Abstract
Nucleic acid-based therapies are emerging as a promising approach for treating a wide range of diseases, including cancer, genetic disorders, and infectious diseases. However, the delivery of nucleic acids to target cells remains a significant challenge due to their large size, negative charge, and susceptibility to degradation. Biomaterials have the potential to overcome these barriers and enable efficient and safe delivery of nucleic acids. Engineering biomaterials for nucleic acid-based therapies involves the design and optimization of materials and formulations that can protect nucleic acids from degradation, deliver them to desired target cells, facilitate cellular uptake, and promote endosomal escape. This book chapter is focused on the engineering of such materials from a chemical, formulation, and manufacturing point of view. The chapter begins by providing an introduction of nucleic acid therapies and the challenges associated with delivering them to the correct target cells. The chapter then explores various biomaterials that have been developed for nucleic acid delivery with a particular focus on lipids and polymers. The properties of these biomaterials are described, and their advantages and limitations are discussed. Next, the chapter delves into the engineering approaches used to modify their physical and chemical properties to enhance efficacy and specificity. The chapter also covers the manufacturing, characterization, and evaluation of these biomaterials for nucleic acid delivery, including physiochemical characterization, such as sizing, surface charge, and pKa evaluation. In vitro and in vivo experimental set-ups to assess biocompatibility, stability, and gene transfection efficiency are also discussed. The chapter also emphasizes the importance of regulatory compliance and safety considerations in the development of clinical-grade biomaterials. Finally, the chapter concludes by highlighting the main nucleic acid therapeutics that have been already marketed and that are in development with a particular focus on siRNA, mRNA, and CRISPR applications.
Graphical Abstract

Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction to Nucleic Acid-based Therapies
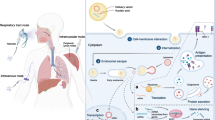
Gene therapy involves the delivery of therapeutic nucleic acids (DNA or RNA) into a patient’s cells allowing to (i) increase target gene expression, (ii) silence the expression of the disease-causing gene, or (iii) modify a target gene by inserting, removing or entirely replacing it. Historically gene therapy refers to a gene transfer by introducing a DNA molecule into the cells but over the years it has expanded to delivery of other types of nucleic acids with different mechanisms of gene regulation, e.g. RNA interference or gene editing. Nucleic acid drugs allow specific control of gene and protein expression and are on the path to becoming a major platform in drug development alongside small molecules and other biologics. Those currently used for therapy, include plasmid DNA (pDNA), short interfering RNAs (siRNA), antisense oligonucleotides (ASOs), and messenger RNA (mRNA), as well as gene editing tools. Transient silencing of the desired gene can be achieved by neutralizing its mRNA transcript using siRNA or ASOs while the delivery of mRNA or pDNA allows producing any therapeutic protein inside the target cells. The mechanisms of action, regulation of gene and protein expression of RNA are summarized in Fig. 1. As of 2022, there were 16 drug products utilizing therapeutic delivery of RNAs (9 ASO, 5 siRNA, 2 mRNA) and 27 products exploiting viral gene delivery or the use of genetically modified cells, such as chimeric antigen receptor (CAR-T) cells, that are also considered a form of gene therapy.
All types of nucleic acids share physicochemical properties which impede their uptake across biological membranes and lead to their rapid clearance from circulation, including high molecular weight (pDNA > 1000 kDa, mRNA 300–5,000 kDa, siRNA ~14 kDa, and ASOs 4–10 kDa), the hydrophilic nature, and the negative charge. These molecules are also prone to enzymatic degradation by endo and exonucleases (e.g. ribonucleases or deoxyribonucleases) that cleave the phosphodiester bond or catalyse the removal of nucleotides from the polynucleotide chain resulting in their relatively poor half-life in the circulation (most stable: DNA > ASO > siRNA > mRNA). To exert their biological function most RNA-based drugs need to reach the cytoplasm of the cell while ASO due to their small size can penetrate through both cellular and nuclear membranes allowing them to interact with both cytoplasmic and nuclear targets. pDNA requires entry to the nucleus which creates an additional delivery barrier for this type of therapy. At the cellular level, cellular uptake and escape from the endosomal compartment are essential for their therapeutic activity and remain one of the main challenges for nucleic acid delivery. However, before even reaching their intracellular destination, nucleic acid drugs have to avoid non-specific tissue distribution and reach the diseased cells, escape renal clearance, and survive the hostile extracellular environment after systemic or local administration.
Overcoming these delivery barriers is critical to facilitate the clinical translation of nucleic acid-based therapies and requires the development of effective and safe delivery systems. The first class of vectors adopted to deliver nucleic acids was viruses since they are naturally able to transfect their genetic material (DNA or RNA) into cells. Despite a number of products based on viral vectors having been approved for clinical use, this class of carriers is facing challenges related to complex manufacturing, immunogenicity, and pre-existing immunity towards disease-causing viruses (e.g. adenovirus) which may limit their ability for repeated dosing. For some types of viral vectors potential for integration into the genome (e.g. lentivirus), and constraints on the size of DNA payload (e.g. adeno-associated virus) can also narrow the scope of possible applications. For this reason, nucleic-acid delivery approaches based on non-viral vectors have been developed which include the use of nanoparticles composed of organic and inorganic materials, extracellular vesicles, and nucleic-acid conjugates, which have been discussed in detail elsewhere [1, 2]. In general, non-viral delivery vectors are easy to scale up and have the capacity to address many limitations of viral vectors, mainly concerning manufacturing and safety. A range of biocompatible materials have been designed to complex, encapsulate and deliver therapeutic nucleic acids and among them, the most widely adopted are lipids and polymers which will be the main focus of this chapter.
