Abstract
An important component of human analysis of medical images and their context is the ability to relate newly seen things to related instances in our memory. In this paper we mimic this ability by using multi-modal retrieval augmentation and apply it to several tasks in chest X-ray analysis. By retrieving similar images and/or radiology reports we expand and regularize the case at hand with additional knowledge, while maintaining factual knowledge consistency. The method consists of two components. First, vision and language modalities are aligned using a pre-trained CLIP model. To enforce that the retrieval focus will be on detailed disease-related content instead of global visual appearance it is fine-tuned using disease class information. Subsequently, we construct a non-parametric retrieval index, which reaches state-of-the-art retrieval levels. We use this index in our downstream tasks to augment image representations through multi-head attention for disease classification and report retrieval. We show that retrieval augmentation gives considerable improvements on these tasks. Our downstream report retrieval even shows to be competitive with dedicated report generation methods, paving the path for this method in medical imaging.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
The promise of automated deep learning systems to assist radiologists is enormous. At the moment, important milestones, such as better consistency or even better performance have been achieved on an increasing number of use-cases [18, 37]. A source of inspiration in further improvement of these efforts is the way humans register and analyze images, which for deep learning has shown to be effective in the past [17, 37].
In any analysis, a doctor provides the memory and knowledge to place what is currently seen in the context of what has been seen before. In principle this can be compared to what implicitly happens at scale in any deep learning method. A doctor’s analysis is not implicit though. Their analysis process can be described and verified. We wonder whether (medical) deep learning methods could benefit from an explicit memory/knowledge infusion.
Making deep learning methods more explicit in terms of using past observations has already been studied in Natural Language Processing (NLP), in the form of retrieval augmentation [14, 21]. Supplementing data by retrieving relevant retrieved information can lead to performance gains [4]. This process can be thought of to work as both an enrichment and regularization process. A benefit of retrieval augmentation is that context from a trusted knowledge source is used as a supplement [13, 29]. The versatility of retrieval augmentation, which essentially provides a non-parametric memory expansion, is gaining traction in the multi-modal field [4, 28].
Multi-modal data modalities typically have different strengths leading to a strong and a weak data modality [37]. For instance, radiology reports generally contain richer and more complete information than X-rays, since the report is essentially a clinician’s annotation [24]. With retrieval augmentation information can be transferred explicitly from the strong to the weak modality.
A reason retrieval augmentation methods are not yet adopted for medical applications lies in the weakness of retrieval methods for the medical domain. Retrieval in the general domain is focused on global image regions [8, 16] whereas in medical images global features, such as body/organ structure are similar across patients. Meanwhile more fine-grained aspects are more discriminating as disease indicators, but are easily overlooked. The need for fine-grained results makes medical image retrieval magnitudes more complex.
We propose X-Ray Task Retrieval Augmentation (X-TRA), a framework for retrieval augmentation in a multi-modal medical setting, specifically designed for X-ray and radiology report analysis. To do so we introduce a cross-modal retrieval model and retrieval augmentation method. We make the following contributions.
-
We propose a CLIP-based multi-modal retrieval framework with a dedicated fine-tuning component for efficient content alignment of medical information which improves state-of-the-art results in multi- and single-modal retrieval on radiology images and reports.
-
We introduce a multi-modal retrieval augmentation component for disease classification and report retrieval pipelines.
-
We show that our method (1) reaches state-of-the-art performance both in multi-label disease classification and report retrieval. (2) Our report retrieval is competitive with dedicated report generation methodologies. (3) We show the cross-dataset versatility and the limitations of our method.
2 Related Work
Multi-modal Alignment. The introduction of Transformers for natural language processing (NLP) accelerated the development of integrated vision-language (VL) alignment models suitable for various VL-tasks, such as ViLBERT [19], LXMERT [30] and SimVLM [33]. These methods provide alignment on region to sentence- or word-level scale. The next step in multi-modal alignment was made by methods using contrastive learning combined with substantially larger datasets. Examples are CLIP [27] and ALIGN [10] which significantly outperform existing methods by using datasets for training consisting of 400M and 1.8B VL-pairs respectively. Domain-specific versions of CLIP, which is open-source, have been fine-tuned with additional data, such as PubMedCLIP [3].
