahn WC, Stockwell BR (2003) Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell 3:285. https://doi.org/10.1016/S1535-6108(03)00050-3
Eagle K, Jiang Y, Shi X et al (2022) An oncogenic enhancer encodes selective selenium dependency in AML. Cell Stem Cell 29:386. https://doi.org/10.1016/j.stem.2022.01.003
Gardner GL, Moradi F, Moffatt C et al (2022) Rapid nutrient depletion to below the physiological range by cancer cells cultured in Plasmax. Am J Physiol Cell Physiol 323:C823. https://doi.org/10.1152/ajpcell.00403.2021
Ge EJ, Bush AI, Casini A et al (2022) Connecting copper and cancer: from transition metal signalling to metalloplasia. Nat Rev Cancer 22:102
Ghelichkhani F, Gonzalez FA, Kapitonova MA et al (2022) Selenoprotein S: A versatile disordered protein. Arch Biochem Biophys 731:109427
Gouge J, Vannini A (2018) New tricks for an old dog: Brf2-dependent RNA polymerase III transcription in oxidative stress and cancer. Transcription 9:61
Harish A, Caetano-Anollés G (2012) Ribosomal history reveals origins of modern protein synthesis. PLoS One 7:e32776. https://doi.org/10.1371/journal.pone.0032776
Hayek H, Eriani G, Allmang C (2022) eIF3 interacts with Selenoprotein mRNAs. Biomol Ther 12:1268. https://doi.org/10.3390/biom12091268
Hill KS, McDowell A, McCorkle JR et al (2021) Keap1 is required for artesunate anticancer activity in non-small-cell lung cancer. Cancers (Basel) 13:1885. https://doi.org/10.3390/cancers13081885
Hou W, Xu H (2022) Incorporating selenium into heterocycles and natural products-from chemical properties to pharmacological activities. J Med Chem 65:4436
Howard MT, Moyle MW, Aggarwal G et al (2007) A recoding element that stimulates decoding of UGA codons by Sec tRNA [Ser]Sec. RNA 13:912. https://doi.org/10.1261/rna.473907
Ingold I, Berndt C, Schmitt S et al (2018) Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 172:409. https://doi.org/10.1016/j.cell.2017.11.048
Jankowski CS, Rabinowitz J (2022) A selenium-iron axis dictates cancer cell sensitivity to pharmacologic ascorbate. FASEB J 36. https://doi.org/10.1096/fasebj.2022.36.s1.r2913
Jehan C, Cartier D, Bucharles C et al (2022) Emerging roles of ER-resident selenoproteins in brain physiology and physiopathology. Redox Biol 55:102412
Jenkins T, Gouge J (2021) Nrf2 in cancer, detoxifying enzymes and cell death programs. Antioxidants 10:1030
Jeter JM, Bowles TL, Curiel-Lewandrowski C et al (2019) Chemoprevention agents for melanoma: a path forward into phase 3 clinical trials. Cancer 125:18
Jia L, Qian SB (2021) A versatile eIF3d in translational control of stress adaptation. Mol Cell 81:10. https://doi.org/10.1016/j.molcel.2020.12.016
Kalimuthu K, Keerthana CK, Mohan M et al (2022) The emerging role of selenium metabolic pathways in cancer: new therapeutic targets for cancer. J Cell Biochem 123:532
Karlenius TC (2011) Regulation of the Thioredoxin system under hypoxia and different oxygen conditions. Griffith University
Kayrouz CM, Huang J, Hauser N, Seyedsayamdost MR (2022) Biosynthesis of selenium-containing small molecules in diverse microorganisms. Nature 610:199. https://doi.org/10.1038/s41586-022-05174-2
Kline CD, Anderson M, Bassett JW et al (2022) MITF is regulated by redox signals controlled by the Selenoprotein Thioredoxin reductase 1. Cancers (Basel) 14:5011. https://doi.org/10.3390/cancers14205011
Lamper AM, Fleming RH, Ladd KM, Lee ASY (2020) A phosphorylation-regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science 1979:370. https://doi.org/10.1126/science.abb0993
Lee YC, Tang YC, Chen YH et al (2003) Selenite-induced survival of HuH7 hepatoma cells involves activation of focal adhesion kinase-phosphatidylinositol 3-kinase-Akt pathway and Rac1. J Biol Chem 278:39615. https://doi.org/10.1074/jbc.M304095200
Lee JH, Park KJ, Jang JK et al (2015) Selenoprotein S-dependent selenoprotein K binding to p97(VCP) protein is essential for endoplasmic reticulum-associated degradation. J Biol Chem 290:29941. https://doi.org/10.1074/jbc.M115.680215
Lee YS, Kalimuthu K, Seok Park Y et al (2020) Ferroptotic agent-induced endoplasmic reticulum stress response plays a pivotal role in the autophagic process outcome. J Cell Physiol 235:6767. https://doi.org/10.1002/jcp.29571
Lei P, Bai T, Sun Y (2019) Mechanisms of ferroptosis and relations with regulated cell death: a review. Front Physiol 10:139. https://doi.org/10.3389/fphys.2019.00139