2 Design of Biomaterials for Nucleic Acid Delivery
Biomaterials used in the formulation of nanocarriers for nucleic acid delivery must meet a series of important characteristics to protect and safely deliver the cargo to the correct site [3]. First, they must allow loading of nucleic acids into a delivery system capable of protecting them from the nucleases present in the environment and in the body. Preferentially, the biomaterials should be able to form nanometric colloids (i.e. nanoparticles) that can be safely injected intravenously and effectively enter the cell membranes. The nanoparticles should be stable both in simple (e.g. phosphate-buffered saline (PBS), tris(hydroxymethyl)aminomethane (Tris) buffer) and complex fluids (e.g. blood) to avoid aggregation-related toxicity and they should possess narrow size distribution to assure uniform biodistribution in the body. Moreover, the biomaterial should be able to induce the escape of the nanoparticles from the intracellular compartments such as endosomes and then, it should allow the release of the cargo once reached the target site (e.g. either the cytoplasm or the nucleus). Ideally, the nanocarrier should degrade into safe and biocompatible components to avoid accumulation and toxicity. It is also of primary importance that the nanoparticles are masked from the immune system to allow the re-dosing of the therapeutic and avoid allergic reactions. Moreover, they should be able to mainly target the desired type of cells in the desired organs to avoid off-target toxicity. In the end, the biomaterial must also be easy to produce, to characterize, and to scale-up to avoid reproducibility problems and complex quality control procedures that can affect the availability of the final product on the market (i.e. the so-called “drug shortages”). All these characteristics are difficult to obtain with only one type of biomaterial and, for this reason, the nanoparticles used in gene delivery often consist of multiple components.
In general, the priori design of nucleic acid delivery systems that possess tissue specificity, high delivery efficacy, and safety is challenging due to the many requirements and the complexity of the nanoparticle-biological environment interactions. For this reason, there is not a “one-size-fits-all” solution for every therapeutic application and optimization of the formulation composition and/or of the physiochemical characteristics of the single formulation components is generally always required. Therefore, it is crucial to identify and optimize the parameters that have the highest impact on the different NP characteristics relevant to their performance, such as NP size and surface composition. This is usually achieved by the synthesis of a chemically diverse library of biomaterials and by the optimization of the final formulation, e.g. the composition of each single component. In the contest of such multidimensional optimization problems, the use of advanced mathematical tools, such as design of experiment (DOE) and machine learning, can accelerate the discovery and selection of the lead candidate [4].
2.1 Lipids
Lipid nanoparticles represent the most advanced class of nucleic-acid delivery carriers as evidenced by the recent clinical approvals, including the first siRNA drug Onpattro in 2018 for the treatment of hereditary transthyretin-mediated amyloidosis (hATTR amyloidosis) and two mRNA-based COVID-19 vaccines (BNT 162b and mRNA-1273) in 2020/21. The lipid nanoparticles (LNPs) generally consist of four different components: a cationic or ionizable lipid, a helper lipid, cholesterol, and a PEGylated lipid (Fig. 2).
(a) Lipid-based structures can include micelles, which consist of a lipid monolayer, or liposomes, which consist of a bilayer. Lipid nanoparticles are composed of multiple lipid layers as well as microdomains of lipid and nucleic acid. (b) LNPs often consist of cholesterol, a helper lipid, a PEG-lipid, and (c) a cationic or ionizable lipid. (d) The molar ratios of the four components making up the FDA-approved Acuitas/BioNTech/Pfizer COVID vaccine (BNT 162b) and Patisiran (Onpattro). (Adapted from Paunovska et al. [3])
The cationic or ionizable lipid represents one of the most important components of the formulation and it is responsible for the loading of the cargo into the NPs via ionic complexation between the positively charged polar head of the lipid and the negatively charged phosphate groups present in the backbone of the nucleic acids. In contrast, the two alkyl tails of the lipid are responsible for holding together the nanoparticle via hydrophobic effect and to interact and fuse with biological membrane bilayers to promote cellular uptake and endosomal escape. The nature of the positively charged head group and, in particular, the acid dissociation constant (pKa) of the amine groups plays a vital role in the endosomal escape, efficacy, and toxicity of the lipid nanoparticles. Lipid heads that contain chemical groups with pKa > 7.4, such as primary, or quaternary ammonium groups, confer a permanent cationic nature to these ionizable lipids in relevant physiological conditions. For this reason, although this class of cationic lipids can achieve high cargo loading efficiency, their permanent cationic nature can induce potential toxicity via perturbation of cellular membranes and hampers the release of the cargo from the nanoparticles by a strong electrostatic binding. For this reason, ionizable lipids with pKa around 5.5–6.5 have been found to be the most effective thanks to their ability to load nucleic acids during formulation at low pH (usually around pH = 3.5–5), to avoid toxicity at physiological conditions (pH = 7.4) due to their neutral charge, and then to induce endosomal escape when the nanoparticles reach the more acidic endosomal environment (pH = 5–6.5). Another important characteristic of these ionizable lipids is the presence of biodegradable chemical groups, such as esters or amides, that can be hydrolysed in the biological milieu by different enzymes (e.g. lipases or esterases) into shorter and safer compounds to avoid accumulation into the body and the associated toxicity.
While cholesterol is used to modulate the membrane fluidity and stability, the helper lipids are generally neutrally charged lipids that are primarily used to modulate the physiochemical properties of the lipid nanoparticles through the alteration of the geometry of the lipid layer, in particular its curvature and planarity. Helper lipids can be classified according to the cross-sectional area of the lipid head group and the lipid tail as cylindrical, cone, and inverse cone shapes [5].
In the end, PEGylated lipids are lipids that contain hydrophilic polyethylene glycol as hydrophilic head and are used to provide a hydrophilic surface coat able to increase colloidal stability and reduce the interaction of the LNPs with serum proteins. This hydrophilic shell is responsible for extending the LNPs circulation half-life into the blood and to avoid the clearance from the mononuclear phagocytic system. However, there is increasing evidence that PEG can induce the formation of anti-PEG antibodies. These antibodies can lower the efficacy of subsequent administered doses of the LNPs in the so-called accelerated blood clearance effect or, in the worst case scenario, they can cause allergic reactions [6]. For this reason, researchers are focused on finding PEG alternatives [7].