Retrieval Augmentation. The origin of retrieval augmentation lies in the NLP field. It was created to fully utilise the power of large datasets. With retrieval augmentation we are not only dependent on a parametric model, but can also supplement data as a non-parametric component. Previous methods have shown the simple yet effective and versatility working of retrieval augmentation in a number of applications [5, 13, 29]. Retrieval in Medical Imaging. Up until recently the only retrieval methods in medical imaging were tailored hand-crafted methods [16]. With access to large datasets and pre-trained methods the balance shifted towards making automated retrieval methods [6, 26]. Especially in the histopathology and radiology domain major strides were made with retrieval methods [2, 8]. The use of text to improve image retrieval has been adopted for improving chest X-ray retrieval. Yu et al. [35] use CNN and word2vec features for multi-modal alignment and retrieval. Zhang et al. [36] approach this problem with a hash-based retrieval method.
Retrieval for Chest X-ray Analysis. Common tasks in chest X-ray analysis are disease classification and report generation [1, 11, 15]. Using retrieval for report generation has been a common approach. The approaches often entail the use of retrieved information as an input or template for a decoder which crafts a custom report [23, 32, 34]. Augmentation of chest X-ray tasks with synthetically generated diffusion-based images was shown to be possible [1], however the clinical use of non-genuine images can lead to complications and is not undisputed [37].
3 Methods
Our method is composed of two separate parts (Fig. 1). The first part is the alignment of the two modalities and construction of the retrieval model. The second part uses the output of the retriever as a non-parametric component in (cross-modal) retrieval augmentation to enhance the downstream tasks.
We consider a dataset \(\varTheta ^{N}_{\{\textbf{x},\textbf{r}\}}\) consisting of pairs containing an X-ray (\(\textbf{x}_i\)) and radiology report (\(\textbf{r}_i\)). To align these modalities we make use of the powerful CLIP vision-language aligner. Our objective is to minimize the distance between \(\textbf{x}\) and \(\textbf{r}\), to make cross-modal tasks possible. These aligned features will be used for retrieval augmentation to do multi-label classification and report retrieval as downstream tasks.
3.1 Stage I: Multi-modal Content Alignment
We leverage the pre-trained features from CLIP for initial feature representations. However, there is a domain shift between the natural image data CLIP is trained on and medical images we want to use in our method. Medical images can be visually very similar, while holding drastically different information. Small localized markers can be indicators for disease. In natural images global representations are more decisive and thus more suitable for unsupervised contrastive alignment. Alignment in CLIP goes as follows [27],
We need to overcome the obvious domain shift between medical images and the natural images on which CLIP is trained. Therefore, we require a more specific type of fine-tuning that is especially geared towards content-based extraction. We introduce the following loss, requiring a global class label for each dataset. With this fine-tuning step we are creating a supervised content-based alignment method with content classifier C:
This content based alignment loss should improve the alignment of detailed content-level details over the global visual appearance of the image.
Creating a Retrieval Index. At retrieval time we need to retrieve images that have a high similarity with query images. To efficiently do so we make use of Facebook AI Similarity Search (FAISS) [12]. This retrieval tool efficiently performs nearest-neighbour similarity search. After multi-modal alignment we encode our data to a FAISS index I conditioned on the training dataset. We can construct indices that only retrieve images (\(I^{\textbf{x}}\)), only reports (\(I^{\textbf{r}}\)), or both (\(I^{\textbf{x}\textbf{r}}\)).
Given a query \(\mathcal {Q}_s\) in source modality s, we can obtain its k neighbours of target modality t through:

this can be either \(\textbf{x}\), \(\textbf{r}\) or both. Once retrieval index I is trained based on the newly aligned training dataset we can consider the retriever as a non-parametric component which retrieves information from a fixed dataset in the subsequent retrieval augmentation steps. Note that during testing time, a query from the test set will be used to retrieve neighbours from the training set.
3.2 Stage II: Retrieval Augmentation
The purpose of retrieval augmentation is to effectively leverage similar representations to adopt a more informative representation of a given input, with our already trained retrieval index we retrieve similar representations.
To obtain a richer representation of \(\textbf{x}_i\), we retrieve intra-  and inter-modal neighbours
and inter-modal neighbours  from \(I^{\textbf{x}}\) and \(I^{\textbf{r}}\) respectively. To integrate the retrieved neighbouring samples, we can use various fusion methods [25]. The simplest one is concatenation:
from \(I^{\textbf{x}}\) and \(I^{\textbf{r}}\) respectively. To integrate the retrieved neighbouring samples, we can use various fusion methods [25]. The simplest one is concatenation:  . A more suitable method is multi-head attention (MHA) which is able to capture the long range dependencies between the original image and the retrieved information [31]:
. A more suitable method is multi-head attention (MHA) which is able to capture the long range dependencies between the original image and the retrieved information [31]:

3.3 Downstream Tasks
We are tackling two common tasks in chest X-ray analysis. These are multi-label disease classification and report retrieval. For this last task our objective is to show how well a retriever can perform on the report generation task. We measure performance by comparing task performance of \(\textbf{x}^{TRA}\) in comparison to \(\textbf{x}\).