Li Z, Ferguson L, Deol KK et al (2022) Ribosome stalling during selenoprotein translation exposes a ferroptosis vulnerability. Nat Chem Biol 18:751. https://doi.org/10.1038/s41589-022-01033-3
Malinouski M, Hasan NM, Zhang Y et al (2014) Genome-wide RNAi ionomics screen reveals new genes and regulation of human trace element metabolism. Nat Commun 5:3301. https://doi.org/10.1038/ncomms4301
Marciel MP, Hoffmann PR (2019) Molecular mechanisms by which Selenoprotein K regulates immunity and cancer. Biological Trace Element Research 192:60–68. https://doi.org/10.1007/S12011-019-01774-8
Moncan M, Mnich K, Blomme A et al (2021) Regulation of lipid metabolism by the unfolded protein response. J Cell Mol Med 25:1359
Park JM, Kim DH, Na HK, Surh YJ (2018) Methylseleninic acid induces NAD(P)H: Quinone oxidoreductase-1 expression through activation of NF-E2-related factor 2 in Chang liver cells. Oncotarget 9:3014. https://doi.org/10.18632/oncotarget.10289
Rashidieh B, Molakarimi M, Mohseni A et al (2021) Targeting brf2 in cancer using repurposed drugs. Cancers (Basel) 13:3778. https://doi.org/10.3390/cancers13153778
Rehman G, Shehzad A, Khan AL, Hamayun M (2014) Role of AMP-activated protein kinase in cancer therapy. Arch Pharm (Weinheim) 347:457–468
Rong Y, Gao J, Kuang T et al (2021) DIAPH3 promotes pancreatic cancer progression by activating selenoprotein TrxR1-mediated antioxidant effects. J Cell Mol Med 25:2163. https://doi.org/10.1111/jcmm.16196
Seibt TM, Proneth B, Conrad M (2019) Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic Biol Med 133:144
Shahdadfar A, Frønsdal K, Haug T et al (2005) In vitro expansion of human mesenchymal stem cells: choice of serum is a determinant of cell proliferation, differentiation, gene expression, and transcriptome stability. Stem Cells 23:1357. https://doi.org/10.1634/stemcells.2005-0094
Shchedrina VA, Everley RA, Zhang Y et al (2011) Selenoprotein K binds multiprotein complexes and is involved in the regulation of endoplasmic reticulum homeostasis. J Biol Chem 286:42937. https://doi.org/10.1074/jbc.M111.310920
Stoytcheva ZR, Berry MJ (2009) Transcriptional regulation of mammalian selenoprotein expression. Biochim Biophys Acta Gen Subj 1790:1429
Sun X, Ou Z, Chen R et al (2016) Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 63:173. https://doi.org/10.1002/hep.28251
Turanov AA, Kehr S, Marino SM et al (2010) Mammalian thioredoxin reductase 1: roles in redox homoeostasis and characterization of cellular targets. Biochem J 430:285. https://doi.org/10.1042/BJ20091378
Varlamova EG, Goltyaev MV, Turovsky EA (2022) The role of Selenoproteins SELENOM and SELENOT in the regulation of apoptosis, ER stress, and calcium homeostasis in the A-172 human glioblastoma cell line. Biology (Basel) 11:811. https://doi.org/10.3390/biology11060811
von Krusenstiern AN, Robson RN, Qian N et al (2023) Identification of essential sites of lipid peroxidation in ferroptosis. Nat Chem Biol 19:719. https://doi.org/10.1038/s41589-022-01249-3
Voorde Vande J, Ackermann T, Pfetzer N et al (2019) Improving the metabolic fidelity of cancer models with a physiological cell culture medium. Sci Adv 5:eaau7314. https://doi.org/10.1126/sciadv.aau7314
Wu W, Li D, Feng X et al (2021) A pan-cancer study of selenoprotein genes as promising targets for cancer therapy. BMC Med Genet 14:78. https://doi.org/10.1186/s12920-021-00930-1
Yagoda N, Von Rechenberg M, Zaganjor E et al (2007) RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 447:865. https://doi.org/10.1038/nature05859
Yang WS, Stockwell BR (2008) Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol 15:234. https://doi.org/10.1016/j.chembiol.2008.02.010
Acknowledgments
Dr. Kalimuthu Kalishwaralal, MK Bhan Young Researcher Fellowship for 2020–2021 (Ref: No, HRD-12/4/2020-AFS-DBT) awarded by DBT, India. We thank Dr. Ramakrishnan Muthuswamy, Associate Professor, Nanjing Forestry University, China, for the help rendered in image generation using Bio Render software.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Kalishwaralal, K. et al. (2023). Selenium Metabolic Pathway in Ferroptotic Cell Death. In: Tang, D. (eds) Ferroptosis in Health and Disease. Springer, Cham. https://doi.org/10.1007/978-3-031-39171-2_17
Download citation
DOI: https://doi.org/10.1007/978-3-031-39171-2_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-39170-5
Online ISBN: 978-3-031-39171-2
eBook Packages: MedicineMedicine (R0)