The nature and relative amount of all these components affect also the physiochemical properties of the LNPs, such as size, charge, and surface properties. In particular, the composition of the LNP surface plays an important role in their ability to deliver the cargo to specific sites. Once the LNPs are in contact with complex biological fluids, different biomolecules start to adsorb on their surface generating a protein coat that alters how the nanoparticles interact with the biological environment, i.e. the immune systems and cells in different tissues. This so-called protein corona is ultimately responsible for the trafficking of the LNPs into different cells and organs via the so-called “passive targeting”. As an example, the adsorption of the apolipoprotein E (ApoE) on the surface of LNPs is responsible for their delivery to hepatocytes in the liver. Similarly, the biological milieu can affect the composition of the protein corona and therefore change the LNPs tropism. As an example, higher concentration or absence of specific biomolecules caused by the presence of a disease can alter the biodistribution of nanoparticles by modifying the endogenous trafficking pathways. Another common strategy (the so-called “active targeting”) to improve the on-target capability is to introduce on the LNP surface ligands (e.g. antibodies) that can specifically bind to biomolecules or receptors overexpressed on the surface of specific cells.
2.2 Polymers
Polymers adopted for gene delivery applications share many traits of the ionizable lipids used in the formulation of lipid nanoparticles: (i) the presence of ionizable amine groups to promote nucleic acid complexation and to improve endosomal escape; (ii) the presence of hydrophobic groups to increase the NP colloidal stability; and (iii) the presence of hydrolysable groups to increase biodegradability and therefore reducing toxicity. In general, biomaterial hydrophobicity, pKa, and biodegradability are among the most important design parameters to obtain effective and safe gene delivery systems [8]. The biggest difference between these two classes of biomaterials relies on the fact that while ionizable lipids generally consist of a single molecule, polymers are composed of a repetition of one or more different types of subunits. This represents a double-edged sword that increases the versatility and chemical space of these biomaterials, but at the same time increases the complexity and may hamper the reproducibility of these systems. The multimeric nature of these materials allows to introduce additional parameters that can be used to optimize the nucleic acid delivery performance. Namely, the molecular weight of the polymer (i.e. the overall number of monomeric units in the polymer chain) and the charge density (i.e. the composition and distribution of different monomeric units in the polymer chain) are two important parameters that can be balanced to obtain high nucleic acid loading efficiency, nanoparticles stability, efficient endosomal escape, and reduced cytotoxicity. In a similar manner, the topological structure of the polymer (such as linear, branched, and brush-like structures) can be also tuned to obtain more stable and efficient carriers. As an example, dendrimers are orderly branched polymers with a spherical structure that are proven effective in delivering mRNA, even though they are difficult to synthesize. Since polymers consist of a distribution of different chemical species with different monomer composition, sequence, and length, the manufacturing process plays a fundamental role to control the main properties and to assure batch-to-batch reproducibility (see Sect. 3.2). As an example, it would be important to obtain polymer with narrow molecular weight distribution or, in other words, with a dispersity (Đ) close to 1. Dispersity is defined as the ratio between the polymer weight-average molecular weight (Mw) and number-average molecular weight (Mn) and it is a measure of how heterogenous is a distribution of molecules in terms of molecular weight (i.e. length in case of polymers). A polymer with a Đ = 1 is monodisperse and contains only polymer chains of the same length. In contrast, a polymer with a Đ > 1 is dispersed and contains polymer chains of different lengths.
The history of polymers resembles the history of cationic/ionizable lipids, i.e. from the use of permanently charged species, then moving to the use of ionizable amine groups, and, in the end, including biodegradable moieties to boost efficacy and safety [9]. In fact, the first generation of polymers consists of permanently charged polycations that are structurally composed of one or more type of monomer, such as polyamino acids (e.g. poly-L-lysine and poly-L-ornithine). In the second generation, ionizable amine groups were included, such as in the case of linear and branched polyethyleneimine (PEI). PEI and its derivatives are polymers with abundant amino groups with different pKa and, for this reason, they provide both high loading efficiency and strong endosomal escape. However, as in the case of the first generation, these polymers have limited biodegradability, and, for this reason, they have also poor biocompatibility and relatively high cytotoxicity.
In the latest generation, labile chemical linkages are introduced in the main backbone of the polymers to increase biodegradability. Among them, the most adopted linkages are ester bonds due to their ability to degrade in physiological conditions via hydrolysis and esterases. Example of these polymers include poly-(β-amino-esters) (PBAEs), poly(amine-co-esters) PACEs, amino-polyesters (APEs), and charge-altering releasable transporters (CARTs).
3 Examples of Polymers for Nucleic Acid Delivery
Polymers are highly valued in biology and medicine due to their versatility and unique properties resulting from their chain molecular structure. They have been widely used for the development of delivery systems for a variety of therapeutic payloads, including small molecules, proteins, and nucleic acids. Biodegradable polymers can enhance the biocompatibility of the delivery system via degradation into natural and non-toxic metabolites. Both synthetic and naturally derived biodegradable polymers hold the potential for the development of safer nucleic acid therapeutics.
3.1 Natural and Synthetic Polymers
Natural and nature-derived polymers are composed of building blocks that occur in nature and can often be extracted from natural sources, e.g. plants, shrimp shells, etc. Nature-derived polymers used for nucleic acid delivery are often composed of amino acids (polypeptides, e.g. poly-l-lysine (PLL)) or sugar molecules (polysaccharides, e.g. chitosan). These polymers are usually biodegradable. Chitosan and PLL polymers have the ability to complex nucleic acids via electrostatic interaction with their cationic amine groups, however, the design constrained to natural building blocks can contribute to their low transfection efficiency and cytotoxicity, while relatively high dispersity may complicate manufacturing these polymers.
Synthetic polymers are built from fully synthetic monomers which often do not exist in nature. These polymers are generally used to expand the design features and improve on properties of natural polymers. With the ability to select from variety of synthetic monomers they can be tailored to meet desired properties such as mechanical strength, self-assembly, degradability, stimuli responsiveness, dispersity, and drug cargo release but the use of synthetic building blocks may pose challenges with biocompatibility.