A useful property of our retrieval index would be usability of an pre-trained model across datasets. Three clinically relevant scenarios for this are: From scratch training on the new dataset, frozen usage of the trained retrieval model and fine-tuning of the existing retrieval model with another image-report dataset.
3.4 Datasets
The primary dataset to which our method is applied is MIMIC-CXR (200k image-report pairs) [11]. Disease labels for each pair are extracted from the report through a rule-based extraction method [9]. To evaluate the versatility and cross-domain capabilities of our method, we use the small openI (4k image-report pairs) [20] and image-only CheXpert (200k images) [9] datasets. Official train-test splits are used.
3.5 Experimental Setup
As pre-processing step, the X-ray images are normalized and standardized by rescaling with center-cropping to scale \(256\times 256\), from which images of size \(224\times 224\) are sampled. The maximum number of tokens for representing radiology reports in the text encoder is set to 256. Three different VL models are used as encoders. At first a CNN-BERT model, composed of a DenseNet121 image encoder and a ClinicalBERT [7] text encoder. Given the strong performance of large vision-language models we also use CLIP (ViT-32 image encoder and text encoder) [27] and its medically fine-tuned equivalent PubMedCLIP [3]. This model is fine-tuned using the Radiology Objects in COntext (ROCO) dataset [22].
Multi-modal alignment is implemented as a single pass through a two-layer ReLu activated MLP, with dimension \(z_{enc}\), a dropout rate of 0.5, and layer normalization. \(z_{enc}\) is the output dimension of the encoder. We implement C as a three layer classifier head with dimensions \(\{z_{enc},256,14\}\). During retrieval we make use of \(k=10\) retrieved neighbours. To prevent overfitting, early stopping with a tolerance of 3 is applied to all training operations.
4 Results
4.1 Cross-Modal Retrieval
We are comparing the performance of our retrieval method against previous methods in Table 1 in terms of class-based mean average precision (mAP). Due to the powerful alignment of CLIP and tailor made fine-tuning we are outperforming all existing retrieval approaches for radiology images and/or reports by a large margin. The performance difference with similarly fine-tuned encoder-decoder combination DenseNet121 and ClinicalBERT further underwrites the power of CLIP in building a strong retrieval method, specifically on cross-domain retrieval. Interestingly, we observe that PubMedCLIP is not outperforming CLIP. This can be explained by a domain shift between MIMIC-CXR and ROCO, together with the ability of CLIP to generalize well out-of-domain [27]. In our downstream tasks image-based retrieval is most important, which is performing similar on inter- and intra-modal retrieval tasks.
4.2 Multi-label Disease Classification
Disease classification results in terms of AUC in Table 2 show that retrieval augmentation gives a clear improvement across different disease classes. It is interesting to see that we find a positive, albeit weak, correlation (R\(\approx \)0.60) between the increase in class AUC performance and retrieval mAP. Moreover, the performance gain from retrieval augmentation (\(0.80\rightarrow 0.85\)) is similar to additional training with synthetic diffusion-generated X-rays (\(0.80\rightarrow 0.84\)) [1]. The benefit of our method is that the supplemented information originates from the trusted dataset itself and is not synthetically generated.
4.3 Report Generation
In retrieval augmented report retrieval we show interesting performance on the report generation metrics compared to a selection of previous methods. While it should not be expected that simple retrieval outperforms dedicated report generation methods we are able to provide a result that can be considered competitive (Table 3). On the METEOR and ROUGE metric we are even outperforming most existing methods. The metrics reflect that the strength of report retrieval is in the global representation of the report. Our retriever is fine-tuned to retrieve samples with equivalent label spaces, hence good results on metrics that reward global similarity. An interesting outlook is the application of this method in a dedicated report generation framework which could boost performance further.
4.4 Cross-Dataset
By evaluating the cross-dataset scenarios (Table 4) with the CheXpert and openI datasets we can conclude that transferability to images from other domains is limited. However we do see that if retrieval augmentation is not useful, it can be ignored by the model and will not be detrimental for performance. The domain shift between different chest X-rays is a remaining problem [24]. Currently the most practical solution for this problem is the addition of a fine-tuning step.
Cross-domain results on open-I show that learning across modalities is possible with fine-tuning. When adding the openI dataset to the existing retrieval index, we can integrate the existing index with this new dataset. We can see that X-TRA benefits openI in this setting. In the updated retrieval index 23% of the retrieved information originates from openI and 77% from MIMIC-CXR.
