Abstract
In December 2019, a series of acute respiratory illnesses were first reported in central China. Investigations have led to the identification of a novel coronavirus (2019-nCoV), subsequently designated as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), as the causative agent of the so-called coronavirus disease 2019 (COVID-19). Since its emergence, SARS-CoV-2 has spread rapidly across the globe, resulting in the current ongoing COVID-19 pandemic, which has claimed the lives of millions of people throughout the world and continues to do so. Beginning with a brief overview of different historical and contemporary theories of infectious diseases, this chapter moves on to review the most recent literature on the origin, structure, pathogenesis, host immune responses, viral evasion of the host immunity, and mutated variants of SARS-CoV-2. In addition, patients’ clinical characteristics and risk factors, clinical trials, preventative measures, and the COVID-19 death toll among different countries are discussed. We also overview the utilization of various technologies in the battle against the pandemic, the impact of the pandemic on clinical research and trials, medical insurance, biomedical waste (BMW) generation and management, and the clinical lessons learned.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
In recent decades, coronaviruses (CoVs) have been responsible for major worldwide outbreaks, including the 2002 severe acute respiratory syndrome (SARS) and the 2012 Middle East respiratory syndrome (MERS) (Liu et al., 2020a). In December 2019, a strain of CoV containing a not-previously-known genome sequence was first detected in Wuhan City, Hubei Province of China, and was hence named the 2019-nCoV; however, it was later called SARS-CoV-2 (Dhama et al., 2020). Although SARS-CoV-2 is considered to be less pathogenic, compared to the previously known MERS-CoV or SARS-CoV, it is more transmissible and, as a result, has led to the current ongoing COVID-19 pandemic (Dhama et al., 2020). In the chapter to follow, we begin with some historical and epidemiological information to call attention to the past understandings and theories of infectious diseases. Further, we review the most recent literature on various human coronaviruses (hCoVs), including SARS-CoV-2 and its origin, structure, mechanisms of cell entry, host immune evasion, mutations, and the emerged variants, as well as the COVID-19 risk factors, signs, and symptoms, clinical trials, preventative measures (e.g., quarantine and social distancing), the death toll, and the role of innovative technologies (e.g., artificial intelligence, etc.) in controlling the pandemic.
Moreover, we discuss COVID-19 impact on biomedical wastes (BMWs) generation and management, medical insurance, and research, among others. We ultimately end the chapter by highlighting some of the clinical lessons learned from the pandemic. We have used several databases, such as PubMed, Google Scholar, ResearchGate, and Cochrane Library, to collect the relevant literature, using the search terms “SARS-CoV-2 pathogenesis,” “SARS-CoV-2 origin,” “SARS-CoV-2 variants,” “COVID-19 and genetic polymorphism,” as well as “COVID-19 risk factors” and “COVID-19 clinical symptoms,” “COVID-19 radiological findings,” “COVID-19 histopathological features,” and so forth. It is noteworthy to mention that COVID-19 and its causative virus are new and unknown phenomena, and numerous ongoing research are being conducted across the globe, which will contribute to the current understanding. As a result, this chapter is only a glimpse of the medical perspectives of COVID-19 and is not representative of all related published papers.
Historical Views and Theories of Infectious Diseases
Since the beginning of time, humans have always coexisted with diseases, which have sparked creative thinking in disease prevention and treatment (Tulchinsky & Varavikova, 2014a). Childhood fever has been the most serious issue afflicting humans, and in the last 10,000 years, mortality by the age of 15 has been about 50%, with fever being by far the most common cause of death, killing far more people than war and famine (Casanova & Abel, 2013). Throughout history, several theories have been postulated in order to explain the etiologies of infectious diseases. In order to explain phenomena such as disease outbreak that lacked a clear scientific or psychological explanation, people in pre-germ theory societies often invoked the presence of supernatural forces, energies, power, or spirits in order to explain phenomena such as disease outbreak that lacked a clear scientific or psychological explanation (Bastian et al., 2019). The primitive mystical or religious theories stated that diseases are either brought about by demonic spirits or delivered by the gods as a way of punishment for committed sins and thus must be subdued via exorcism (Karamanou et al., 2012). For instance, in Ancient Rome, Febris (fever) was the goddess who protected people against fever and malaria and had three temples located in Palatine Hill, Vicus Longus, and Sacra, and demonology was highly valued in ancient Persia (Karamanou et al., 2012). Using mystical or religious rituals, as well as herbal remedies, shamans or witch doctors sought to cure maladies (Tulchinsky & Varavikova, 2014a). The term “moral vitalism” is given by Bastian et al. (2019) to describe such beliefs in contaminating and contagious evil forces, and moral vitalists believed that individuals are susceptible to being possessed (infected) by evil spirits and that these forces are communicable (transmitted) among people (Bastian et al., 2019).
The pre-Socratic philosophers from the sixth century BC heralded the beginning of an era in science during which it was claimed that the environment had a significant impact on health and illness (Karamanou et al., 2012). The Hippocrates’ treatise Airs, Waters, Places by Hippocrates linked a variety of symptoms and diseases, including malaria, catarrh, and diarrhea to geographical and meteorological conditions, such as climate changes impact on stagnant water or marshy area, which eventually evolved further into the “miasma theory” of contagious disease (Karamanou et al., 2012). According to this theory, diseases are produced by the noxious vapor from decaying organic matter that contaminates the air, and in order to prevent such infectious diseases, sanitary measures such as clearing the streets of trash, sewage, animal corpses, and wastes were required (Tulchinsky & Varavikova, 2014a). Traditional Persian Medicine described flu-like respiratory pandemics using the terms, such as háwāy-e vábāī (polluted air) or “polluted wind,” from which numerous infectious diseases (e.g., smallpox, typhoid, plague, and respiratory illnesses), and their associated symptoms (i.e., fever, dyspnea, palpitations, and syncope), as well as high death rate, would result; thus, in order to control such pandemics, Persian Medicine recommended distancing oneself from the pandemic area and practicing self-quarantine (Iranzadasl et al., 2021). Avicenna, the Persian medicine scholar, acknowledged the contagiousness of tuberculosis (TB), the transmission of illnesses via water and soil, and the connection between psychology and health (Hajar, 2013), and in his book, The Canon of Medicine, he uses the word vábā (a term for cholera in modern medicine) to refer to the pandemic spread of any diseases in general (Iranzadasl et al., 2021). During the sixteenth century, the “contagious theory” of infectious diseases was postulated by Girolamo Fracastoro, which stated that illnesses are spread by contaminated fomites, such as clothing that have come into contact with the infected individual, or by transmissible chemicals (i.e., not living microorganisms) evaporating and diffusing through the atmosphere (i.e., seed-like entities, seminaria, or germs); each condition had its own distinct germ multiplying in host’s tissue producing disease by chemical putrefactive changes, and the transmission of certain diseases, such as syphilis and gonorrhea, was only possible through direct close contact, while others such as TB and smallpox are capable of traveling through air and be transmitted (Karamanou et al., 2012). Antonie van Leeuwenhoek’s invention of the microscope in 1676 enabled the first visual observation, description, and discovery of “little animals”—bacteria and protozoa (Tulchinsky & Varavikova, 2014a).
Prior to the clinical-pathological paradigm in Parisian hospitals during the French Revolution, physicians struggled to come up with precise descriptions and a suitable nosology for the various types of fever they encountered, and any early accounts failed to address the issue of whether fever and disease were intrinsic or extrinsic (Casanova & Abel, 2013). Later scientists such as Antoine Lavoisier, François Magendie, and Claude Bernard opposed vitalism theory by stating that laws of physics apply to all living organisms and that any alteration in physiology brings about the diseases, and that diseases including fever are intrinsic; however, the internal environment—“milieu intérieur”—of living organisms protect them against environmental changes (Casanova & Abel, 2013). Moreover, work by other scientists, such as Semmelweis and Lister, elucidated the mechanisms these microorganisms spread and further enhanced the science of public health and preventative medicine (Ryan, 2004); however, despite their excellent findings, they were unable to convince people that fevers were transmissible (Casanova & Abel, 2013).
The breakthrough came in around 1870 when the French scientist Louis Pasteur, who lost three of his children to fever, first postulated the “germ theory” (Casanova & Abel, 2013), which states that living microorganisms are the etiology of infectious diseases (Karamanou et al., 2012). German scientist Robert Koch (1843–1910) was the one who further expanded the germ theory by claiming that there exists a specific microorganism for every infection, and this was the beginning of the modern concept of disease transmission (Karamanou et al., 2012). Koch, together with Loeffler (1852–1915), described the “Koch postulate,” which explains the relationship between bacteria and diseases by stating that a diseased person must have a higher number of the infectious agent compared to non-diseased ones; such agents can be isolated and grown in culture; healthy individuals must lack such specific agent; microorganism introduction and inoculation elicit the same disease in healthy people, from whom the same microorganism can be re-cultured and re-isolated (Karamanou et al., 2012). During the twentieth century, more details were discovered regarding the structure, physiology, genetics, and molecular basis of different microorganisms, including bacteria, fungi, parasites, and, eventually, viruses, leading to the development of antimicrobial medication and the subsequent emergence of antimicrobial resistance among pathogens (Ryan, 2004). However, the fundamental conundrum in the field of infectious disease is the enormous clinical variability among individuals throughout the course of an infection (Casanova & Abel, 2013). The germ theory failed to explain why microbes predominantly cause asymptomatic infections and why there is interindividual variability of clinical presentation and outcome, ranging from asymptomatic, carriers, symptomatic, to fatal (Casanova & Abel, 2013). This led to the emergence of four other complementary and overlapping theories, including immunological, microbiological, and genetic, among others (Casanova & Abel, 2013). Previously, using attenuated microbes to immunize children against cholera and anthrax, Louis Pasteur prevented such infections, which subsequently resulted in the implicit idea that previous natural infection by a less virulent pathogen or lower quantity of the same pathogen would result in natural acquired immunity, which would protect or help the afflicted person survive against future infection (Casanova & Abel, 2013). Pasteur’s vaccine discovery, together with three other groundbreaking discoveries, including antigen-specific antibody (Ab) responses,Footnote 1 serological diagnosis,Footnote 2 and sero-therapy,Footnote 3 by Paul Ehrlich, Fernand Widal, and Charles Richet/Emil von Behring, respectively, were the foundation of the immunological theory of infectious diseases. However, while these observations were able to account for interindividual variability partially, they were unable to explain the so- called infection enigma, that is, why the most virulent pathogen can be harmless to some individuals, yet the least virulent ones can be lethal to others (Casanova & Abel, 2020). For instance, findings by Charles Nicolle’s that some infected individuals with typhus pathogen may remain healthy and asymptomatic, yet still able to transmit the disease (Casanova & Abel, 2013), or in the infamous Lübeck disaster, only 72 of the 251 neonates died after being accidentally given a vaccine contaminated with highly virulent TB bacteria (Fox et al., 2016). These could be explained in part by the microbiological and immunological theories, which take into account microbial and human host variability. For example, microorganism’s inheritedFootnote 4 or acquired virulence,Footnote 5 and route of entry, or the host inherited or acquired immunodeficiencies, all of which alter infection outcome (Casanova & Abel, 2020). The pathogenicity of a microorganism is measured by its virulence, which refers to the organism’s capacity to produce a disease and is influenced by a variety of parameters, including the quantity of microorganism present, route of entry into the host body, host immune system response, as well as pathogen virulence factors (Sharma et al., 2016). These virulence factors enable microorganism to enter the host body, evade host defense mechanisms, and produce disease (Sharma et al., 2016). During the period 1920–1949, human geneticists came to the conclusion that an individual’s genetic makeup has a significant impact on his or her susceptibility and resistance to an infectious disease. This was the beginning of the germline genetic theory of infectious diseases, which was developed on the basis of studies on TB infection in twins (Casanova & Abel, 2013), that found the concordance of certain infections is much higher in monozygotic twins than in dizygotic twinsFootnote 6 (Casanova & Abel, 2018). In addition, a simple or complex pattern of genes inheritance may result in susceptibility or resistance to certain illnesses. For example, the one infection-multiple genes phenomena (Casanova & Abel, 2007) describes that a single gene mutation (Mendelian traits) will produce rare primary immunodeficiency, which is often linked with numerous and recurring infections caused by weakly virulent opportunistic microorganisms (Picard et al., 2006). On the other hand, the one infection-multiple genes refers to the polygenic inheritance of several susceptibility genes, resulting in common infections (Casanova & Abel, 2007). For instance, multigene mutations of membrane attack complex (MAC) of complement pathways of the innate immunity, and IL-12- and IL-23-dependent interferon-gamma (IFN-γ)-mediated pathways, will predispose individuals to recurrent invasive bacterial infections by Neisseria species (e.g., meningitis by Neisseria meningitidis), and weakly virulent mycobacteria or Mycobacterium bovis bacillus Calmette-Guerin (BCG) vaccines, respectively (Picard et al., 2006). Similarly, X-linked recessive lymphoproliferative disease (XLP) due to mutations of genes responsible for natural killer (NK) and CD8+ cytotoxicFootnote 7 activation pathways will increase susceptibility to Epstein-Barr virus infection (Picard et al., 2006). In contrast, resistance to human immunodeficiency virus-1 (HIV-1) infection is induced by mutations in the chemokine receptor (CCR5) (Picard et al., 2006). It was not until the early 1950s that the study of human genetics of infectious diseases transitioned into the current molecular and cellular era (Casanova & Abel, 2013).