3.2 Approaches for the Synthesis of Cationic Polyesters
Polyesters are synthetic polymers widely used in drug delivery systems due to several favourable traits such as relatively low toxicity, biodegradability, biocompatibility, and most importantly they can be obtained using Generally Recognized as Safe (GRAS) molecules. Examples of clinically approved polyesters include polylactide (PLA), polyglycolide (PGA), polycaprolactone (PCL), and polylactide-co-glycolide (PLGA). However, the amine-containing polyesters are the main class of cationic polyesters that have been developed for nucleic acid delivery and the majority are synthesized using one of these three strategies: (1) poly-condensation of diol and dicarboxylic acid derivatives, (2) Michael addition of diacrylates and diamines, or (3) ring opening polymerization (ROP) of cyclic monomers (e.g. lactones) (Fig. 3).
-
1.
Poly-condensation is a step-growth polymerization which involves the polymerization of diol and dicarboxylic acid or diester-containing monomers through the elimination of a small molecule, such as water or alcohol, to form a long-chain polymer. PACEs are cationic polyesters with low charge density usually synthesized by polycondensation reaction via enzymatic copolymerization of hydrophobic diesters and amine-containing diol monomers [10]. This process is usually carried out under vacuum with controlled temperature. Drawbacks of this method include relatively low molecular weight of the synthesized polymer with broad molecular weight distribution (Đ: 1.5–2.5) resulting in the poor control over polymerization process. This method also precludes the use of monomers containing primary or secondary amines from the composition of the resulting polymer. This is because primary and secondary amines can interfere in the formation of ester bonds during the condensation process owing to the formation of amide bonds which is favoured over ester bonds.
-
2.
Cationic polyesters can also be synthesized by Michael addition which is also a step-growth polymerization and involves the combination of a diamine with a diacrylate such as bis-acrylates or bis-methacrylates. This is a simple and one-step polymerization method which provides access to large chemical space of monomers compatible with both primary and secondary amines. This method has been used to synthesize PBAEs which endow materials with tunable properties including degradation, mechanics, hydrophilicity, and swelling [11]. One of the major drawbacks of this method is the lack of control over the polymerization process (polyaddition mechanism) especially for branched materials, resulting in a broad molecular weight distribution.
-
3.
ROP is a versatile method for synthesizing polyesters with well-defined molecular weight and low dispersity by opening the ring of a cyclic monomer followed by transesterification reaction between monomer and a polymer chain with a reactive hydroxyl group. These reactions are often catalysed by organic or metal-based catalysts. APEs are cationic polymers synthesized via controlled ROP of lactones with tertiary amino-alcohols, offering a one-step process with the ability to control the degree of polymerization [12]. Similar to poly-condensation, this method also precludes the use of monomers containing primary or secondary amines. Charge-altering releasable transporters (CARTs) is another class of cationic polymers used for nucleic acid delivery that can be synthesized via ROP of cyclic carbonates with secondary amines and, moreover, they can be designed to be pH-sensitive for controlled release at specific pH conditions. This method allows the use of monomers containing secondary amines but requires additional steps for amine protection and deprotection. Limitations of ROP include sensitivity to contamination with water and solvents containing hydroxyl group, limited selection of catalysis and monomers, and potential risk of racemization.
In addition to the described polymerization strategies, synthesis of cationic polymers in nucleic acid delivery can also be achieved using more advanced methods such as reversible addition fragmentation chain transfer (RAFT) polymerization and atom transfer radical polymerization (ATRP). These methods have several benefits including capacity for continued chain growth, the ability to control the polymerization process while maintaining a low dispersity and high end-group fidelity, ease of functionalization, and the ability to create a diverse range of structures by utilizing a broad range of monomers and initiators [13]. However, in addition to the requirement for rigorous oxygen exclusion the challenge with these techniques is that the carbon backbones of the prepared polymers are not degradable which limits their use for biomedical applications.
4 Manufacturing of Carriers for Nucleic Acid Delivery
4.1 The Importance of Good Manufacturing Practice (GMP)
The manufacturing of gene delivery carriers typically involves several steps such as synthesis, purification, and characterization of delivery system components (e.g. biomaterial and nucleic acids), nanoparticles formulation, quality control, sterilization, packaging, and storage. The exact process may vary depending on the type of delivery system and its intended use. All the components of the delivery systems for clinical use must be produced according to Good Manufacturing Practice (GMP) quality standards and maintained in sterile condition essential to ensure the safety and efficacy of the therapy. GMP describes the production standards that must be met to minimize risks, waste, and production losses and to ensure that medicines are manufactured and quality controlled according to set standards. All drugs intended for the European Union (EU) market must comply with GMP requirements that ensure that they meet the standards for market authorization.
4.2 Example of Manufacturing Process for mRNA Therapeutics
Production of cationic polymers and lipids for nucleic acid delivery is a key aspect of the manufacturing process. These materials should be relatively easy to synthesize at large scale (e.g. grams to kilograms), purify, and characterize, and should meet GMP standards for commercial production. Compared to cationic polymers, synthesis of lipids often requires multiple steps that can compromise product yield, but their advantage is a defined chemical structure and molecular weight, making them easier to characterize and control quality of the final product. Lipid nanoparticles are being used to manufacture siRNA-based drug Onpattro against hereditary thransthyrein amyloidosis and mRNA-based COVID-19 vaccines (Comirnaty, Spikevax) which represent the first examples of RNA-based therapeutics.
Manufacturing process of mRNA drugs involves several steps, including (1) pDNA manufacturing, (2) mRNA synthesis and purification, and (3) mRNA-LNP formulation and purification (Fig. 4).
-
1.
pDNA template production involves cloning a gene of interest into a plasmid vector, growing the plasmid-containing bacteria, lysing the cells to release the pDNA, purification to remove impurities and contaminants, conducting quality control, formulating, and lyophilizing the pDNA. It is a complex process done under GMP guidelines.