4.5 Ablation Studies
We study the effect of the components in our retrieval augmentation method in Fig. 2. Specifically we look at the influence of each component in content- and CLIP based alignment. Interestingly, the composition of data modalities in retrieval augmentation does not have a big effect, since the retriever has similar results in inter- and intra-modal retrieval. In case randomly selected data is used instead of retrieved information, we achieve comparable results compared to our method without X-TRA. This is in accordance with cross-modal results, showing that if X-TRA supplemented information is not useful, it can be ignored. Using a partial retrieval index we can conclude that X-TRA can be useful with a small retrieval index, however performance reaches optimal levels when \(N>100k\).
4.6 Insight and Limitations
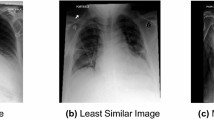
Qualitative results from our retrieval method for 2 different query images is shown in Fig. 3. We retrieve from the image index and report index. The retrieved images match well in terms of labels attributed to them, showing that our fine-tuning is preventing the retrieval of images that are only globally similar.
Fine-tuning of the entire CLIP model to domain-specific data is an interesting prospective. Potentially this can further improve the performance of our retrieval model. However, as we have shown in this paper regarding the performance of CLIP against PubMedCLIP, the loss of generalization can also be detrimental. In future studies this an promising avenue to explore.
5 Conclusion
In this work we present X-TRA, a simple yet effective method to improve multiple tasks on radiology images. Our method is composed of a content alignment and a retrieval augmentation step. With a new label-based alignment loss we are able to leverage pre-trained CLIP features to create a powerful cross-modal retrieval model. The general CLIP model appears to be more useful for our retrieval model than the slightly out-of-domain medically fine-tuned PubMedCLIP. We use this retrieval model to improve chest X-ray analysis through retrieval augmentation. With this we are adding an enrichment and regularization component that improves both multi-label disease classification and report retrieval by up to over 5%. On this last task we are even showing to be competitive with dedicated report retrieval methods. It opens up possibilities for retrieval augmentation as a generic tool in medical imaging.
References
Chambon, P., et al.: RoentGen: vision-language foundation model for chest x-ray generation. arXiv preprint arXiv:2211.12737 (2022)
Endo, M., Krishnan, R., Krishna, V., Ng, A.Y., Rajpurkar, P.: Retrieval-based chest x-ray report generation using a pre-trained contrastive language-image model. In: Machine Learning for Health, pp. 209–219. PMLR (2021)
Eslami, S., de Melo, G., Meinel, C.: Does CLIP benefit visual question answering in the medical domain as much as it does in the general domain? arXiv e-prints arXiv:2112.13906 (Dec 2021)
Gur, S., Neverova, N., Stauffer, C., Lim, S.N., Kiela, D., Reiter, A.: Cross-modal retrieval augmentation for multi-modal classification. In: Findings of EMNLP 2021, pp. 111–123 (2021)
Guu, K., Lee, K., Tung, Z., Pasupat, P., Chang, M.: Retrieval augmented language model pre-training. In: ICML, pp. 3929–3938 (2020)
Hu, B., Vasu, B., Hoogs, A.: X-MIR: explainable medical image retrieval. In: WACV, pp. 440–450 (2022)
Huang, K., Altosaar, J., Ranganath, R.: ClinicalBERT: modeling clinical notes and predicting hospital readmission. arXiv preprint arXiv:1904.05342 (2019)
Ionescu, B., et al.: Overview of the ImageCLEF 2022: multimedia retrieval in medical, social media and nature applications. In: CLEF, pp. 541–564 (2022)
Irvin, J., et al.: CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. In: AAAI, vol. 33, pp. 590–597 (2019)
Jia, C., et al.: Scaling up visual and vision-language representation learning with noisy text supervision. In: ICML, pp. 4904–4916 (2021)
Johnson, A.E., et al.: MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 6(1), 317 (2019)
Johnson, J., Douze, M., Jégou, H.: Billion-scale similarity search with GPUs. IEEE Trans. Big Data 7(3), 535–547 (2019)
Komeili, M., Shuster, K., Weston, J.: Internet-augmented dialogue generation. In: ACL, pp. 8460–8478 (2022)