The “major histocompatibility complex” (MHC) genes were discovered in the 1930s in the context of posttransplantation tissue rejection; however, it was not until many decades later that the function of proteins encoded by MHC genes, in the adaptive and innate immune responses, was recognized (Mak et al., 2014) (Blackwell et al., 2009). Inherited in a Mendelian fashion, the human MHC is also called human leukocyte antigen (HLA) (Choo, 2007), with loci including MHC-I (HLA-A, -B, -C, -E, -F, and -G), II (HLA-DR, -DQ, -DM, and -DP), and III (tumor necrosis factor (TNF), complement factors C2 and C4b) (Blackwell et al., 2009). HLA genes are known to be the highest polymorphic human genes, especially at the antigen-binding site, thus altering the binding specificity to antigens (Choo, 2007). These polymorphisms and variants are believed to be evolutionary selected in order to present antigens of the most prevalent infectious pathogen in various geographic regions (Choo, 2007). MHC-I, located on almost all nucleated cells, facilitates innate and adaptive immune response against intracellular pathogens, such as viruses (Blackwell et al., 2009). On the other hand, MHC-II is expressed on antigen presenting cells (APC) (e.g., B-lymphocytes, dendritic cells (DC), macrophages, etc.) and is primarily engaged in defense against extracellular microbes, including bacteria and parasites (Abbas et al., 2015). Furthermore, due to their involvement in both the innate and adaptive immune responses, human HLA alleles play critical roles in host susceptibility to autoimmune disorders, diabetes, ischemic heart disease, as well as a wide range of infections (Blackwell et al., 2009) and the severity of such infections (Naemi et al., 2021). For instance, individuals with heterozygous HLA-I alleles are less likely to acquire AIDS following HIV infection and thus have lower mortality, whereas those with HLA-II alleles heterozygosity have a greater chance of clearing and overcoming hepatitis B virus infection (Blackwell et al., 2009).
Epidemiology
The occurrence spectrum of an infectious disease in a population can be described using several terms, including sporadic, endemic, epidemic, or pandemic. A “sporadic” disease occurs randomly and at irregular times (Straif-Bourgeois et al., 2014). Hippocrates was the first to use the terms “epidemic” and “endemic” (Swaroop, 1957). “Endemic” either refers to an area where a disease is common (“endemic area”) or to a disease that is common and that exists at a constant rate among certain populations (“endemic disease”) (Swaroop, 1957). “Epidemic” is used when a disease occurs at a higher rate than is typically expected in a specific area, compared to the previously observed baseline level, and if the disease spreads to several countries affecting a large number of people worldwide, it is referred to as a pandemic (Tulchinsky & Varavikova, 2014b). Several major pandemics have occurred throughout history (Table 1), killing millions worldwide (LePan, 2020). The recent COVID-19 pandemic, which started as an epidemic in Wuhan city, China, in December 2019 (Khafaie & Rahim, 2020), was declared a public health global emergency on January 31, 2020, and later a global pandemic on March 11, 2020, by the World Health Organization (WHO) (Dhama et al., 2020). As of October 01, 2021, the COVID-19 pandemic has affected 233,503,524 individuals and killed 4,777,503 people, worldwide (WHO, 2021a).
Human Coronaviruses (hCoVs)
Coronaviruses (CoVs) are enveloped non-segmented positive single-stranded ribonucleic acid (+ssRNA) viruses, which contain the largest genome among RNA viruses and are surrounded by crown-like surface projections under the electron microscope (EM) (Ye et al., 2020). They belong to the Coronaviridae family, which is divided into the subfamily Coronavirinae, which is in turn subdivided into four CoV genera, alpha-CoV, beta-CoV, gamma-CoV, and delta-CoV (Shors, 2021). Alpha and beta-CoVs are known to infect mammals (including bats, humans, etc.), while gamma and delta-CoVs infect birds and mammals (Shors, 2021). There are seven known hCoVs up until now, two in the alpha-CoV (HCoV-229E and HCoV-NL63) and five in the beta-CoV genera (HCoV-OC43, HCoV-HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2) (Ye et al., 2020). The low-pathogenic hCoVs predominantly cause mild (except in infants, elderly, and immunocompromised patients) upper respiratory tract (URT) infections (i.e., the common cold), while the highly pathogenic hCoVs (SARS-CoV, MERS, and SARS-CoV-2) cause lower respiratory tract (LRT) infections (i.e., pneumonia), as well as gastroenteritis, nephritis, hepatitis, etc. (Shors, 2021). Interestingly, for thousands of years, CoVs have been known to be transmitted from species to species, allowing for the emergence of pathogenic hCoVs (Ye et al., 2020). The RNA recombinationFootnote 8 is extremely common among various strains of CoVs, which results in host ranges being expanded, and new CoV with higher pathogenesis and virulence being emerged (Wang et al., 2021a). For instance, this may occur if species carrying distinct CoVs come into close contact and exchange their viruses, and SARS-CoV-2 might have emerged during such occurrences (Singh & Yi, 2021).
SARS-CoV-2 Virion Structure
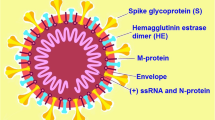
The spherical or ellipsoid SARS-CoV-2 virion seen under the EM is approximately 70–110 nm (Menter et al., 2020). It contains 4 structural glycoproteins, spike (S), membrane (M), nucleocapsid (N), and envelope (E), as well as 9 accessory proteins and 16 nonstructural proteins (NSPs); the E and M glycoprotein make up the viral envelope, while the N-glycoprotein is bound to viral RNA genome (Al-Horani et al., 2020). The S-glycoprotein is composed of two subunits, S1 (the receptor-binding fragment) and S2 (the fusion fragment) (Zhang et al., 2021). S1 is composed of four domains, including N-terminal domain (NTD), receptor-binding domain (RBD), and C-terminal domains (CTD1 and CTD2), while S2 is made up of fusion peptide (FP), fusion-peptide proximal region (FPPR), heptad repeat 1 (HR1), central helix (CH), connector domain (CD), heptad repeat 2 (HR2), transmembrane segment (TM), and the cytoplasmic tail (CT) (Zhang et al., 2021). The S glycoprotein is a trimer attached to the viral membrane by its transmembrane fragment, and its apex is composed of three RBDs, forming up and down conformations indicating receptor-accessible and or receptor-inaccessible states, respectively (Zhang et al., 2021). The surface of the spike protein is heavily covered by N-linked glycan molecules derived from the host cells (Watanabe et al., 2020), and it is the S-glycoprotein protruding from the viral envelope that gives each virion a crown-like appearance (corona—Latin for crown), and hence the name coronavirus (Sahu et al., 2021). Figure 1 shows the Schematic diagram and electron microscopic image of coronaviruses (CoV), such as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).
Schematic diagram and electron microscopic image of coronaviruses (CoV), such as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2). (a) Virion structure of coronaviruses (CoV), such as SARS-CoV-2, which contains four structural glycoproteins, including spike (S), membrane (M), nucleocapsid (N), envelope (E). The viral positive single-stranded ribonucleic acid (+ssRNA) is associated with N-glycoproteins. (b) The four envelope structural glycoproteins and their topology (c) CoV (e.g., SARS-CoV-2) and the S-glycoprotein crown-like surface projections under the electron microscope. (d) The S-glycoprotein is composed of S1 and S2 subunits; S1 is made up of four domains, N-terminal domain (NTD), receptor-binding-domain (RBD), and two C-terminal domains (CTDs), while S2 is made up of fusion peptide (FP), fusion-peptide proximal region (FPPR), heptad repeat-1 (HR1), central helix (CH), connector domain (CD), heptad repeat-2 (HR2), transmembrane (TM), and the cytoplasmic tail (CT) domains. The S1/S2–S2′ is the cleavage site for viral entry into host cell. The surface of the spike protein is heavily covered by N-linked glycan molecules (tree-like structures) derived from the host cells and protrude from the viral envelope giving each virion a crown-like appearance under the EM. Note: SARS-CoV-2 lack the hemagglutinin-esterase (HE), which is present in some Beta-CoVs. (Sources: (a–c) Ujike and Taguchi (2015); (d) Zhang et al. (2021))
Viral Entry
The spike glycoprotein recognizes the angiotensin-converting enzyme-2 receptor (ACE2-R), which is expressed on various host cell membranes (Alanagreh et al., 2020). According to one research, small intestine, testicular, renal, cardiac, thyroid, and adipose cells had the greatest levels of ACE2 expression, whereas the blood, spleen, bone marrow, brain, muscular, and vascular endothelial cells had the lowest levels of ACE2 expression. On the other hand, cells other tissues, such liver, colon, bladder, and suprarenal gland were shown express moderate level of ACE2-R (Li et al., 2020b). Interestingly, in spite of the fact that lung inflammation is the most common symptom in COVID-19 patients, ACE2-R was found to be moderately expressed in pulmonary tissues (Li et al., 2020b).
Viral membrane and host cell membrane fusion occur upon binding of the S1-RBD to ACE2-R, followed by the conformational changes in the S2 subunit (Xia et al., 2020). Viral entry requires S-protein cleavage and activation (i.e., priming) at S1/S2 region, which occurs through two different pathways, namely, the direct fusion or the endocytotic entry pathway. In the direct fusion pathway, the priming occurs by viral use of the host transmembrane protease serine 2 (TMPRSS2) and/or furin, while the clathrin-mediated endocytosis takes place intracytoplasmically and by the action of furin and later cathepsin B and L in the acidic environment of endolysosome (Al-Horani et al., 2020). Both of these pathways result in the intracytoplasmic release of the viral RNA, followed by genomic replication, translation, and the subsequent release of new virions (Al-Horani et al., 2020). These two pathways are shown in Fig. 2. The presence of furin-cleavage site leads to a more effective viral entry into human cells, which may account for the increased infectivity of SARS-CoV-2 (Kaina, 2021). Recently, it is revealed that viral entry can also occur when the already-cleaved SARS-CoV-2 at the furin cleavage site binds neuropilin-1 (NRP1) on olfactory endothelial and epithelial cells (Cantuti-Castelvetri et al., 2020). Another study reported a new possible mode of entry via high-density lipoprotein (HDL) scavenger receptor B type 1 (SR-B1) expressed on cells of LRT, as well as retina, testis, ovaries, metabolic organs, and other extrapulmonary organs, and that the co-expression of ACE2-R and SR-B1 makes these cells more susceptible to SARS-CoV-2 infection (Wei et al., 2020).
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) cell entry pathways. Notes: SARS-CoV-2 host cell entry occurs via two pathways, endocytosis or direct fusion, upon binding of viral spike (S) protein to host cell angiotensin-converting enzyme 2 (ACE2) receptors. Both pathways result in intracytoplasmic viral genome release (2′ and 5). The endocytosis pathway is mediated by furin-induced spike cleavage and subsequent cathepsin L or B (not shown) and acidic (H+) endolysosome action (1–4), while direct fusion is mediated by host transmembrane protease serine (TMPRSS2) and/or furin (1′), leading to spike cleavage. (Modified from: Al-Horani et al. (2020))
Life Cycle
Viruses are obligate intracellular microorganisms, and in order to translate, replicate, and assemble into new virions, they must use host cell machinery (Banerjee et al., 2020) and thus halt the host cell protein synthesis (Lapointe et al., 2021). SARS-CoV-2 has an unusually large genome (~29.0–30.2 kb) (Al-Horani et al., 2020), composed of six open reading frames (ORF); at the 5′-end is the ORF1a-ORF1b region which makes up two-thirds of the genome, while the other one-third of the genome is at the 3′-end, containing the remaining ORFs (Alanagreh et al., 2020). Upon SARS-CoV-2 entry and viral RNA release into the host cell cytoplasm, the ORF1a-ORF1b regions encode two polypeptides (pp1a, pp1ab), which are further cleaved by viral proteinase (i.e., Papain-like protease [PL-pro aka NSP3] and chymotrypsin-like proteinase [3CL pro aka NSP5]) into a total of 16 nonstructural proteins (NSP1-NSP16) (Sahu et al., 2021). Other ORF regions encode viral accessory proteins, such as ORF3a, 6, 7a, 7b, 8, and 10, as well as the structural glycoproteins (Majumdar & Niyogi, 2020). The viral RNA and the N-protein are synthesized and associated together in the host cell cytoplasm, and the nucleocapsid is then assembled with the M, E, and S proteins and bud into RER-Golgi lumen, forming mature virions which are eventually released from the host cell (Patocka et al., 2021). Figures 3 and 4 represent SARS-CoV-2 genome replication and life cycle, respectively.
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) genome replication. Notes: (A) SARS-CoV-2 genome (~30 kb) (B) The genome contains the 5′-end ORF1a and ORF1b regions, encoding two polypeptide (pp1a, pp1ab) that are further cleaved into nonstructural proteins (nsp1–nsp16) by papain-like protease (PLpro aka. nsp3) and chymotrypsin-like proteinase (3CLpro aka. nsp5) (cleavage sites are shown with black/grey triangles), and the 3′-end encoding the structural glycoproteins, spike (S), envelope (E), memberane (M), nucleocapsid (N) (red), and accessory proteins, 3a-10 (gray). Abbreviations: RdRp, RNA-dependent RNA polymerase (aka. nsp12); Hel/MTase, helicase and RNA ATPase (aka. nsp13); ExoN/N7-MTase, exonuclease methyl transferase (aka. nsp14); NendoU, endoribonuclease (aka. nsp15); 2′-O-MTase, 2′-O-methyl transferase (aka. nsp16). (Sources: Romano et al. (2020))
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) life cycle. Notes: Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) binds angiotensin-converting enzyme 2 (ACE2) receptor and enters host cells via two pathways: endocytosis (endosome-cathepsin L) and direct fusion (TMPRSS2). Viral positive sense RNA is released and replicated and translated into viral proteins (N, E, S, and M) in the cytoplasm. The replicated viral RNA and the N-protein are associated together in host cell cytoplasm (forming nucleocapsid), while S, M, and E proteins are translated and undergo posttranslational modification (i.e., surface glycosylation) in the host endoplasmic reticulum (ER)/Golgi intermediate compartment (ERGIC); the nucleocapsid is then assembled with the M, E, and S proteins and bud into RER-Golgi lumen forming mature virions which are eventually released from the host cell via exocytosis. Abbreviations: ACE2, angiotensin-converting enzyme 2; TMPRSS2, transmembrane protease serine 2; RBD, receptor-binding domain; RNA, ribonucleic acid; ORF, open reading frame. (Source: Duan et al. (2020))
Transmission Mode
Viruses causing respiratory tract infections are mainly transmitted via direct physical contact, indirect fomites, droplets, and/or airborne (Leung, 2021). Traditionally, droplets were identified as being too large (i.e., larger than 5 μm) to remain in the air and thus unable to traverse distances greater than 1 m, thus requiring close proximal contact between the infected carrier and the susceptible host (Fennelly, 2020). In contrast, the small airborne agents (usually less than 5 μm) would travel distances greater than 1 m and maintain their infectivity and virulence while suspended in the air (Fennelly, 2020). Originally, the person-to-person transmission of SARS-CoV-2 was thought to be impossible (Kaina, 2021). It was later shown to have the ability to be transmitted by direct contact and large droplets but was subsequently discovered to have the potential for airborne transmission (Patel et al., 2020). The airborne transmission was especially reported in places with ventilation-induced airflow (Tellier et al., 2019) or during procedures like endotracheal intubation, bronchoscopy, and manual ventilation (Jayaweera et al., 2020).