-
2.
mRNA molecules are synthesized using a pDNA template through an enzymatic reaction known as in vitro transcription (IVT). Purification of the synthesized mRNA usually involves tangential flow filtration (TFF) followed by sterile filtration. High-performance liquid chromatography (HPLC), anion exchangers, cellulose purification, and hydrogen bonding are other systems often used for purification of mRNA products.
-
3.
The final step in the manufacturing process involves formulation of mRNA into lipid nanoparticles. This process usually involves the rapid mixing of mRNA/buffer and lipid solutions to create spontaneous self-assembly of encapsulated mRNA-LNPs formulation. The next step is diafiltration/concentration TFF which eliminates the residual solvent and concentrates the formulation, followed by sterile filtration to ensure that it is free of any contaminants before it is packaged and released for use.
4.3 Formulation Methods for Nanoparticle Manufacturing
Adequate control over the chemistry of cationic biomaterial and formulation parameters such as nucleic acid/polycations ratio, nanocarrier composition (e.g. helper lipids, PEG-lipid, cholesterol, surfactants (e.g. Pluronic)), and formulation method are critical for ensuring a high degree of reproducibility and consistent performance of nucleic acid delivery system. Optimization of the formulation requires changing the ratio of nucleic acid to ionizable lipid/polymer, modifying excipients compositions, and the appropriate selection of solvents and buffers for each of the components. To facilitate this, a reproducible method of mixing lipophilic (e.g. lipids, polymer) and hydrophilic (nucleic acid) components to control self-assembly into the uniform particles driven by electrostatic and hydrophobic interactions is necessary to ensure batch-to-batch consistency. This process should also be scalable from millilitres to hundreds of litres, to minimize the need for process re-development during translation to the clinic.
Different methods have been used for the formulation of nucleic acid complexes with cationic polymers or lipids, which include low energy methods taking advantage of simple self-assembling of nanoparticle components facilitated by mixing, vortexing or microfluidics. Ethanol injection method is one of the most frequently used techniques to produce nanocarriers by rapidly diluting ethanolic solution of lipids/polymers into an aqueous medium containing nucleic acid. In contrast, high energy methods may require additional energy input in the form of heat (e.g. electrostatic spray-drying) or pressure (e.g. dry lipid film hydration followed by extrusion or sonication to control the size of nanoparticles) to form uniform nanoparticles. Simple mixing and vortexing methods are generally used in the lab for small-scale production. However, ethanol injection method coupled with microfluidics are widely used in industry and for pre-clinical animal studies. Microfluidics is a scalable, robust, and highly reproducible formulation technique. It involves mixing of the aqueous phase containing nucleic acid and organic phase containing lipids/polymers via a micromixer (Fig. 5). Well-defined LNPs with tunable properties (e.g. different LNP size) can be produced by controlling the operating parameters which include continuous flow, diffusion distance, flow rate ratio, controlled mixing time, and temperature control, resulting in high reproducibility. The microfluidics technique allows for rapid optimization of LNP manufacturing conditions, reproducibility, and ease of scaling up, provide the exceptional contribution to the LNP-based nucleic acid delivery technology and their applicability of translation from laboratory to practical applications.
5 Analytical Methods to Control Quality of Nucleic-Acid Delivery Systems
5.1 Characterization of Physicochemical Properties
Physicochemical properties of the nucleic acid nanocarriers, such as particle size, size distribution, and surface charge, are commonly characterized using techniques such as dynamic light scattering (DLS), transmission electron microscope (TEM), nanoparticle tracking analysis (NTA), and electrophoretic light scattering (ELS). DLS measures the hydrodynamic diameter and size distribution of the nanocarriers (in the size range of 10–1000 nm), while particle tracking analyses the movement of individual particles in solution to provide information about their size, concentration, and size distribution. The particle surface charge is evaluated via ELS and it is represented by zeta potential which is a measure of the difference in potential between the bulk fluid (in which a particle is dispersed) and the layer of fluid containing the oppositely charged ions (associated with the nanoparticle surface). The electrostatic or charge attraction/repulsion between nanoparticles is one of the main parameters known to affect nanoparticle colloidal stability. Polydispersity index (PDI) reflects size distribution and PDI values between 0.05 and 0.2 indicate monodisperse particle population. Aggregation of particles leads to increase in their size and can indicate poor stability. Particle morphology can be investigated using cryogenic electron microscopy (cryo-TEM) or atomic force microscopy (AFM). Cryo-TEM captures the images of samples embedded into a thin layer of non-crystalline ice to provide the information about the nanoparticle internal structure. Monitoring those physicochemical properties is important since particles larger (>500 nm) or highly charged (> +/−20 mV) tend to be quickly cleared from circulation due to the uptake by mononuclear phagocyte system (MPS). Both concentration and nucleic acid encapsulation can be measured using nucleic acid-binding fluorescent dyes (e.g. Ribo/PicoGreen) or liquid chromatography methods (e.g. ion-pair reversed-phase chromatography (IP-RP) or size exclusion chromatography (SEC)) and are critical to determine the therapeutic dose. Nucleic acid encapsulation refers to entrapment of the nucleic acid cargo into nanoparticles and is measured after disruption of the particles, usually using a detergent (e.g. Triton X-100) or an organic solvent. Monitoring physicochemical properties allows to assess the quality of nucleic acid delivery system and ensure batch-to-batch reproducibility.