Lewis, P., et al.: Retrieval-augmented generation for knowledge-intensive NLP tasks. NeurIPS 33, 9459–9474 (2020)
Li, J., Li, S., Hu, Y., Tao, H.: A self-guided framework for radiology report generation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 588–598 (2022)
Li, Z., Zhang, X., Müller, H., Zhang, S.: Large-scale retrieval for medical image analytics: a comprehensive review. Med. Image Anal. 43, 66–84 (2018)
Litjens, G., et al.: A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017)
Liu, X., et al.: A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit. Health 1(6), e271–e297 (2019)
Lu, J., Batra, D., Parikh, D., Lee, S.: ViLBERT: pretraining task-agnostic visiolinguistic representations for vision-and-language tasks. NeurIPS 32 (2019)
OpenI: Indiana University - chest x-rays (PNG images). https://openi.nlm.nih.gov/faq.php
Pasupat, P., Zhang, Y., Guu, K.: Controllable semantic parsing via retrieval augmentation. In: EMNLP, pp. 7683–7698 (2021)
Pelka, O., Koitka, S., Rückert, J., Nensa, F., Friedrich, C.M.: Radiology Objects in COntext (ROCO): a multimodal image dataset. In: Stoyanov, D., et al. (eds.) LABELS/CVII/STENT -2018. LNCS, vol. 11043, pp. 180–189. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01364-6_20
Pino, P., Parra, D., Besa, C., Lagos, C.: Clinically correct report generation from chest x-rays using templates. In: International Workshop on Machine Learning in Medical Imaging, pp. 654–663 (2021)
Pooch, E.H., Ballester, P.L., Barros, R.C.: Can we trust deep learning models diagnosis? The impact of domain shift in chest radiograph classification. arXiv preprint arXiv:1909.01940 (2019)
Priyasad, D., Fernando, T., Denman, S., Sridharan, S., Fookes, C.: Memory based fusion for multi-modal deep learning. Inf. Fusion 67, 136–146 (2021)
Qayyum, A., Anwar, S.M., Awais, M., Majid, M.: Medical image retrieval using deep convolutional neural network. Neurocomputing 266, 8–20 (2017)
Radford, A., et al.: Learning transferable visual models from natural language supervision. In: ICML, pp. 8748–8763 (2021)
Ramos, R., Martins, B., Elliott, D., Kementchedjhieva, Y.: SmallCap: lightweight image captioning prompted with retrieval augmentation. arXiv preprint arXiv:2209.15323 (2022)
Siriwardhana, S., Weerasekera, R., Wen, E., Kaluarachchi, T., Rana, R., Nanayakkara, S.: Improving the domain adaptation of retrieval augmented generation (RAG) models for open domain question answering. arXiv preprint arXiv:2210.02627 (2022)
Tan, H., Bansal, M.: LXMERT: learning cross-modality encoder representations from transformers. In: EMNLP, pp. 5100–5111 (2019)
Vaswani, A., et al.: Attention is all you need. NeurIPS 30 (2017)
Wang, J., Bhalerao, A., He, Y.: Cross-modal prototype driven network for radiology report generation. In: Avidan, S., Brostow, G., Cissé, M., Farinella, G.M., Hassner, T. (eds.) Computer Vision (ECCV 2022). LNCS, vol. 13695, pp. 563–579. Springer, Cham (2022). https://doi.org/10.1007/978-3-031-19833-5_33
Wang, Z., Yu, J., Yu, A.W., Dai, Z., Tsvetkov, Y., Cao, Y.: SimVLM: simple visual language model pretraining with weak supervision. In: ICLR (2021)
Yang, X., Ye, M., You, Q., Ma, F.: Writing by memorizing: hierarchical retrieval-based medical report generation. In: ACL, pp. 5000–5009 (2021)
Yu, Y., Hu, P., Lin, J., Krishnaswamy, P.: Multimodal multitask deep learning for x-ray image retrieval. In: de Bruijne, M., et al. (eds.) MICCAI 2021. LNCS, vol. 12905, pp. 603–613. Springer, Cham (2021). https://doi.org/10.1007/978-3-030-87240-3_58
Zhang, Y., Ou, W., Zhang, J., Deng, J.: Category supervised cross-modal hashing retrieval for chest x-ray and radiology reports. Comput. Electr. Eng. 98, 107673 (2022)
Zhou, S.K., et al.: A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc. IEEE 109(5), 820–838 (2021)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
van Sonsbeek, T., Worring, M. (2023). X-TRA: Improving Chest X-ray Tasks with Cross-Modal Retrieval Augmentation. In: Frangi, A., de Bruijne, M., Wassermann, D., Navab, N. (eds) Information Processing in Medical Imaging. IPMI 2023. Lecture Notes in Computer Science, vol 13939. Springer, Cham. https://doi.org/10.1007/978-3-031-34048-2_36
Download citation
DOI: https://doi.org/10.1007/978-3-031-34048-2_36
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-34047-5
Online ISBN: 978-3-031-34048-2
eBook Packages: Computer ScienceComputer Science (R0)