The reproduction number (R0), defined as the number of newly infected cases produced by each infected individual, is used to determine the transmissibility of a virus, which for SARS-CoV-2 was first estimated by the WHO to be between 1.4 and 2.5 (Rahman et al., 2020a). Other values of R0 have been reported, ranging from 2.2 to 6.47, especially in the initial phase of the pandemic (Shaw & Kennedy, 2021). A value of one or less for R0 suggests that the total number of new infections is gradually declining, and the pandemic will ultimately resolve; however, R0 greater than one implies that the virus is spreading rapidly, and more public health measures are required to limit its transmission (Rahman et al., 2020a). Moreover, using different R0 values from several countries (Brazil, Japan, Iran, Italy, and South Korea), Rahman et al. (2020a) estimated the mean R0 for SARS-CoV-2 to be 2.71.
There is, however, significant variability in the transmissibility of the virus. One observed that 69% of infected individuals do not infect other people, while 15–24% of cases accounted for 80% of all SARS-CoV-2 transmission (Adam et al., 2020). However, recent studies suggest that several factors play a role in the transmission, including contact patterns, viral load, and environmental factors. For example, it is reported that within the same household, individuals with very close contacts had an increased risk of transmission (43.4% for spouse vs. 18.3% for other contacts); the viral transmission is found to be higher within the first 5 days of symptoms onset due to higher viral load; and based on contact tracing studies, the indoor transmission is reported to be approximately 19-fold greater than outdoor transmission (Cevik et al., 2020). Moreover, based on the findings that certain countries with high temperatures and humidity, like Brazil, India, and Malaysia, are seeing more cases, compared to countries with lower temperatures, such as Japan and South Korea, such environmental variables are thought to have an impact on the spread of SARS-CoV-2 (Islam et al., 2020a). Moreover, in contrast to SARS-CoV, which mainly replicates in the lung alveolar cells and macrophages, SARS-CoV-2 replication predominantly occurs in the epithelial cells of the URT, making it more transmissible (V’kovski et al., 2020), as new virions are actively shed from the nasopharynx (Cantuti-Castelvetri et al., 2020). Furthermore, recent study has also shown that the SARS-CoV-2 spike glycoprotein has a 10–20 times higher affinity for the ACE2-R receptor when compared to SARS-CoV, which contributes to its greater infectious capacity (Alanagreh et al., 2020).
More recently, United Health ProfessionalsFootnote 9 have referred to the COVID-19 pandemic as the “Biggest Health Scam of the 21st Century.” They reject the claim that SARS-CoV-2 is highly transmissible, saying that one infected person can only transmit the virus to only two or three other individuals, which makes the virus moderately contagious, as opposed to someone who is infected by the extremely transmissible measles virus who can infect up to 20 people. They further argue that there are other infectious diseases that infect and kill more people worldwide, yet they are underreported by the media compared to COVID-19 (United Health Professionals, 2021). For instance, the influenza virus infects and kills 1 billion (30 times more than SARS-CoV-2) and 650,000 people a year, respectively (i.e., globally infects 3 million and kills 2000 people daily), and TB bacteria that infects and kills 10.4 and 1.8 million people annually, respectively (i.e., infects 30,000 and kills 5000 people daily worldwide) (United Health Professionals, 2021).
Other possible routes of transmission have also been reported for SARS-CoV-2, including fecal-oral, vertical, etc. For example, compared to SARS-CoV-1, SARS-CoV-2 has higher affinity to bind intestinal ACE2-R, thus more transmissible, and also difficult to rule out fecal-oral transmission (Gavriatopoulou et al., 2020). Recently, fecal-oral transmission has been considered a possibility after an investigational study (Xu et al., 2020) revealed persistent positive real-time reverse transcription-polymerase chain reaction (RT-PCR) result from rectal swabs of eight out of ten infected children even after negative nasopharyngeal swabs RT-PCR. Vertical transmission refers to prenatal transplacental or intrapartumFootnote 10 maternal-fetal transmission, and although it is uncommon, it has been reported; however, these terms are misleading, and it has been recommended to use more accurate terms for fetal or newborn infection, such as intrauterine transplacental or neonatal, acquired intrapartum neonatal, or acquired postpartum neonatal infection (Konstantinidou et al., 2021).
Immunology
Immune Response Against Viruses
The nonspecific host innate immune system is the first line of defense against microbes, including viruses, followed by activating the more specific adaptive immune system (Abbas et al., 2015). The pathogen-associated molecular patterns (PAMPs) (e.g., viral RNA, etc.) are recognized by various host innate immunity pattern recognition receptors (PRR), such as toll-like receptors (TLRs), leading to the downstream signaling cascade and the activation of interferon regulatory factor 3/7 (IRF3/7) and NF-κB pathways (Lei et al., 2020). For instance, segments of ssRNA of RNA viruses are recognized by TLR7 and 8 leading to the production of type I and type III interferons (IFNs) as well as pro-inflammatory mediators via IRF7 and NF-κB pathways, respectively (Lester & Li, 2014). In fact, when it comes to ssRNA viral infections, greater levels of TLR7 expression may result in a better prognosis since it stimulates a stronger immune response (Khanmohammadi & Rezaei, 2021). Furthermore, in both experimental organisms and human investigations, it is reported that TLR3 deficiency is linked with increased vulnerability to RNA virus infection (Dhangadamajhi & Rout, 2021).
Viral infections, including CoVs, will also activate another component of the innate immunity called the inflammasome, a multi-protein complex composed of the sensor protein NLR (e.g., NLRP3, etc.), or an adaptor protein ASC, and caspase-1 (de Rivero Vaccari et al., 2020). These cascades will result in the synthesis of pro-inflammatory cytokines, interleukin (IL)-1, 6, 8, 12, TNFα, IFN-III (Belizário, 2021), IL-1β, IL-18, and type-I IFN (IFN-α/β) (Lee et al., 2020), as well as a form of cell death called pyroptosis (de Rivero Vaccari et al., 2020). In addition, pyroptosis integrated with other inflammatory cell death pathways is also activated, leading to cell death via PANoptosis (i.e., pyroptosis, necroptosis, and apoptosis). However, these cell death pathways act as a “double-edged sword” with both anti-inflammatory and pro-inflammatory effects; the former is typically beneficial in restricting viral replication and facilitating viral clearance, while the latter will release more intracellular cytokines and PAMPs, leading to cytokine storm and extensive tissue damage (Lee et al., 2020). Another major component of innate immunity is the complement system, composed of various transmembrane and soluble serum proteins, which are activated by viral antigens or by attached Abs to viruses (Abbas et al., 2015). These proteins neutralize viruses by various mechanisms, including the formation of a MAC that mediates lysis of viruses or via viral opsonization,Footnote 11 promoting phagocyte recruitment to the site of infection (Abbas et al., 2015). The opsonization results in the formation of neutrophilic extracellular traps (NETs) that will lead to another type of programmed cell death, termed NETosis, while the recruitment of other inflammatory cells contributes to more pro-inflammatory cytokine production, as well as create a pro-thrombotic state via damaging the vascular endothelial cells (Java et al., 2020).
In an ideal situation, the components of the host’s innate immunity immediately recognize the virus and release cytokines (within a few hours) which limit intracellular viral replication and recruit other immune cells, creating an antiviral state that will eventually prime the adaptive immune system (Sette & Crotty, 2021). IFN-I recruits and activates other innate immunity cells, such as dendritic cells (DC) and natural killer (NK) cells, neutrophils, monocytes, and macrophages, as well as the repertoire of T and B lymphocytes (cells of the adaptive immune system) (Subbian, 2021), which stimulate the production of other cytokines (e.g., IFN-γ, a type-II IFN) (Costela-Ruiz et al., 2020). The APCs of innate immunity present the viral antigens to CD8+ cytotoxic T cells (CTLs) or CD4+ T-helper lymphocytes (Th1 and Th2 cells) via MHC-I and II molecules, respectively, resulting in the formation of long-lasting antigen-specific memory Th-cells and CTLs (Belizário, 2021). The produced innate cytokines will mostly shift the balance toward Th1 cells, specific to intracellular pathogens, like viruses (Belizário, 2021). In addition, B-cells are also activated indirectly by CD4+ cells or directly by viral antigens, leading to the formation of long-term memory B-plasma cells that will secrete neutralizing Abs, including high-avidity immunoglobulin (Ig)-M and high-affinity IgG, 3–5 days and 2 weeks postinfection, respectively (Belizário, 2021). The clearance of all viral infections depends on the more specific adaptive immune response and its components (Sette & Crotty, 2021), which upon activation, will typically increase host lymphocytes count; however, the failure of proper adaptive immune response will result in a state of constitutively active innate immunity with a detrimental impact on multiple organs (Moutchia et al., 2020).
Immunology
Immune Response Against Viruses
The nonspecific host innate immune system is the first line of defense against microbes, including viruses, followed by activating the more specific adaptive immune system (Abbas et al., 2015). The pathogen-associated molecular patterns (PAMPs) (e.g., viral RNA, etc.) is recognized by various host innate immunity pattern recognition receptor (PRR) (e.g., TLR, RIG-I-like), leading to the downstream signaling cascade and the activation of interferon regulatory factor 3/7 (IRF3/7) and NF-κB pathways (Lei et al., 2020). Viral infections, including CoVs, will also activate another component of the innate immunity called the inflammasome, a multi-protein complex composed of the sensor protein NLR (e.g., NLRP3, etc.) or an adaptor protein ASC, and caspase-1 (de Rivero Vaccari et al., 2020). These cascades will result in the synthesis of pro-inflammatory cytokines, interleukin (IL)-1, 6, 8, 12, TNFα, IFN-III (Belizário, 2021), IL-1β, IL-18, and type-I IFN (IFN-α/β) (Lee et al., 2020), as well as a form of cell death called pyroptosis (de Rivero Vaccari et al., 2020). In addition, pyroptosis integrated with other inflammatory cell death pathways is also activated, leading to cell death via PANoptosis (i.e., pyroptosis, necroptosis, and apoptosis). However, these cell death pathways act as a “double-edged sword” with both anti-inflammatory and pro-inflammatory effects; the former typically beneficial in restricting viral replication and facilitating viral clearance, while the latter will release more intracellular cytokines and PAMPs, leading to cytokine storm and extensive tissue damage (Lee et al., 2020). Another major component of innate immunity is the complement system, composed of various transmembrane and soluble serum proteins, which are activated by viral antigens or by attached Abs to viruses (Abbas et al., 2015). These proteins neutralize viruses by various mechanisms, including the formation of a MAC that mediates lysis of viruses or via viral opsonization,Footnote 12 promoting phagocyte recruitment to the site of infection (Abbas et al., 2015). The opsonization results in the formation of neutrophilic extracellular traps (NETs) that will lead to another type of programmed cell death, termed NETosis, while the recruitment of other inflammatory cells contribute to more pro-inflammatory cytokine production, as well as create a pro-thrombotic state via damaging the vascular endothelial cells (Java et al., 2020).
In an ideal situation, the components of the host’s innate immunity immediately recognize the virus and release cytokines (within a few hours) which limit intracellular viral replication, recruit other immune cells, creating an antiviral state that will eventually prime the adaptive immune system (Sette & Crotty, 2021). IFN-I recruits and activate other innate immunity cells, such as DC and NK cells, neutrophils, monocytes, and macrophages, as well as the repertoire of T and B lymphocytes (cells of the adaptive immune system) (Subbian, 2021), which stimulate the production of other cytokines (e.g., IFN-γ, a type-II IFN) (Costela-Ruiz et al., 2020). The APCs of innate immunity present the viral antigens to CD8+ cytotoxic T cells (CTLs) or CD4+ T-helper lymphocytes (Th1 and Th2 cells) via MHC-I and II molecules, respectively, resulting in the formation of long-lasting antigen-specific memory Th-cells and CTLs (Belizário, 2021). The produced innate cytokines will mostly shift the balance toward Th1 cells, specific to intracellular pathogens, like viruses (Belizário, 2021). In addition, B-cells are also activated indirectly by CD4+ cells or directly by viral antigens, leading to the formation of long-term memory B plasma cells that will secrete neutralizing Abs, including high-avidity immunoglobulin (Ig)-M and high-affinity IgG, 3–5 days and 2 weeks postinfection, respectively (Belizário, 2021). The clearance of all viral infections depends on the more specific adaptive immune response and its components (Sette & Crotty, 2021), which upon activation, will typically increase host lymphocytes count; however, the failure of proper adaptive immune response will result in a state of constitutively active innate immunity with a detrimental impact on multiple organs (Moutchia et al., 2020).