5.2 Measurement of the Acid Dissociation Constant (pKa)
pKa of polymer/lipid ionizable head groups affects the surface charge and ionization behaviour of the nanocarrier which can influence nucleic acid encapsulation, delivery efficacy and nanocarrier bio-distribution. pKa indicates the pH at which 50% of the headgroups exist in ionized (positively charged) state. The apparent pKa value is linked to the nanocarrier composition and is strongly influenced by noncovalent interactions such as dielectric constant, ionic strength, π–π stacking, hydrophobic interactions, and nature of neighbouring charges. The apparent pKa of the nanocarriers is relatively lower as compared to the calculated pKa value of individual ionizable biomaterial. Ideally, the apparent pKa value of the nanocarrier should align with the pH of the endosomal compartment to facilitate effective escape of nucleic acid cargo. The acid-base titration (or potentiometric titration method), fluorescent 2-(p-toluidino)-6-naphthalene sulfonic acid (TNS) and z-potential methods can be used for the measurement of the apparent pKa of nanocarriers (Fig. 6). In the acid-base titration method, titration of the nanocarriers (in an acidic solution) is carried out using NaOH or KOH solution to determine the pH in the middle of the two equivalence points. TNS displays strong fluorescence after binding with cationic headgroups and the pKa of the nanocarrier is determined as the pH at the half maximum value of the fluorescence. The pKa of the nanocarrier can also be calculated by measuring the zeta potential as a function of pH.
5.3 Evaluation of the Performance of Nucleic Acid Nanocarriers
To facilitate effective nucleic acid delivery, the nanocarrier must be (1) taken up by the cells, (2) escape from the endocytic compartment, and (3) release the therapeutic cargo into the cytoplasm allowing gene expression or gene silencing. The uptake is usually evaluated by fluorescence microscopy or flow cytometry which provides insight into how effectively the nanocarrier is able to cross the cell membrane and enter the cell. Nucleic acid conjugated with fluorophore (e.g. Cy5, Alexa Fluor 488) or nanocarrier labelled using lipophilic dyes (e.g. DiI, DiD) allow tracking the delivery and the fate of nanocarrier components. Flow cytometry is a quantitative method that can measure the fluorescence signal in the single cell and allows separation of different cell populations based on their size and internal complexity (e.g. granularity) rendering it particularly suitable for analysis of complex tissue samples. Intensity of the fluorescence signal reflects the extent of nanocarrier uptake providing a high-throughput and quantitative analysis but compared to fluorescence microscopy this method lacks intracellular resolution. Monitoring of the endosomal release of the nucleic acid requires the use of confocal microscopy that allows to image narrow optical section inside the cell (e.g. 0.5 um thick) combined with immuno-fluorescent staining for marker of endosomal rupture (e.g. Galectin 9) or staining of acidic compartments within a cell (e.g. LysoTracker).
Efficacy of mRNA/DNA delivery can be measured by analysing expression of a reporter gene in the transfected cell such as green fluorescent protein or firefly luciferase or desired therapeutic gene. Efficacy of gene silencing with siRNA is usually assessed by a real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) and Western Blot to determine the levels of mRNA transcript and protein knockdown in the transfected cells. Evaluation of the nucleic acid delivery in the cell culture models that lack the complexity of the whole organism offers a limited indication of the in vivo performance therefore pre-clinical efficacy evaluation in animal models such as rodents and non-human primates is needed prior to clinical trials involving human subjects. In vivo testing can also provide information about safety of the therapeutic nucleic acid delivery by assessing various parameters including organ function, body weight loss, and markers of inflammatory response.
6 Therapeutic Applications of Nucleic Acid Delivery
Whole genome sequencing enabled identifying the genetic roots of many diseases bringing us closer to developing personalized treatments based on precise control of gene and protein expression with nucleic acid-based drugs. This therapeutic strategy holds the promise to address a wide range of diseases, including genetic disorders, cancer, cardiovascular, and infectious diseases. Currently approved drug products that utilize non-viral vectors focus predominantly on systemic and local delivery of therapeutic RNA molecules (siRNA, mRNA). Lipids-based nanocarriers are presently the most clinically advanced platform for RNA delivery but the available drug products (Onpattro, Spikevax, Comirnaty) are limited to targeting cells in easily accessible tissues such as liver hepatocytes after systemic administration or muscle and dendritic cells after intramuscular administration. Polymers show the potential to facilitate RNA delivery to the lungs and gastrointestinal tract tissues, however, no products have been approved to date. Encouraging data on PBAE-mediated delivery of mRNA to lungs via inhalation of nebulized nanocarriers as well to gastrointestinal tissue via oral administration have been reported in large animals (non-human primates and pigs) [14, 15]. To fully unlock the therapeutic potential of nucleic acid base therapies, it is therefore critical to develop efficient delivery systems suitable for administration via parenteral, oral, and inhalation routes.
6.1 Short Interfering RNAs (siRNA)
Short interfering RNAs are double-stranded RNAs, 21–23 base pairs in length, that can selectively bind and degrade complementary mRNAs leading to transient silencing of protein expression. siRNAs are loaded onto the RNA-inducing silencing complex (RISC) that facilitates the cleavage of target mRNA. This conserved mechanism is known as RNA interference and its discovery was recognized with a Nobel Prize in physiology and medicine in 2006. After almost two decades of research, siRNA-based therapies represent one of the most clinically advanced platforms for RNA drugs. Onpattro was the first siRNA drug to reach the market in 2018 and was directed against the dysfunctional transthyretin gene underlying a rare genetic disease hereditary transthyretin-amyloidosis causing the buildup of amyloid in tissues and organs. Onpattro contains siRNA which silence the expression of TTR mRNA and its corresponding protein formulated into lipid nanoparticles (size range 60–100 nm) composed of a blend of four lipid excipients: DLin-MC3-DMA; PEG2000-C-DMG; DSPC; and cholesterol. These lipids protect siRNA from degradation by endo- and exo-nucleases in the circulatory system and facilitate delivery to the liver hepatocytes. Presence of ionizable lipid DLin-MC3-DMA is important for particle formation, endosomal release of the siRNA and coating of the LNP by apolipoprotein E which facilitates binding to the low-density lipoprotein receptor on hepatocytes. Following the success of Onpattro several siRNA drugs have been approved targeting liver and cardiovascular diseases, including Givlaari (acute hepatic porphyria), Oxlumo (primary hyperoxaluria type 1), Amvuttra (hATTR) and Leqvio (primary hypercholesterolaemia). Latest siRNA drugs shifted from using LNPs to N-acetylgalactosamine (GalNAc) conjugated siRNA that show high affinity binding to the asialoglycoprotein receptor (ASGR) expressed on hepatocytes. Due to their small size and good stability, siRNA conjugates allow subcutaneous administration and require less frequent dosing that improves patient compliance. However, future applications outside the liver will require the discovery of suitable cellular receptors and new types of biomaterials to design targeted nanoparticles and conjugates.