Immune Response in COVID-19 Patients, SARS-CoV-2 Pathogenesis and Evasion of Host Immune Responses
It is reported that the innate and adaptive immunity in COVID-19 patients is dysregulated. This dysregulation is thought to cause a delayed IFN-I antiviral response (Subbian, 2021), resulting in overactive innate immunity and underactive adaptive immunity, leading to an extensive cytokine storm state (Moutchia et al., 2020). Autopsies of patients who died of COVID-19 showed high viral loads in the respiratory tract, as well as other tissues, implying ineffective immune responses (Mathew et al., 2020). The hallmark of severe COVID-19 is low lymphocyte count (lymphopenia), with low CD4+ and CD8+ T-cell counts (Cox & Brokstad, 2020), which implies a defective adaptive immune response (Neumann et al., 2020). It is demonstrated that lymphocyte depletion predominantly affects CD8+ T cells (Mathew et al., 2020). It is believed that the direct binding of SARS-CoV-2 to ACE2-R on cells of the reticuloendothelial system, such as spleen and lymph nodes, leads to lymphoid follicles atrophy and thus lymphocytes depletion (Gubernatorova et al., 2020). Studies on the humoral and cellular response in COVID-19 patients demonstrated contradictory findings. For instance, a study on 96 critically ill ICU-admitted patients identified three heterogeneous phenotypes: type 1 phenotype (35% of patients) had a deficient humoral immune response (i.e., lymphopenia with low NK and B cells and low Igs), but preserved T-lymphocyte count, and a moderate level of Il-6 and IL-1β; type 2 (21% of patients) demonstrated a hyper-inflammatory response and cytokine release syndrome (CRS) (IL-1β, Il-6, IL-8, and TNF⍺) with decreased CD4+ and CD8+ T cells, and discrete elevated soluble complement MAC (C5b-9); type 3 was the complement-dependent response patients, with high C3 level, profound C5b-9 elevation (Dupont et al., 2020).
Another research by Gao et al. (2021a) demonstrated a “dichotomous pattern” of the humoral and cellular immune response, which induced in asymptomatic/mild or moderate/severe COVID-19 cases. They indicated that peripheral blood of such cases contains low SARS-CoV-2-specific IgG, as S1- or S2-specific B-cell responses are transient, with no formed long-lived memory B cells, and IgG-secreting plasma cells; however, they reported a profound and sustained IFN-γ-secreting CD4+ Th and CD8+ cell response in these patients, all of which implying the failure to mount humoral immunity in the presence of a strong cellular immunity that might probably prevent them from progressing to severe COVID-19. On the other hand, moderate and severe COVID-19 patients had defective Th1 and IFN-γ-producing CD8+ T-cell responses, while they produced more sustained B cells and humoral responses (Gao et al., 2021a).
In addition, genes for TLR-7 and 8 are located on X chromosome, which may explain such gender-dependent immune response to SARS-CoV-2 (Khanmohammadi & Rezaei, 2021). In fact, when it comes to ssRNA viral infections, greater levels of TLR7 expression may result in a better prognosis since it stimulates a stronger immune response (Khanmohammadi & Rezaei, 2021). This is supported by a case series involving a pair of previously healthy young brothers from two unrelated families who developed severe COVID-19, requiring mechanical ventilation in the ICU, in which distinct loss-of-function variants in the TLR-7 gene X- were identified (van der Made et al., 2020). Moreover, a greater testosterone level is also responsible for the increased synthesis of TLR4 in men, which may account for the higher levels of pro-inflammatory cytokines (e.g., IL-6) in men as compared to females (Khanmohammadi & Rezaei, 2021). Moreover, significant positive correlation was reported with TLR3 deficiency and mutation (rs3775291) with SARS-CoV-2 susceptibility and COVID-19 mortality, but no correlation was found with percentage recovery of patients (Dhangadamajhi & Rout, 2021). In addition, such deficiency and mutant allele in TLR3 raise the chance of developing pulmonary hypertension and diabetes which increase the likelihood of progressing to severe COVID-19 and eventual dying in such individuals (Dhangadamajhi & Rout, 2021).
The pathogenesis of SARS-CoV-2 depends on S-protein interaction with host cells ACE2-R, expressed on type II alveolar epithelial cells (responsible for surfactant synthesis and regeneration of epithelial cells in damaged lungs) (Ortega et al., 2020), as well as many other human cells, including pulmonary macrophages, cardiovascular, intestinal epithelial, renal tubular, testicular, and brain cells, among others (Verdecchia et al., 2020). Severe COVID-19 is believed to occur via viral-induced direct cytotoxic damage, imbalance of the renin-angiotensin-aldosterone system (RAAS), and dysregulation of the immune system (Gupta et al., 2020). SARS-CoV-2 direct infection and replication in lung type II alveolar and vascular endothelial cells and its subsequent release and spread will infect and activate lung immune cells (i.e., macrophages, neutrophils, DC, etc.), leading to release of IL-6, IL-1, TNF, and other pro-inflammatory cytokines, and further viral spread (Gubernatorova et al., 2020). Moreover, the presence of IL-1β, IL-18, and LDH (a marker of cell death) in the sera of COVID-19 patients is believed to be due to the activation of inflammasome (Rodrigues et al., 2020). It is known that IL-1β and TNFα are the principal activators of IL-6, all of which play a critical role in CRS (Zhang et al., 2020a). IL-6 further induces the liver to synthesize acute phase reactants (APRs), such as C-reactive protein (CRP), serum amyloid A (SAA), fibrinogen, haptoglobin, and α1-antitrypsin, while decrease fibronectin, albumin, and transferrin synthesis (Costela-Ruiz et al., 2020). Lung injury and function loss are attributed to elevated IL-1 (IL-1α), while IL-1β is believed to be responsible for COVID-19 hypercoagulation state and disseminated intravascular coagulation (Costela-Ruiz et al., 2020). The high blood glucose level in COVID-19 patients is caused by elevated IL-2 levels; G-CSF and GM-CSF stimulate bone marrow hematopoietic stem cells to undergo proliferation and maturation into monoblasts, promonocytes, monocytes, macrophages, eosinophils, neutrophils, monocytes, DC, etc. (Costela-Ruiz et al., 2020). MCP-1 mediates inflammatory cell infiltrates in various tissues by recruiting and regulating monocytes, memory T cells, and NK cells migration, while HGF is released by necrotic tissue (Costela-Ruiz et al., 2020). The imbalance in RAAS in COVID-19 patients is due to SARS-CoV-2-induced ACE2-R downregulation, which leads to an increase in angiotensin-II (AT-II), which has vasoconstrictive, pro-inflammatory, and pro-thrombotic and tissue remodeling effects (Verdecchia et al., 2020). Moreover, increased production of clotting factors (e.g., Factor VIII) and certain auto-Abs, such as anticardiolipin (aCL) and/or anti-β2 glycoprotein 1 (aβ2GP1) auto-Abs, have been shown to contribute to the hypercoagulable state in critically ill COVID-19 patients (Halpert & Shoenfeld, 2020). In a normal immune response against viruses, cytokine storm is resolved; however, in severely ill patients, this state persists, leading to thrombo-inflammation, and disseminated intravascular coagulation (DIC) (Gupta et al., 2020), tissue injury, multi-organ failure (MOF), and eventually death (Olbei et al., 2021). Moreover, it is reported that complement system activation, in combination with neutrophilia and dysregulated NETosis, is linked with ARDS, hyper-inflammation, and microthrombi formation leading to MOF (Java et al., 2020).
A virus must possess a minimum of one mechanism to evade the human immune responses in order to be able to cause disease; otherwise, it will cause no harm (Sette & Crotty, 2021). It is reported that CoVs escape the host’s innate immune response during the first 10 days of infection, which leads to extensive systemic inflammation (cytokine storm) and high viral load as a result of robust viral replication, release, and spread (Sa Ribero et al., 2020). Studies are underway, but it is believed that SARS-CoV-2 has the same mechanisms as SARS-CoV for the evasion of host immune responses (Nikolich-Zugich et al., 2020). It has been demonstrated that viral NSPs and structural and accessory proteins disturb host innate immune response (Lei et al., 2020), with ORF3b, ORF6, and N protein of SARS-CoV-2 inhibiting IFN-type I synthesis by counteracting the IRF3 and NF-κB signaling pathways (Lee et al., 2020). It is also believed that in coronavirus-infected pulmonary cells, the PAMP-PRR interactions will activate the inflammasome via ORF3a, ORF8b, and E protein. Moreover, NSP1 is the first protein to be encoded by the SARS-CoV-2 genome, which is believed to bind to host cell 40S ribosomal subunit and prevent host cell protein translation (Lapointe et al., 2021), and hence inhibiting type-1 IFN synthesis (McGill et al., 2021). Furthermore, NSP3 is thought to be responsible for the weakening of the host IFN-I immune response by cleaving the IFN-stimulated gene (Yoshimoto, 2021). The posttranslational modification of the SARS-CoV-2 genome by NSP13-NSP16 allows the virus to escape the host innate immune response recognition, while the heavy glycosylation of spike is also responsible for the peptide folding and further evasion (Yoshimoto, 2021). In addition, the ORF3a accessory protein and nsp6 are reported to decrease the size of autophagosome or prevent its maturation, respectively, thus inhibiting the host cell autophagy mechanism toward infected cells (Miao et al., 2021).
Griffin et al. (2021) have divided stages of COVID-19 into different periods (pre-exposure, incubation, viral replication/detectable viral replication, and the inflammatory periods) and phases (symptomatic, early inflammatory, secondary infection, the multisystem inflammatory, and tail phase). The pre-exposure period ends when a susceptible individual is exposed to SARS-CoV-2, followed by the incubation period beginning at the time of exposure (TE) which results in an asymptomatic carrier state in the majority of people. However, if infection occurs, the detectable viral replication phase starts at the time of detectable viral replication (TDVR) when viral copies start rising. The viral symptom phase corresponds to the peak of viral RNA copies, which is at the time of symptom onset (TS), followed by the early inflammatory phase (7–14 days after TS) at time of early inflammation (TEI). The coagulopathy, as well as a rise in inflammatory markers (cytokines, D-dimer, etc.), starts at TEI. The cytokine storm will result in microvascular endothelial dysfunction, thrombosis, and later macrovascular manifestations. A minimum of one thrombotic complication (TC) was reported in 22.7% of cases within the first 14 days of ICU admission, and 52% of these developed pulmonary embolism (PE), which was also similar to the previous result of 42.7% and 16.7% of TC and PE, respectively (Tacquard et al., 2021). If untreated, the secondary infection phase can occur at the time of secondary infection (TSI), which is due to immune dysregulation and result in fungemia, bacteremia, and the development of pneumonia and other bacterial superinfections (Griffin et al., 2021). The next phase, a hyperinflammatory state called multisystem inflammatory phase beginning at TMI (time of multisystem inflammation), is when the IgG level is at its maximum and the secondary bacterial infection and autoimmune features are manifested (Griffin et al., 2021).
Other researchers have classified COVID-19 stages differently. For example, in the three-stage disease classification, stage I is associated with mild disease and is when the innate and adaptive immunity is activated. This stage corresponds to TLR-3,7, and 8 stimulation; IgM and IgG Abs production against S and N protein; and the onset of signs and symptoms of fever, dry cough, and lymphopenia. Stage I will progress to stage II if the host is not able to eliminate SARS-CoV-2 (e.g., in elderly and those with comorbidities), which will then spread and involve multiple organs (Ortega et al., 2020). In this stage, referred to as macrophage activation syndrome (i.e., hyper-inflammatory response and cytokine storm), the patient will present with dyspnea (IIA) and severe hypoxia (IIB), as well as detectable radiological findings, abnormal liver function test, lymphopenia, and elevated levels of APRs. If therapeutic measures are not effective, stage III will culminate with severe inflammatory response syndrome (SIRS), shock, and MOF, including acute respiratory distress syndrome (ARDS) (Ortega et al., 2020).
The pathophysiology behind COVID-19 extrapulmonary manifestations might predominantly be through widely expressed ACE2-R in various tissues, direct viral cytotoxic effect, or molecular mimicry, among others. For instance, the high expression of ACE2-R in cardiac and smooth muscle cells, as well as fibroblasts and endothelial cells, is responsible for SARS-CoV-2 direct extrapulmonary and atypical symptoms (Gupta et al., 2020). Moreover, the molecular mimicry between SARS-CoV-2 spike glycoprotein and human proteins is reportedly the pathomechanism behind autoimmune diseases seen in COVID-19 patients. For example, the shared peptide sequence between a peptide in S-protein (the major SARS-CoV-2 antigen) and human proteomes will result in the already-formed immune responses against the virus to also cross-react with these human proteins leading to the manifestations of autoimmune disorders (Kanduc & Shoenfeld, 2020). Similarly, molecular mimicry is hypothesized to facilitate peripheral neuropathy since SARS-CoV-2 surface glycoproteins are identical to human neural tissue glycoconjugates (Ramani et al., 2021). The pathophysiology for muscle involvement (e.g., autoimmune myositis and rhabdomyolysis) in COVID-19 patients has also been suggested to be the result of homology between SARS-CoV-2 antigens and human myocytes (Ramani et al., 2021). It has recently been found that several auto-Abs (e.g., antinuclear, anti-β2 glycoprotein-1 Abs, anticardiolipin Abs, etc.) are produced in SARS-CoV-2-infected patients resulting in new-onset autoimmune diseases, including Guillain-Barré syndrome, Miller Fisher syndrome, antiphospholipid syndrome, immune thrombocytopenic purpura, systemic lupus erythematosus, KD, large vessel vasculitis/thrombosis, psoriasis, and type I diabetes mellitus (DM), among others (Halpert & Shoenfeld, 2020). It is revealed that HLA gene polymorphism is responsible for such autoimmune diseases, and hence auto-Abs are developed in genetically susceptible individuals (e.g., those with HLA-DRB1, etc.) (Halpert & Shoenfeld, 2020).
The HLA alleles and COVID-19 severity might also be related, based on the previously reported relationship between these alleles and the severity of clinical manifestations of SARS cases, and the fact that in silico, the affinity of SARS-CoV-2 peptide varies for each HLA alleles (Amoroso et al., 2020). Research that investigated the relationship between the HLA genotype polymorphism and the severity of COVID-19 among 95 patients reported a high frequency of HLA class I, including HLA-B*51 in those who had fatal COVID-19 infections and that of HLA-B*35 in patients with mild infection (Naemi et al., 2021). Even though limited data is available regarding HLA class II, the same study found a high frequency of HLA-DRB1*13 in the fatal group, compared to the mildly infected patients. Comparing HLA alleles between healthy individuals and COVID-19 cases, as well as non-survived and survived patients, Lorente et al. (2021) reported higher HLA-A*32 in healthy individuals and higher HLA-A*03, HLA-B*39, and HLA-C*16 in COVID-19 patients; however, HLA-A*11, HLA-C*01, and HLA-DQB1*04 were found to be greater in non-surviving patients. Additionally, due to some unknown mechanisms, a positive correlation was reported between polymorphisms in CCR5 (i.e., deletion mutation) and SARS-CoV-2 infection and death (Mehlotra, 2020). In addition, the possibility of correlations between TMPRSS2 and ACE2 DNA polymorphisms with COVID-19 susceptibility and severity and outcomes was proposed in a comparative genetic study of 81,000 human genomes (Hou et al., 2020). For instance, the level of TMPRSS2, which is expressed on type I alveolar epithelial cells, is elevated with aging, and ACE2 polymorphisms and cardiovascular and pulmonary diseases (risk factors for COVID-19) are linked. This may explain the decreased risk of SARS-CoV-2 in infants and children relative to adults (Hou et al., 2020).
SARS-CoV-2 Origin
The origin of the majority of hCoVs is considered to be bats or rodents (natural hosts), where they are maintained and propagated yet remain nonpathogenic; they then spill over to the human host (and become pathogenic) via an amplifying intermediate reservoir host within which the virus undergoes transient replication (Shors, 2021). The intermediate host(s) are known for some hCoVs, while it is unknown for others. For instance, the CoVs of the 2003 SARS and 2012 MERS pandemics are believed to have been transmitted via the civet and camel as their intermediate hosts, respectively (Shors, 2021). As previously mentioned, the recombination among various strains of CoVs is reported to lead to the emergence of a novel virus, such as SARS-CoV-2 (Singh & Yi, 2021). Throughout their evolution, the genetic diversity of beta-CoVs, such as SARS-CoV-2, is increased via mutations and recombination, which is also reported to occur within other species (Rastogi et al., 2020). For example, evidence has shown sequence identity between SARS-CoV-2 and horseshoe bat CoV (RaTG13), as well as Malayan pangolins (Singh & Yi, 2021). As a result, bat and pangolin are considered to be the natural and intermediate hosts of SARS-CoV-2, respectively (Singh & Yi, 2021). In fact, the SARS-CoV-2 genome is thought to be a “mosaic” genome, made up of fragments from at least two previously known CoVs (Sallard et al., 2021), and is assumed to be likely a recombinant of those zoonotic viruses (Rastogi et al., 2020).
The unique feature of SARS-CoV-2, compared to any other alpha and beta-CoVs, is the presence of furin-cleavage site, as well as six major amino acid sequences in the RBD domain that is optimized for binding to the human-like ACE2-R (Andersen et al., 2020). It is reported that the SARS-CoV-2 genome is 96% identical to RaTG13, with the RBD domains being only 85% similar, sharing just one of the six major amino acid sequences (Rastogi et al., 2020). On the other hand, RBD regions of the SARS-CoV-2-related virus in pangolin share 92.4–99.8% sequence identity with the RBD of SARS-CoV-2 (Rastogi et al., 2020). Moreover, some studies indicate that all six main amino acids in the RBD regions of SARS-CoV-2 are identical to those in pangolin CoV (Andersen et al., 2020), whereas other studies claim this to be five out of six (Rastogi et al., 2020). This is supported by analyzing pangolin samples from two separate provinces in China, where researchers were able to identify two distinct clusters of SARS-CoV-2-related viruses, one of which shared greater amino acid identity (97.4%) with SARS-CoV-2 in RBD than did the bat CoV RaTG13 (89.2%); however, bat CoV shared more sequence identity (89.2%) with other non-RBD genome regions of SARS-CoV-2 than did pangolin CoV (Han, 2020). Furthermore, since bats have been ecologically separated from the human population, it is possible that SARS-CoV-2 has acquired its adaptive modifications in an intermediate host (e.g., pangolin) prior to its transmission to human (Rastogi et al., 2020). Further support pointing to pangolin as the intermediate host comes from pangolins or their scales being consumed as a source of food or in traditional Chinese medicine, respectively (Shors, 2021). Similarly, analysis of lung samples from two pangolins that died of pulmonary fibrosis, and the subsequent identification of CoVs that were nearly 90.5% and 91% similar to SARS-CoV-2, provided more evidence for this hypothesis (Shors, 2021). However, it is important to note that both bat and pangolin CoVs lack a furin-cleavage site (Andersen et al., 2020). In addition, concluding wild pangolins to be the intermediate host for SARS-CoV-2 is still controversial since the pangolins used in research studies were those from illegal smuggling activities and not wild ones (Singh & Yi, 2021). In addition, in order to get a more accurate estimate of the similarity and the “time to the most recent common ancestor (tMRCA)” of two different CoV strains (e.g., SARS-CoV-2 and bat CoV), it is preferable to utilize synonymous mutations, which are more prevalent in the genome since they are less likely to be subject to natural selection as they do not alter the properties of resulting proteins (Singh & Yi, 2021). For example, comparing such mutations, only 83% similarity is seen between bat RaTG13 CoV and SARS-CoV-2, and thus implying a distant relationship, compared to the initial report of 96% (Singh & Yi, 2021).
Sallard et al. (2021) explained that the similarity between pangolins CoV and SARS-CoV-2 is still considerably lower than the 99.52% similarity reported in the previously known SARS-CoV and its last intermediate host during the previous past zoonotic transmissions. In addition, they stated that human ACE2-R utilized by SARS-CoV-2 is more identical to farm animal proteins than that of wild pangolins and bats. On the other hand, genetic findings unequivocally suggest that SARS-CoV-2 is not generated from any previously known viral backbone (Andersen et al., 2020). Additionally, if pangolin is assumed to be the intermediate host, then the first detected case of SARS-CoV-2 infection would have to have acquired the virus when coming in contact with the intermediate host sold at Wuhan market; in fact, the first infected case did not even visit the market, which possibly excludes pangolin as the reservoir (Sallard et al., 2021).
In the absence of an intermediate host, some scientists have speculated that SARS-CoV-2 could have been synthetically developed in a laboratory, while others suggested it might have been adapted to laboratory animals or to a human, while it was being cultured on human cells (for study purposes) and have accidentally escaped these laboratories (Sallard et al., 2021). Additionally, CoVs are listed in Group 3 of potential bioterrorism agents that require Biosafety Level 3 (BSL-3) laboratories, where generally airborne agents that potentially cause fatal infections are kept (Kaufer et al., 2020). Hence, two circulating conspiracy theories have accused the USA or China of genetically engineering SARS-CoV-2 (Nie, 2020). Further controversies were brought about when the US CDC reported the presence of SARS-CoV-2 Abs in the blood of individuals from France, Italy, and the USA long before the virus was identified in Wuhan (Lew, 2020), as well as when CDC director Robert Redfield in a video interview stated that patients who were previously thought to have died of influenza might, in fact, have died from COVID-19 (Hall, 2020).
SARS-COV-2 Variants
Owing to their lack of proofreading capacity and as part of their evolution to increase genetic diversity, RNA viruses persistently go through recombinations and mutations (Rastogi et al., 2020). The change in the amino acid sequence of the viral protein is referred to as mutation (Lauring & Hodcroft, 2021), and one of the most significant ways in which viruses evolve in nature, is considered to be nucleotide substitution (Phan, 2020). These substitution mutations can be non-synonymous, resulting in the alteration of an amino acid sequence of a protein, as opposed to synonymous ones (silent mutations), which cause no such changes (Chu & Wei, 2019). The synonymous mutations are heavily influenced by the viral mutation rate (Singh & Yi, 2021), and in general, mutations could occur upon human-to-human or human-to-animal viral transmission (Garry, 2021) or due to chronic infections of immunocompromised patients (Williams & Burgers, 2021). Viruses with different genomic sequences are called variants,Footnote 13 and when viral variants have a clearly distinct phenotype, including antigenicity,Footnote 14 transmissibility, or virulence, they are called strains (Lauring & Hodcroft, 2021).
The mutation rate of SARS-CoV-2 is around 23.6 mutations per year (Yao et al., 2020), resulting in the accumulation of mutations at 9.8 × 10−4 substitutions per site annually (Khateeb et al., 2021). Throughout the pandemic, several SARS-CoV-2 variants have evolved and are continuously emerging and spreading throughout the globe (Centers for Disease Control and Prevention (CDC), 2021b). The most variable region of SARS-CoV-2 to undergo mutational changes, including deletions, mutations, and recombination, is the S-protein region (Singh & Yi, 2021), which can alter viral infectivity or reactivity to neutralizing Abs (Li et al., 2020c). However, it is also stated that a single mutation in spike is not likely to cause resistance to neutralizing Abs, as the surface area of RBD is large enough for the Abs to bind (Sette & Crotty, 2021). Other regions, including structural protein (e.g., N) and NSP regions (i.e., ORF1a, ORF1b, ORF3, ORF8) of the SARS-CoV-2 genome, have also been reported to accumulate mutations (Wang et al., 2021a). The spike substitution mutations can occur in the RBD (S1 subunit) and non-RBD domains, as well as the S1/S2 furin-cleavage site. For example, the major substitutions in the RBD (N501Y, E484Q, E484K, T478K, L452R, K417T, K417N) as well as non-RBD (D614G)Footnote 15 regions will increase SARS-CV-2 immune evasion (both host and vaccine-acquired immunity) and affinity toward human ACE2-R (Khateeb et al., 2021). Table 2 summarized major SARS-CoV-2 spike protein mutations and their effects.
The SARS-CoV-2 Interagency Group (SIG) has established a classification system that categorizes SARS-CoV-2 variants into three categories, namely, variant of interest (VOI), variant of concern (VOC), variant of high consequence (VOHC), and variants being monitored (VBM). A variant is termed a VOI when any mutations in the viral genome might reduce neutralization by Abs (produced from previous infection or vaccine), decrease treatment efficacy, affect the diagnostic tests, or possibly increase transmissibility or disease severity (CDC, 2021a). On the other hand, the SARS-CoV-2 variant is classified as a VOC when genetic mutations result in substantial evidence of high transmissibility, severe COVID-19 (hospitalization or death), significantly low Ab neutralization, decreased treatment efficacy, and/or failure of viral detection by diagnostic tests. In contrast, a variant of high consequence, which has not yet been detected for SARS-CoV-2, as of July, 2021, is one that has obvious evidence of a significant reduction in the efficacy of public health preventative measures and medical interventions, compared to the previously known variant, mandating its report to WHO (CDC, 2021a). VBM refers to those variants for which there is adequate evidence of high transmissibility, increased disease severity, and obvious or potential effect on approved therapeutic measures, but which are not currently circulating in the USA and do not represent a major and immediate danger to public health. Any of the VOI and VOC may later be placed in this category, if their proportions have decreased significantly and consistently over time and they no longer represent a significant threat to public health in the USA (CDC, 2021a).
In late January and early February 2020, a new D614G mutation in the non-RBD region of spike appeared (Khateeb et al., 2021; WHO, 2021b). This was the first detected mutation of concern, which has spread globally, and by the end of June 2020 was present in the majority of circulating SARS-CoV-2 variants worldwide (Hossain et al., 2021), that is 99% of all variants (Khateeb et al., 2021). This mutated virus is reported to have increased affinity for olfactory epithelium (Khateeb et al., 2021), be ten times more infectious than the original virus (Li et al., 2020c), with higher ACE2-R affinity, viral load, and hence more transmissibility; however, it has no impact on disease severity (Zhang et al., 2020b) or on the efficacy of therapeutic drugs, diagnostic tests, vaccines, and public health preventative strategies (WHO, 2020). It is also reported that the affinity for ACE2-R is not limited to human but can also target other species, including horseshoe bat, Malayan pangolin, cat, and dog (Wang et al., 2021a). However, it is still unclear which animal is capable of successfully transmitting SARS-CoV-2 to humans. For instance, more than 40 bat species susceptible to SARS-CoV-2 were recently identified in the USA. It is also stated that cat can acquire SARS-CoV-2 and transmit it to other cats, while ferrets develop URT infections but are unable to transmit the virus within their species (Solis & Nunn, 2021).
Between August and September 2020, a mink-associated variant named “Cluster 5” emerged in Denmark and the Netherlands, with the RBD mutations Y453F (the most widespread), del69_70, I692V, and M1229I (Lauring & Hodcroft, 2021). This variant has also been shown to include additional RBD mutations, such as F486L and N501T, which together with Y453F enhance viral affinity to both human and mink ACE2-R, thus making SARS-CoV-2 adaptable to both host species (Salleh et al., 2021). It was originally believed that such mutations would result in viral Ab-neutralization escape (Goodman & Whittaker, 2021); however, recent research in mice models suggests that they have no impact on the neutralizing Abs, and moreover, this variant is not circulating anymore and has already disappeared (Salleh et al., 2021). Following the discovery of an ORF8-deficient lineage with N501T mutations among humans and farmed-mink in Denmark, it has been suggested that ORF8-deficient lineages, which may have emerged as a result of the rapid transmission of SARS-CoV-2 within the mink population, are capable of interspecies spillover (Sharun et al., 2021).