6.2 Messenger RNA
mRNA delivery allows to transiently express the desired therapeutic protein, including secreted, intracellular and transmembrane proteins, inside the host cells. Advantages of mRNA over pDNA include rapid and transient protein production allowing control of the therapeutic dose, no risk of insertional mutagenesis, and potentially greater efficacy with non-viral delivery by virtue of mRNA cytoplasmic activity. Uridine-rich mRNA sequences have been identified as a key activator of toll-like receptors and cytosolic pattern recognition receptors such as retinoic acid-inducible gene I (RIG-I) hampering the therapeutic use of mRNA. Discoveries related to nucleoside modification lead to replacing uridine with modified nucleosides (e.g. pseudouridine) which has proven effective in immune evasion without reducing mRNA translation. In addition, extensive purification of double-strand RNA (dsRNA) fragments during mRNA manufacturing, e.g. using HPLC leads to further reduction of the immunogenicity of mRNA-based drugs and improves protein production. mRNA-based drugs have a wide range of therapeutic applications, which include prophylactic and therapeutic vaccines, protein-replacement therapy aimed at rare genetic diseases, and gene editing. Naked (unformulated) mRNA has been rarely used therapeutically owing to its large size and susceptibility to degradation therefore nanoparticle-formulated mRNA has been the main method of choice for mRNA delivery. The LNPs in mRNA COVID-19 vaccines consist of four main components: a neutral phospholipid (DSPC), cholesterol, a polyethylene-glycol (PEG)-lipid, and a new generation of ionizable cationic lipid (ALC-0315 or SM-102). mRNA vaccines developed against SARS-Cov2 (Comirnaty and Spikevax) are the first mRNA-based drugs on the market that have been administered to over 5 billion people globally since 2021. The vaccine is administered by intramuscular injection and requires 2 doses 3–4 weeks apart. mRNA LNPs are captured by antigen-presenting cells at the injection site and are transported to a draining lymph node, while mRNA is translated into virus spike protein which is presented to immune cells (T-cells) and activates both cellular and humoral responses. The success of mRNA vaccines established it as a safe and effective therapeutic modality and has laid the foundation for the development of a new class of mRNA-based prophylactic and therapeutic vaccines for various indications, including cancer and infectious diseases.
6.3 CRISPR
Clustered regularly interspaced short palindromic repeats (CRISPR) technology is a simple yet powerful tool for editing genomes. It allows to alter DNA sequences and modify gene function and could potentially be used to treat or prevent diseases such as muscular dystrophy, sickle cell anaemia, transthyretin amyloidosis and familial hypercholesterolemia. CRISPR–Cas systems rely on Watson–Crick base-pairing between a single guide RNA (sgRNA) and a corresponding genomic DNA target site for binding of endonuclease CRISPR-associated protein 9 (Cas9) and cleavage of the target sequence, to introduce a double-stranded break (DSB) into a DNA molecule. DSBs can be repaired by the cells using non-homologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ results in stochastic insertions and deletions (“indels”) causing permanent gene knockout, whereas HDR occurs in the presence of a DNA template containing homology to regions flanking the DSB site, leading to the incorporation of desired changes encoded in the repair template into the genome. Cas9 of Streptococcus pyogenes (SpCas9) is the enzyme that is most commonly used for genome editing and genetic manipulation using CRISPR–Cas, but a growing collection of engineered RNA-guided enzymes (e.g. base and prime editing) is expanding the genome-manipulation toolbox. Therapeutic use of CRISPR-based tools focuses on genome editing leading to gene knock-out or base editing resulting in the correction of a single nucleotide mutation. The large size of Cas9 (>160 kDa) and the need for a short exposure to limit non-specific editing events pose a significant delivery challenge. Most clinically advanced approaches of in vivo gene editing utilize the co-delivery of mRNA encoding Cas9 and sgRNA formulated into liver-targeting lipid nanoparticles.
References
Elsharkasy OM et al (2020) Extracellular vesicles as drug delivery systems: why and how? Adv Drug Deliv Rev 159:332–343
Kulkarni JA et al (2021) The current landscape of nucleic acid therapeutics. Nat Nanotechnol 16:630–643
Paunovska K, Loughrey D, Dahlman JE (2022) Drug delivery systems for RNA therapeutics. Nat Rev Genet 23:265–280
Narayanan H et al (2021) Design of biopharmaceutical formulations accelerated by machine learning. Mol Pharm 18:3843–3853
Kulkarni JA, Cullis PR, van der Meel R (2018) Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther 28:146–157
Sellaturay P, Nasser S, Islam S, Gurugama P, Ewan PW (2021) Polyethylene glycol (PEG) is a cause of anaphylaxis to the Pfizer/BioNTech mRNA COVID-19 vaccine. Clin Exp Allergy 51:861–863
Hoang Thi TT et al (2020) The importance of poly(ethylene glycol) alternatives for overcoming peg immunogenicity in drug delivery and bioconjugation. Polymers (Basel) 12
Kim HJ, Kim A, Miyata K (2022) Synthetic molecule libraries for nucleic acid delivery: design parameters in cationic/ionizable lipids and polymers. Drug Metab Pharmacokinet 42:100428
van den Berg AIS, Yun CO, Schiffelers RM, Hennink WE (2021) Polymeric delivery systems for nucleic acid therapeutics: approaching the clinic. J Control Release 331:121–141
Kauffman AC et al (2018) Tunability of biodegradable poly(amine- co-ester) polymers for customized nucleic acid delivery and other biomedical applications. Biomacromolecules 19:3861–3873
Liu Y, Li Y, Keskin D, Shi L (2019) Poly(beta-Amino Esters): synthesis, formulations, and their biomedical applications. Adv Healthc Mater 8:e1801359
Kowalski PS et al (2018) Ionizable amino-polyesters synthesized via ring opening polymerization of tertiary amino-alcohols for tissue selective mRNA delivery. Adv Mater:e1801151
Perrier SB (2017) 50th anniversary perspective: RAFT polymerization – a user guide. Macromolecules 50:7433–7447
Rotolo L et al (2022) Species-agnostic polymeric formulations for inhalable messenger RNA delivery to the lung. Nat Mater
Abramson A et al (2022) Oral mRNA delivery using capsule-mediated gastrointestinal tissue injections. Matter
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Quiz
Quiz
-
Question 1: Which of the following nucleic acids require entry into the nucleus to exert their therapeutic function?