In September 2020, having acquired 17 mutations, indicating a considerable period of evolution and natural selection, perhaps in a host with chronic SARS-CoV-2 infection, the UK VoC 202012/01 (B.1.1.7 lineage aka. Alpha variant) was detected (Lauring & Hodcroft, 2021). The mutations include 14 non-synonymous point mutations, and 3 deletions, with 8 of these being in the S-protein, and the variant is estimated to have a 43–90% higher R0 than the previous variants (Davies et al., 2021). The Alpha variant contains D614G, 69–70del, and 144del in NTD, N501Y in RBD, and P681H at the furin cleavage site, among others (Wang et al., 2021b). The deletion (del69_70) mutation is reported to affect the performance of real-time RT-PCR diagnostic tests (WHO, 2021b) and to help the virus evade the host immune responses. In addition, the N501Y mutation increases viral human-murine ACE2-R affinity (Rambaut et al., 2020) and infects children easily (Hayashi et al., 2021), while the P681H that is exponentially increasing worldwide may enhance systemic infection (Maison et al., 2021). Moreover, this variant is reported to increase hospitalization and disease severity, as well as produce 50% enhanced transmissibility, yet has no impact on neutralization by mAbs, or Abs from vaccinated or convalescent sera (CDC, 2021b). Furthermore, there is speculation that variant B.1.1.7 is responsible for the cases of myocarditis in pets; however, there is little evidence to support this hypothesis (Sharun et al., 2021). Moreover, according to retrospective observational studies, there is 35% higher risk of death linked to the Alpha variant (Farinholt et al., 2021). In addition, in a recent study, the effectiveness of NVX-CoV2373 (by Novavax), a protein subunit vaccine containing the S protein from the original Wuhan virus, against B.1.1.7 variant in 18–84 years old individuals is 85.6%, compared to 95.6% for the original Wuhan virus (Gómez et al., 2021).
In January of 2021, two new variants (Lineage B.1.427/B.1.429), with mutations at NTD (S13I and W152C) and RBD (L452R and D614G) regions, were first detected in California and have rapidly spread across the USA, as well as many other countries (McCallum et al., 2021), hence were initially considered VOCs in March 2021 (Martin Webb et al., 2021). A VOC, classified as the B.1.351 lineage (variant Beta), was first identified in South Africa, which possesses N501Y, K417N, and E484K, and is 50% more transmissible, with significant resistance to polyclonal/monoclonal Abs (mAbs), and Abs of convalescent and post-vaccination sera (CDC, 2021a). It is reported that the Beta variant is 6.5 and 8.6-fold more resistant to neutralization by polyclonal Abs obtained from people who have been vaccinated with Pfizer or Moderna, respectively (Gómez et al., 2021). However, the efficacy of NVX-CoV2373 was reported to be 49.4% against B.1.351 variant among more than 4400 participants, and this value increased up to 60% in the preventing of mild, moderate, and se-vere COVID-19 (excluding human immunodeficiency virus (HIV) positive individuals) (Gómez et al., 2021).
The other variant circulating in Brazil (P1 lineage) was first identified in January 2021 in Japan among people who had visited Brazil (Faria et al., 2021), harboring similar mutations to Beta variant (Faria et al., 2021), and is considered a VOC (Gamma variant) due to its potential impact on infectivity, immune escape, and reinfection (Resende et al., 2020). Reinfection is defined as a second positive PCR at least 28 days after the previous positive PCR (Colson et al., 2020). Furthermore, it is believed that IgG to anti-SARS-CoV-2 are unlikely to give long-lasting protection (Fang et al., 2020), and in fact, several reports have already presented cases of SARS-CoV-2 reinfection (e.g., with the Brazilian variant); however, unless the second infection is caused by a different viral variant (Resende et al., 2020), it is still unknown whether a second positive PCR implies reinfection or it is merely the persistence of COVID-19. In case it is truly reinfection, the management of the pandemic would be challenging, and no herd immunity would develop with either natural infection or vaccination (Falahi & Kenarkoohi, 2020). It is noteworthy to mention that the shared mutations between variants P.1, B.1.1.7, and B.1.351 reportedly emerged independently of each other (Faria et al., 2021).
In December 2020 and during the second wave of the pandemic in India, two other variants, including delta (B.1.617.2) and kappa, have emerged (B.1.617.1). They both have E484Q and L452R, while delta also carries T478K mutation, and are considered VOC (Khateeb et al., 2021). Moreover, reports indicate that compared to the alpha variant, the Delta variant is more transmissible (60% more than Alpha variant), increases hospitalization rate, and has intermediate resistance to the vaccine, especially in those who only had their first dose (Callaway, 2021). It must be mentioned that both the CDC (2021a) and Public Health Agency of Canada consider Variant Kappa a VOI (CDC, 2021a; Government of Canada, 2021). A recent study reports that vaccine-induced antibodies provide low immunity to Delta variant.
Harboring common mutations in their S1 subunit, the major circulating SARS-CoV-2 VOC includes alpha, beta, gamma, and delta, each having a global frequency of 48%, 7%, 7%, and 14%, respectively (Khateeb et al., 2021). As of July 21, 2021, there have been several SARS-CoV-2 VOI, including Epsilon (B.1.427 and B.1.429), Zeta (P.2), Eta (B.1.525), Theta (P.3), Iota (B.1.526 and B.1.526.1), Kappa (B.1.617.1), Lambda (C.37), B.1.1.318, and B.1.617.3 (WHO, 2021c). However, as of September 21, 2021, and according to CDC, there is no VOI, and Alpha, Beta, Gamma, Epsilon, Eta, Iota, Kappa, Mu, and Zeta are classified as VBM (CDC, 2021a). On the other hand, as of October 12, 2021, WHO considers both Lambda and Mu a VOI, while the remaining variants (except theta and zeta) are considered variants under monitoring (VUM), which was previously referred to as “Alerts for Further Monitoring” (WHO, 2021c). Evidence of increased transmission is reported in Epsilon, Eta, Iota, and Kappa, while almost all of them might have reduced sensitivity to neutralization by polyclonal Abs in convalescent sera or to mAb therapy (Epsilon) (Government of Canada, 2021) (Table 3 summarizes WHO and PANGO-lineage classification systems of major variants of SARS-CoV-2 and their characteristics).
Clinical Characteristics
Signs and Symptoms
The typical symptoms of COVID-19 appear after an average incubation period (interval between viral exposure to symptom onset) of 2–14 days (Griffin et al., 2021) but can also occur within a minimum of 1 day to a maximum of 20 days postexposure (Qu et al., 2021). SARS-CoV-2 replication starts before the onset of symptoms; thus, in the majority of cases, COVID-19 is recognized when viral RNA copies have already reached their maximum level, which is during the symptom phase at the time of symptom onset (TS) (Griffin et al., 2021). These clinical presentations can range from asymptomatic, mild, moderate, severe, to critical respiratory symptoms (Table 4), but the majority of cases remain asymptomatic or only develop mild symptoms (Gao et al., 2021b).
The initial signs and symptoms of hypoxemia and increased respiratory rate in the early inflammatory phase might require airway support, and if untreated, this can lead to cardiac disorders, renal failure, neurological symptoms, and MOF (Griffin et al., 2021). Hypoxia, a common and yet atypical feature of ARDS seen in the early disease stage, is surprisingly well-tolerated and is thus called “silent hypoxia” (Gavriatopoulou et al., 2020). According to a systematic review and meta-analysis (Rodriguez-Morales et al., 2020), 32.8% of patients developed ARDS with shock (6.2%), and 20.3% needed intensive care unit (ICU) admission, while 13.9% had fatal consequences. Moreover, diaphragm muscle can be involved in COVID-19 patients secondary to critical illness myopathy, ventilation-associated diaphragm dysfunction, phrenic nerve damage, or direct viral injury (as diaphragm express ACE2-R); this will further exacerbate respiratory distress (Ramani et al., 2021). Moreover, severe dyspnea and tachypnea were mostly reported by elderly patients who died of COVID-19, while fever and headache were present mostly in recovered elderly patients (Perrotta et al., 2020).
A systematic review and meta-analysis study (Grant et al., 2020) of 24,410 laboratory-confirmed COVID-19 adults revealed the most prevalent symptoms to be fever (78%), cough (57%), fatigue (31%), dyspnea (23%), rigors (18%), wheeze (17%), myalgia (17%), arthralgia 11%, headache (13%), confusion (11%), and diarrhea (10%), among others. This is consistent with Li et al. (2020a) meta-analysis result that found the most prevalent symptoms, in decreasing order of prevalence, to be fever (88.5%), cough (68.6%), fatigue or myalgia (35.8%), expectoration (28.2%), dyspnea (21.9%), headache or dizziness (12.1%), diarrhea (4.8%), and nausea and vomiting (3.9%). A few cases of acute arthritis secondary to COVID-19 and within a couple of weeks of SARS-CoV-2 infection have also been reported (Ramani et al., 2021).
COVID-19 patients can also present with loss of smell (anosmia) and taste (dysgeusia). In a meta-analysis among 20,451 patients, Ibekwe et al. (2020) reported anosmia and dysgeusia in approximately 49% and 41% of cases, respectively. This was in agreement with a global meta-analysis (Agyeman et al., 2020) among 8438 cases that reported the loss of smell and taste in 41.0% and 38.2% of patients, respectively. Anosmia and dysgeusia are more prevalent in mild and moderate cases, although the latter is more associated with severe disease (Mullol et al., 2020), and both are strongly suggestive of COVID-19 (Griffin et al., 2021).
Compared to mild COVID-19 cases, gastrointestinal (GI) symptoms occur more predominantly in patients with severe COVID-19 (8.1% vs. 23%) (Gavriatopoulou et al., 2020), and the most common reported GI symptoms in decreasing order were anorexia (21%), nausea and/or vomiting (7%), diarrhea (9%), and abdominal pain (3%) (Gupta et al., 2020). A retrospective study reported that 16% of 1141 patients solely developed GI symptoms (Gavriatopoulou et al., 2020), and in other cases, digestive symptoms occurred prior to the typical respiratory signs and fever (Philips et al., 2020). Moreover, it is stated that about 14–53% of critically ill hospitalized patients will have findings of hepatocellular injuries (Gupta et al., 2020). However, there has not been a convincing report on the direct involvement of the hepatobiliary system in COVID-19 cases, and it is believed that hepatic injury is mainly secondary to the disease itself or the result of hepatotoxic therapeutic agents used in these patients (Philips et al., 2020), including remdesivir, lopinavir, and tocilizumab (Gupta et al., 2020). In addition, earlier studies done prior to the pandemic have reported that ACE2-R expression was more than 30-fold higher in the liver of cirrhotic patients, which might explain the increased hepatotropism of SARS-CoV-2 in those with chronic liver disease (Marjot et al., 2021). Furthermore, during the pandemic social isolation, alcohol consumption has increased significantly, leading to alcoholic liver disease (ALD) (Marjot et al., 2021). For instance, it is reported that 17% of individuals who had previously abstained from alcohol but had a history of alcohol use disorder were shown to relapse during lockdown (Marjot et al., 2021). A study conducted at a single center in the UK found that the number of ALD referrals and the percentage of critically ill inpatients with ALD (without COVID-19) increased by more than twofold in June 2020 when compared to June 2019 (Marjot et al., 2021).
Dermatological manifestations of COVID-19 are present in approximately 20% of COVID-19 patients, and the cutaneous lesions range from urticaria, vesicles, purpura, papulosquamous, as well as purpuric eruptions and livedo reticularis (Gavriatopoulou et al., 2020). These skin lesions are thought to be due to hypersensitivity reactions to the SARS-CoV-2 genome, CRS, and vasculitis with micro-thrombi formation (Gupta et al., 2020). The endocrine system can also be affected by SARS-CoV-2; however, the information available on the impact of COVID-19 on the endocrine system, including the hypothalamic-pituitary-adrenal (HPA), is very limited (Alzahrani et al., 2021). It is reported that SARS viruses can impair the HPA axis, and research on the previous SARS pandemic revealed that 40% of cases had signs and symptoms of secondary adrenal insufficiency (SAI) 90 days after their recovery (Alzahrani et al., 2021). Moreover, the molecular mimicry between a SARS-CoV amino acid and adrenocorticotropic hormone (ACTH) results in a host immune response against SARS to cross-react with ACTH residue and induce adrenal insufficiency, hence decreased cortisol level (Alzahrani et al., 2021). Recent autopsy findings of SARS-CoV-2 patients have demonstrated microscopic changes in 46% of these cases, with evidence of suprarenal cortical necrosis, cortical lipid degeneration, focal inflammation, hemorrhage, and vascular thrombosis, were seen in these patients; however, no adrenal insufficiency was reported (Alzahrani et al., 2021).
On the other hand, an investigation on the response of the HPA axis in COVID-19 patients detected no normally expected robust cortisol response in any of the acute COVID-19 patients (Alzahrani et al., 2021). The cortisol and ACTH levels were in the low/normal low, which were diagnostic of SAI, while low dehydroepiandrosterone sulfate (DHEAS) levels in these patients were indicative of chronic ACTH deficiency. Interestingly, with except for one, none of the patients exhibited signs or symptoms of adrenal insufficiency (Alzahrani et al., 2021). It is noteworthy to mention that the nonspecific signs and symptoms of adrenal crisis, such as fever, nausea, vomiting, extreme fatigue, weakness, myalgia, postural hypotension, and abdominal pain, are similar to those of acute COVID-19 and thus are difficult to differentiate (Alzahrani et al., 2021).
COVID-19 can also present as atypical symptoms (Philips et al., 2020), with elderly patients more likely to present with these symptoms, such as falls, reduced mobility, generalized weakness, and delirium (Gan et al., 2020). Several case reports have also presented patients with other rare atypical symptoms, such as persistent hiccups (a reflex inspiratory movement) in patients with no travel history or sick contact. For example, the first reported case was a 62-year-old man with a previous history of hypertension (HTN), diabetes mellitus (DM), and coronary heart disease, who presented with unintentional weight loss and 4-day persistent hiccups but lacked the typical symptoms of cough, fever (though he later developed a fever in the hospital), sore throat, dyspnea, and so on (Prince & Sergel, 2020). In another case report, a 48-year-old man with a history of HTN visited the hospital complaining of persistent hiccups for 1 week, which started after he developed a fever 7 days earlier (Bakheet et al., 2020).
The systemic inflammatory response, CRS, and hypoxemia can also cause arrhythmia, myocardial ischemia, and myocardial infarction (MI), which are exacerbated in those with the previous history of heart conditions (Philips et al., 2020). For example, CRS is thought to disrupt the already existing atherosclerotic plaques via macrophage activation and leukocyte adhesion molecule expressions on vascular endothelial cells. In major arteries of the heart, this leads to an acute coronary syndrome, myocardial ischemia, and MI (Gavriatopoulou et al., 2020). Furthermore, hypotension, which is a clinical hallmark of CRS and sepsis, together with fever and systemic infection, causes an imbalance between the oxygen supply and demand of cardiomyocytes, leading to further cardiac damage and eventual left ventricular systolic dysfunction and cardiogenic shock (Gavriatopoulou et al., 2020).
Other cardiac manifestations included sinus tachycardia or bradycardia, pulseless electrical activity, atrial fibrillation, and atrial flutter (Gupta et al., 2020). Moreover, in a Chinese center, approximately 6% of 187 patients treated for COVID-19 developed sustained ventricular tachycardia (SVT) or ventricular fibrillation (VF) (Guo et al., 2020). The inflammation of cardiac muscle cells (myocarditis) is thought to be responsible for heart dysfunction (Gupta et al., 2020), with ventricular arrhythmias being a common clinical finding of acute myocarditis (Gavriatopoulou et al., 2020). Moreover, some of the therapeutic agents, such as chloroquine (CQ), hydroxychloroquine (HCQ), and azithromycin that are used in COVID-19 patients, are pro-arrhythmic and may result in a long QT interval and the resultant torsade de pointes (Gavriatopoulou et al., 2020). COVID-19 may also affect the reproductive glands, such as male testes via ACE2-R on spermatogonia, leydig, and sertoli cells, leading to testis orchitis and infertility (Deshmukh et al., 2021).
Laboratory Findings and Diagnosis
Severe and critical COVID-19 cases are more likely to have abnormal laboratory parameters (Moutchia et al., 2020), while asymptomatic patients might have both normal and abnormal results (Zhang et al., 2020c). However, the lack of specific laboratory parameters in the early disease stage has made it difficult to establish an early diagnosis (Ebrahimi et al., 2020). A systematic review and meta-analysis among 4663 COVID-19 patients demonstrated the most prevalent abnormal laboratory findings to be an elevated level of CRP, erythrocyte sedimentation rate (ESR), IL-6, and lactate dehydrogenase (LDH), and decreased albumin, eosinophils, and lymphocytes (Zhang et al., 2020d). The elevated level of ESR, CRP, and LDH, as well as ferritin, D-dimer, and fibrinogen in COVID-19 patients implies dysregulation of immunity and hyper-inflammation (Gupta et al., 2020).
Several other pro-inflammatory cytokines and chemokines, including IL-1α, 2, 4, 7, 9, 10, G-CSF, GM-CSF, M-CSF, IP-10, MCP-1, MIP 1-α, HGF, PDGF, and VEGF, were also reported to be elevated in COVID-19 patients (Costela-Ruiz et al., 2020). Moreover, severe COVID-19 patients were shown to have elevated alanine aminotransferase and aspartate aminotransferase (ALT/AST), creatinine kinase (CK), total bilirubin, gamma-glutamyltransferase (GGT), myoglobin, blood urea nitrogen (BUN), and creatinine, while albumin is found to be lower in severe COVID-19 (Danwang et al., 2020). The complete blood count (CBC) in COVID-19 patients demonstrated mild thrombocytopenia, elevated total white blood cells (WBC) counts, with neutrophilia, but lymphopenia; thus increased neutrophil to lymphocyte ratio (NLR) (Gupta et al., 2020). Furthermore, abnormal coagulation markers were reported, including elevated D-dimer, fibrinogen, prothrombin time (PT), and activated partial thromboplastin time (aPTT) (Gupta et al., 2020). However, interestingly, unlike patients with DIC due to bacterial sepsis or trauma who have high PT and aPTT, DIC in severe COVID-19 cases show minimal prolongation of aPTT and/or PT (Wool & Miller 2020). D-dimer is elevated to a greater extent and out of proportion than other parameters such as PT/INR, aPTT, fibrinogen, or platelets (Al-Samkari et al., 2020). Abnormal glucose metabolism in COVID-19 patients presents with high blood glucose level, euglycemic ketosis, and classic diabetic ketoacidosis, while patients with acute kidney injury demonstrated high blood urea nitrogen (BUN), creatinine, proteinuria, hematuria, metabolic acidosis, and electrolyte imbalance (e.g., hyperkalemia, hypo/hypernatremia) (Gupta et al., 2020). Furthermore, interestingly, a study among 129 patients with various COVID-19 reported significantly high levels of lung cancer tumor markers, such as carcinoembryonic antigen (CEA), cytokeratin 19 fragment (CYFRA21-1), neuron-specific enolase (NSE), squamous cell carcinoma antigen (SCCA), and pro-gastrin releasing peptide (ProGRP) (He et al., 2020).
The gold standard test for the diagnosis of SARS-CoV-2 infection is using real-time RT-PCR to detect viral genome from swab samples of deeper anatomical sites, such as nasopharynx, oropharynx, or upper and lower respiratory tract aspirates and bronchoalveolar lavage (Yüce et al., 2021). A successful SARS-CoV-2 infection will result in viral genome replication shortly after viral exposure (i.e., is detectable as early as 1-day postexposure) and peak 3–4 days postexposure; however, the currently used technologies are not able to detect viral replication in the immediate postexposure time, as viral RNA copies can range from undetectable to millions 1–3 days prior to the onset of symptoms (Griffin et al., 2021); thus, false-negative (FN) results might be generated, as RT-PCR sensitivity is not high (Falahi & Kenarkoohi, 2020), and a negative RT-PCR result in the initial stage of infection cannot rule out SARS-CoV-2 infection (Adams et al., 2020). A retrospective study on 280 hospitalized patients diagnosed with COVID-19 reported a positive RT-PCR in 39.6% of them (Özel et al., 2021); as high as 29% FN rate for RT-PCR of nasal swabs samples had been reported (Sasisekharan et al., 2021). Furthermore, the majority of mild COVID-19 patients will have their viral RNA copy number decreased to values under an infectious level by day 10; however, in immunocompromised and severe COVID-19 cases, the value may stay above infectious level until day 20, or in some cases to 3 weeks post-discharge (Griffin et al., 2021).
Other methods, including both lateral flow type assays (LFA) and enzyme-linked immunosorbent type assays (ELISA), can be utilized to detect either serum antiviral Abs (IgM/IgG) or viral antigens (S, M, or N glycoprotein antigens) (Yüce et al., 2021). Abs against SARS-CoV-2 appear 1–2 weeks after the onset of symptoms (Fang et al., 2020), and as previously explained, the humoral response to SARS-CoV-2 demonstrates heterogeneity among different cases. Thus, low levels produced in cases of viral evasion of the immune system (Shang et al., 2020), short-lived Abs in asymptomatic and mild cases, and late Ab response will give FN results (Fang et al., 2020).
Radiological Features
Pulmonary Findings
Computed tomography (CT) is a technique that combines X-rays and computer technology to produce sharp images of the lungs (Zahan et al., 2021). It is the most sensitive modality to detect early COVID-19 pneumonia and is predominantly used for patients with strong clinical suspicion of COVID-19 (Campagnano et al., 2021); however, CT exact sensitivity and specificity for COVID-19 are not known, and currently, it is not recommended for routine screening of COVID-19 pneumonia (Simpson et al., 2020). Furthermore, it is reported that CT has higher sensitivity compared to RT-PCR in suspected COVID-19 patients (Yau et al., 2020), but the patterns found on CT scans are nonspecific and do not differentiate between viral and bacterial pneumonia (Sun et al., 2020). Moreover, it is more accurate than conventional chest X-ray (CXR) in identifying lung abnormalities, especially in cases of false-negative RT-PCR (Zahan et al., 2021). Although more sensitive than other modalities, CT cannot rule in or rule out the diagnosis (Gavriatopoulou et al., 2020).
A systematic review and meta-analysis of chest CT and CXR (Garg et al., 2021) in COVID-19 pneumonia patients reported the pooled prevalence of 66.9% ground-glass opacity (GGO), 32.1% only consolidation, 44.9% GGO plus consolidation, 29.1% crazy paving, 23.6% halo sign, 8.9% nodules, 5.6% pleural effusion, and 2.7% lymphadenopathy on chest CT. Moreover, they found the most common lung areas involved on CT in decreasing order to be peripheral (58.5%), central plus peripheral (19.4%), and central (16%); bilateral lung involvement was seen in 44% of CT scans, while unilateral involvement was seen in 9.1% images (Garg et al., 2021). Regarding the CXR findings, more consolidation (46.9%) than GGO (38.7%) was observed in the radiographs of these patients (Garg et al., 2021); however, CXR has lower sensitivity in detecting abnormalities (Sun et al., 2020). Similarly, in another systematic review and meta-analysis of CT findings (Cao et al., 2020), in 46,959 patients with COVID-19 pneumonia, more bilateral involvements (75.5%) were seen compared to unilateral (20.4%), and the most prevalent CT patterns were GGO (69.9%), irregular or halo sign (54.4%), air bronchogram (51.3%), bronchovascular bundle thickening (39.5%), grid-form shadow (24.4%), and hydrothorax (18.5%); however, some of the used radiological terminology (e.g., grid-form shadows and bronchovascular bundle thickening) are not recommended by the Radiological Society of North America expert consensus statement in the reporting instructions for CT finding of COVID-19. According to another systematic review and meta-analysis, pure GGO is more prevalent in the early disease stage, which later progresses to consolidation, and 76% of patients had multilobar lung involvement, with the right lung being affected more frequently (Awulachew et al., 2020). Figure 5 shows chest CT findings of confirmed COVID-19 patients.
Computed tomography (CT) findings of confirmed coronavirus disease 19 (COVID-19) patients. Note: (a) Pneumonia ground-glass opacity (GGO); (b) GGO and air bronchogram in a patient with severe pneumonia; (c) consolidation in patients with pneumonia; (d) consolidation and air bronchogram in a patient with severe pneumonia; (e) nodular opacities in patient with pneumonia; (f) pleural effusion on right side in a patient with severe pneumonia. (Source: Liu et al. (2020b))
Some COVID-19 patients will also have atypical findings on chest CT. A study among 298 confirmed COVID-19 cases with pneumonia revealed 73.1% had typical CT features, while 21.1% presented with atypical and typical CT patterns. The most prevalent atypical features were pulmonary cysts (9%), pleural effusion 5.7%, nodules 4.3%, bull’s eye/target sign (1.3%), cavitation (1.0%), spontaneous pneumothorax (0.6%), hilar lymphadenopathy (0.6%), spontaneous pneumomediastinum with subcutaneous emphysema (0.3%), as well as halo sign, empyema, and necrotizing pneumonia with abscess (each 0.3% prevalent) (Gurumurthy et al., 2021). Studies have reported contradictory results regarding the CT findings in asymptomatic carriers, which ranged from normal to some GGO patterns (Zhang et al., 2020c).
COVID-19 Reporting and Data System (CO-RADS), a categorical system developed by The Dutch Radiological Society (NVvR), is a useful tool to assess the level of suspicion of pulmonary involvement in COVID-19 patients based on CT findings (Özel et al., 2021). It is revealed that patients with CO-RADS5 had a positive RT-PCR test which was statistically significant, while all cases in the CO-RADS2 category had negative RT-PCR; thus, they concluded that in the presence of a negative RT-PCR, CO-RADS is able to diagnose COVID-19 (Özel et al., 2021). Several other radiological systems were developed to standardize chest CT reporting in suspected COVID-19 cases, each having different sensitivity,Footnote 16 specificity,Footnote 17 positive predictive value (PPV),Footnote 18 and negative predictive value (NPV),Footnote 19 as well as different RT-PCR results. These systems included COVID-19 imaging reporting and data system (COVID-RADS), the RSNA expert consensus statement, and the British Society of Thoracic Imaging (BSTI), all with good performance and interobserver agreements in reporting CT features (Inui et al., 2020).
Lung ultrasound (LUS), such as point-of-care ultrasound (POCUS), has also been used to manage COVID-19 patients either at triage, ED, or ICU (Bhoi et al., 2020). The most common LUS findings are discrete B-lines (in 81.3% of cases), confluent B-lines (in 50%), small subpleural consolidations (usually <3 cm) (in 42.7%), and normal LUS (in 18.8%). The discrete hyperechoic B-lines, which are usually the first signs of COVID-19, are due to increased interstitial fluid in the acute disease stage, while the confluent lines are formed by coalescence of many individual B-lines as the disease progress, and more interstitial fluid is accumulated (Mateos González et al., 2021). Figure 6 shows LUS patterns of COVID-19. LUS was found to be able to detect pulmonary infiltrates more than CXR and with more sensitivity (81% vs. 63%) (Mateos González et al., 2021). The B-lines found on LUS correspond to the GGO on chest CT (Hussain et al., 2020), and the “light beam” artifact (“waterfall” sign) is specific for COVID-19 pneumonia and represents the early GGO appearance on CT (Yau et al., 2020). Other LUS features of COVID-19 are thick irregular pleural lines, mobile air bronchograms (at live scan) with large consolidation, especially in those on mechanical ventilation, indicating ARDS progression or secondary bacterial infection (Hussain et al., 2020). Furthermore, in cases where vital signs are normal, LUS can still identify COVID-19 pneumonia and differentiate viral and bacterial pneumonia (Hussain et al., 2020). If pleural effusion is detected, other differential diagnoses must be considered (e.g., bacterial pneumonia, secondary bacterial infection, or congestive heart failure) (Campagnano et al., 2021). LUS is found to have comparable sensitivity and specificity to CT scans, with the advantage of avoiding radiation exposure (i.e., safe for pregnant patients and children), lowering costs, and decreasing the chance of spreading the virus (due to increased portability) (Schmid et al., 2020).