-
(a)
ASOs
-
(b)
siRNA
-
(c)
mRNA
-
(d)
pDNA
-
(a)
-
Correct Answer(s): (d) – Transcription machinery needed to express exogenous DNA is present in the nucleus therefore a nuclear entry is necessary
-
Question 2: What are the main challenges for nucleic acid delivery at the cellular level?
-
(a)
Renal clearance
-
(b)
Susceptibility to endo and exonucleases
-
(c)
Cellular uptake
-
(d)
Escape from the endosomal compartment
-
(a)
-
Correct Answer(s): (c) and (d) – Renal clearance and susceptibility to endo and exonucleases describe challenges for systemic delivery
-
Question 3: What is the purpose of PEGylated lipids in the formulation of lipid nanoparticles?
-
(a)
To increase the interaction of the LNPs with serum proteins
-
(b)
To decrease the circulation half-life of the LNPs
-
(c)
To reduce the clearance from the mononuclear phagocytic system (MPS)
-
(d)
To prevent the accelerated blood clearance after repeated dosing
-
(a)
-
Correct Answer(s): (c) – PEG makes nanoparticle surface more hydrophilic, masks the surface charge and decreases particle size which in turn makes them less recognizable by the MPS
-
Question 4: The key role(s) of ionizable/cationic lipid in the lipid nanoparticle (LNP) RNA delivery system, include:
-
(a)
Supporting the lipid bilayer
-
(b)
Promoting RNA encapsulation
-
(c)
Facilitating endosomal escape
-
(d)
Preventing LNP binding to serum proteins
-
(a)
-
Correct Answer(s): (b) and (c) – Positive charge of ionizable/cationic lipid helps attract negatively charged RNA and promote interactions with negatively charged lipids in the endosomal membrane
-
Question 5: What is dispersity in the context of polymers?
-
(a)
The ratio between the polymer weight-average molecular weight (Mw) and number-average molecular weight (Mn)
-
(b)
The number of monomeric units in the polymer chain
-
(c)
The overall charge density of the polymer
-
(d)
The hydrophobicity of the polymer
-
(a)
-
Correct Answer(s): (a) – dispersity is a measure of the heterogeneity of a polymer sample and is defined as the ratio of the weight-average molecular weight (Mw) to the number-average molecular weight (Mn)
-
Question 6: Which of the following is true about natural polymers used for nucleic acid delivery?
-
(a)
They are built from fully synthetic monomers
-
(b)
They can be extracted from natural sources
-
(c)
Poly-(β-amino-esters) is an example of natural polymer
-
(d)
They can be composed of sugar molecules
-
(a)
-
Correct Answer(s): (b) and (d) – other answers refer to synthetic polymers
-
Question 7: What is the major drawback of Michael Addition as a method for synthesizing cationic polyesters?
-
(a)
Limited control over polymerization process
-
(b)
Inability to use monomers containing primary or secondary amines
-
(c)
Sensitivity to temperature
-
(d)
Limited selection of catalysts and monomers
-
(a)
-
Correct Answer(s): (a) – The mechanism of Michael Addition polymerization makes it difficult to control the polymer molecular weight and reach high molecular weight
-
Question 8: Which of the following describes the purpose of Good Manufacturing Practice (GMP) in gene delivery carrier manufacturing?
-
(a)
To decrease production losses and waste
-
(b)
To maximize safety and efficacy of therapy
-
(c)
To maintain quality control standards
-
(d)
To ensure compliance with European Union market requirements
-
(a)
-
Correct Answer(s): (a), (b), (c), and (d) – GMP describes the production standards that must be met to minimize risks, waste, and production losses and to ensure that medicines are manufactured and quality controlled according to set standards
-
Question 9: Which of the following methods is used for the formulation of clinically approved RNA-based lipid nanoparticles?
-
(a)
Self-assembly facilitated by simple mixing or vortexing
-
(b)
Ethanol injection method coupled with microfluidics
-
(c)
Electrostatic spray-drying
-
(d)
Dry lipid film hydration followed by extrusion
-
(a)
-
Correct Answer(s): (b) – Ethanol injection method is often used to produce LNPs for clinical use, providing good scalability and control over particles self-assembly, size and polydispersity index.
-
Question 10: Which physicochemical property of the nanoparticle is reflected by the zeta potential?
-
(a)
The size of the nanoparticles
-
(b)
The concentration of nanoparticles
-
(c)
Surface charge
-
(d)
The nucleic acid encapsulation
-
(a)
-
Correct Answer(s): (c) – Zeta potential is the electric potential difference between a dispersed particle and the surrounding liquid and it is a measure of the electrical charge and stability of a colloidal system.
Rights and permissions
Copyright information
© 2023 American Association of Pharmaceutical Scientists
About this chapter
Cite this chapter
Kumar, P., Capasso Palmiero, U., Kowalski, P.S. (2023). Engineering Biomaterials for Nucleic Acid-Based Therapies. In: Domb, A., Mizrahi, B., Farah, S. (eds) Biomaterials and Biopolymers . AAPS Introductions in the Pharmaceutical Sciences, vol 7. Springer, Cham. https://doi.org/10.1007/978-3-031-36135-7_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-36135-7_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-36134-0
Online ISBN: 978-3-031-36135-7
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)