Lung ultrasound patterns of coronavirus disease-2019 (COVID-19). Notes: (a) Normal lung with pleural sliding and A-lines; (b) discrete hyperechoic individual B-lines which are usually the first signs of COVID-19 and are due to increased interstitial fluid in the acute disease stage; (c) confluent B-lines are formed by coalescence of many individual B-lines as the disease progress and more interstitial fluid is accumulated; (d) subpleural consolidations (usually are small and <3 cm). (Source: Mateos González et al. (2021))
Musculoskeletal and Soft Tissue Findings
Electromyography (EMG) and nerve conduction studies can confirm and differentiate between SARS-CoV-2 myopathy and other diseases mimicking myopathy (e.g., motor neuron diseases); however, magnetic resonance imaging (MRI) is the gold standard to identify soft tissue necrosis and to localize pathology site (Ramani et al., 2021). Moreover, diaphragm involvement can be diagnosed and monitored using a fluoroscopy sniff test for real-time diaphragmatic movement and ultrasound to detect muscular atrophy and determine diaphragmatic thickening during inspiration (Ramani et al., 2021).
The MRI findings of acute GBS and MFS are enlargement, with signal hyperintensity, and contrast enhancement of spinal nerve roots, nerve plexus, and cauda equina, while COVID-19 peripheral neuropathy presents as nonspecific hypoechogenicity and hyperintensity on ultrasound and MR neurography, respectively (Ramani et al., 2021). Moreover, SARS-CoV-2-induced or vasopressor-induced (given for hemodynamically unstable patients) gangrene can present with MRI hyperintense signals and absence of enhancement of necrotic tissue, while arthritis and synovitis present as MRI synovial enhancement and ultrasound power Doppler signals (Ramani et al., 2021).
Histopathological Findings in Autopsy or Endoscopic Specimens
Several histopathological, immunohistochemical, and EM findings were reported in autopsy or endoscopic specimens of various tissues from COVID-19 patients, including pulmonary, GI, cardiovascular (CV), endocrine, and genitourinary, among others (Table 5).
Prognostic Value of Clinical and Laboratory Parameters
Fever combined with dyspnea and smoking is the most important prognostic factor for disease progression, while acute cardiac injury, preexisting cardiovascular disease (CVD), DM, respiratory disease, and HTN are reported to be the most significant prognostic factors for mortality rate (Hatmi, 2021). Moreover, CVD and HTN are prognostic factors for disease severity (Hatmi, 2021). It is reported that 52 (27.8%) out of 187 COVID-19 patients in China developed a myocardial injury, confirmed by an increased level of troponin (Guo et al., 2020). Heart failure (HF) was reported in 24.4% of 176 COVID-19 patients confirmed with HF marker, N-terminal pro-b-type natriuretic peptide (NTproBNP), with non-survivors having a higher prevalence compared to survivors (49.4% vs. 3.2%) (Gavriatopoulou et al., 2020).
Several laboratory parameters have been found to have prognostic value in COVID-19 patients. For instance, elevated NLR is associated with severe disease, whereas lymphopenia is correlated with a higher need for mechanical ventilation and death in severe cases (Lee et al., 2020). Similarly, reduced platelet to lymphocyte ratio is predictive of COVID-19 severity and worse outcome, and low platelets also increase hospitalization risk and inpatient mortality (Lee et al., 2020). In another meta-analysis and systematic review, platelet count was reported to be lower in severe patients (177.38 × 109 L−1) than that of critical cases (205.96 × 109 L−1) (Kazemi et al., 2021). Kazemi et al. (2021) also found the mean CRP level to be significantly higher in dead patients vs. recovered ones (85.82 vs. 32.99 mg/L), while the mean values among the invasive-ventilated, no-oxygen therapy, and noninvasive-ventilated groups were reported to be 48.89, 44.03, and 40.58 mg/L, respectively. Moreover, a significant correlation between Hb, platelets, and creatinine levels, and COVID-19 severity, with mean Hb and platelets being higher among critical patients is compared to other patients (Kazemi et al., 2021).
Moreover, the development of ARDS and mortality is associated with elevated ferritin (Lee et al., 2020). High levels of CRP, ferritin, procalcitonin, D-dimer, and neutrophils imply severe pneumonia, and a decrease in these parameters indicates therapy effectiveness; an increase in lymphocyte count is also correlated with a better outcome (Yu et al., 2020).
Furthermore, high admission levels of markers such as cardiac troponin-I ≥21 ng/L, D-dimer ≥1112 ng/mL, CRP ≥10 mg/dL, and LDH ≥334 U/L are directly correlated with short-term mortality (Peiró et al., 2021), while elevated bilirubin is linked to disease severity and progression (Gupta et al., 2020). This was in agreement with another study that reported total bilirubin level and LDH to be significantly elevated in deceased patients compared to survivors (Danwang et al., 2020).
Lung cancer tumor markers, such as CEA, NSE, CYFRA21-1, and SCCA, can predict clinical outcomes and increased mortality (He et al., 2020), and viral load, abnormal ALT/AST, CK, and CXR are also prognostic factors (Yi et al., 2020). In an investigation, it was demonstrated that, compared to mild COVID-19 respiratory samples, those with severe COVID-19 have significantly higher SARS-CoV-2 viral load (Zheng et al., 2020a). Elevated IL-1 (IL-1α) is correlated with high viral load and mortality and is found in severely ill cases, whereas high levels of IFN-γ are reported in COVID-19 patients compared to healthy individuals, which are correlated with higher viral load and lung injury (Costela-Ruiz et al., 2020). Moreover, it is reported that viral load is directly associated with elevated M-CSF, IFN-α2, IL-IL-1α/1β, IL-2, 4, 7, 10, 13, 17, IP-10, G-CSF, IL-12, IFNγ, and HGF but indirectly correlated to PDGF, while scores of lung injury are associated with IL-1α, IL-2, 4, 7, 10, 17 IFNγ, HGF, IFN-α2, IP-10, G-CSF, and M-CSF (Liu et al., 2020c).
Moreover, IL-6 was found in higher levels in those who died than in the recovered cases (Costela-Ruiz et al., 2020), and low CD4+ and CD8+ T cells are seen in severe diseases (Gupta et al., 2020). It is reported that even a moderate elevation in IL-6 (i.e., above 80 pg/mL) implies that the COVID-19 patient is at high risk of respiratory failure, and hence serial laboratory tests to measure IL-6 level might be critical in recognizing disease progression (Gubernatorova et al., 2020). It is reported that the plasma of both ICU and non-ICU patients contained high levels of several cytokines, such as IL-1β, IL-7, IL-8, IL-9, IL-10, G-CSF, GM-CSF, IFNγ, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF, TNFα, and VEGF, while patients who required both ICU admission and oxygen therapy due to severe pneumonia-induced ARDS had higher levels of IL-2, IL-7, IL-10, G-CSF, IP-10, MCP-1, MIP-1α, and TNFα (Kircheis et al., 2020). The result of a study on laboratory diagnosed symptomatic or asymptomatic patients, as well as healthy individuals, demonstrated plasma level of 18 cytokines to be lower in asymptomatic patients, compared to symptomatic patients, while the same level of 32 cytokines was detected in both asymptomatic and healthy individuals (Long et al., 2020).
COVID-19 Complications and Long-Term Effects
For unknown pathomechanisms, some COVID-19 patients, referred to as long hauler COVID, long COVID, or long-term COVID by various sources, will still experience residual and long-term persistence and recurrence of symptoms after the initial 4-week period of the acute phase and during the last phase of COVID-19 that starts at TT (time of the tail onset) in the post-acute tail phase (Griffin et al., 2021). A systematic review and meta-analysis (Lopez-Leon et al., 2021) on 47,910 patients revealed that 80% of patients continue to experience one or more symptoms in long term, with the most prevalent symptoms being fatigue (58%), headache (44%), attention disorder (27%), hair loss (25%), dyspnea (24%), ageusia (23%), anosmia (21%), postexercise polypnoea (21%), arthralgia (19%), cough (19%), sweat (17%), as well as nausea/vomiting, chest pain, memory loss (each 16%), and hearing loss/tinnitus (15%). It is estimated that 6 months after the onset of symptoms, 76% of patients will continue to have a minimum of one adverse symptom, with tiredness or muscular weakness being the most frequent complaint (63%), followed by sleeping difficulties (26%) (Wang et al., 2021a). It has been reported that D614G and possibly N501Y mutation are responsible for such neurological symptoms, as well as anosmia and ageusia (Wang et al., 2021a). Other chronic signs and symptoms such as anxiety, depression, digestive problems, weight loss, skin lesions, pain, sleep disorders, intermittent fever, and high heart rate at rest were also reported (Lopez-Leon et al., 2021). Moreover, the most common persistent abnormal laboratory results in long-term COVID-19 cases were abnormal CXR/CT (34%), D-dimer (20%), as well as elevated CRP (8%), ferritin (8%), procalcitonin (4%), and IL-6 (3%) (Lopez-Leon et al., 2021).
Furthermore, peripheral neuropathy due to prone positioning is a very rare complication in both the perioperative care (in 0.14% patients) and ARDS patients, to a point where the landmark PROne positioning in SEvere Acute respiratory distress syndrome (PROSEVA) trial does not even include it as a complication (Malik et al., 2020). Prone positioning is included in The Surviving Sepsis Campaign COVID-19 guidelines as the mainstay of treatment for moderate to severe ARDS cases and has been reported to significantly lower mortality and improve oxygenation (Coppo et al., 2020). However, a significantly high prevalence of possibly prone positioning-induced peripheral neuropathy has been reported in these patients. Malik et al. (2020) reported peripheral neuropathy diagnosis among 14.5% of COVID-19 patients with ARDS (n = 83), with 91.7% of them having prone positioning in their histories. Upper extremities were most frequently involved in these patients (76.2%), as well as ulnar nerve (28.6%), radial nerve (14.3%), sciatic nerve (14.3%), brachial plexus (9.5%), and median nerve (9.5%). The etiology behind such neural injuries seen in SARS-CoV-2 and other viral infections (e.g., HCV, HIV, and VZV) is still not known and might be the result of prone positioning-induced nerve compression or stretch, as well as postinfectious inflammatory neuropathy, systemic neuropathy, or hematoma-induced nerve entrapment (secondary to anticoagulant therapy) (Fernandez et al., 2021). Osteoporosis and osteonecrosis can be the complication of COVID-19 coagulopathy or corticosteroid treatment (Ramani et al., 2021). Similar to myositis seen in other viral infections (e.g., hepatitis, influenza, or HIV), myositis and its complication, rhabdomyolysis, have been reported in SARS-CoV-2-infected patients (Ramani et al., 2021). Myalgia and weakness in some COVID-19 patients can be due to myositis and/or rhabdomyolysis, which might result in compartment syndrome, and intravascular coagulation, and myoglobinuria-induced acute kidney injury (Ramani et al., 2021).
COVID-19 Clinical Trials
Since the start of the pandemic, approximately 500 randomized controlled trials (RCTs) have been registered globally for therapeutics or postexposure prophylaxes of COVID-19 (Davis et al., 2020). The most common drugs used for the treatment or prophylaxis purposes in decreasing order were HCQ, immunomodulatory drugs, antivirals, angiotensin system antagonists, colchicine, NSAIDs, and tranenxamic acid (Babaei et al., 2020).
HCQ and chloroquine (CQ), two antimalarial and anti-inflammatory medications, were among the earliest drugs used alone or in combination of several trials (Gao et al., 2020) for either prophylaxis or treatment of mild, severe, or critical COVID-19 patients (Babaei et al., 2020). It was found that neither CQ nor HCQ improve outpatient or inpatient outcomes, nor do they reduce infection rates when used as postexposure prophylaxes (Fang et al., 2020). The second category of widely used agents was immunomodulatory, such as mAb (e.g., tocilizumab, which is an IL-6 receptor antagonist), glucocorticoids, immunoglobulins, and IFNs (such as IFNα2b) (Babaei et al., 2020). Glucocorticoids (dexamethasone), alone or with other drugs, were found to decrease mortality (by one-third in those on ventilation and by one-fifth in on oxygen therapy) in critically ill patients; however, it might be dangerous if administered at early disease stage as it disrupts antiviral immune response (Babaei et al., 2020). A retrospective study on the effect of dexamethasone on COVID-19 pneumonia patients demonstrated no significant impact on clinical course, adverse events, or outcome (Hu et al., 2020). It is reported that immunomodulatory agents could increase the risk of bacterial and fungal superinfections, such as secondary invasive pulmonary aspergillosis (Fang et al., 2020). Additionally, combining inhaled IFN-α2b and angiotensin receptor blockers (ARB) was shown to increase inflammatory process resolution (by lowering IL-6) and viral clearance (Zhou et al., 2020a). In a multicenter, open nonrandomized observational study (Pereda et al., 2020) in Cuba, where all laboratory-confirmed COVID-19 cases were treated with intramuscular IFN-α2b (Heberon Alpha R) combined with lopinavir-ritonavir (LPV/RTV) (except those with contraindications), it was concluded that those who received these agents had almost 60 times greater chance of recovery. Other clinical trials that tested the efficacy of antiviral therapy, including LPV/RTV, remdesivir, and umifenovir, found LPV/RTV to have therapeutic potential; however, they found no efficacy when antivirals (or HCQ) were combined with azithromycin (Babaei et al., 2020). Moreover, Babaei et al. (2020) reported umifenovir to be safe, with a higher rate of negative PCR on day 14 of infection, nonetheless, with no evidence of improved outcomes. The “Adaptive COVID-19 Treatment Trial” (ACTT), a double-blind placebo RCT, found that remdesivir decreased recovery time compared to a placebo, prevented disease progression and resulted in a shorter hospital stay and faster discharge (Beigel et al., 2020). A randomized trial of convalescent plasma (that contains Abs of recovered patients) and placebo in severe COVID-19 patients with pneumonia did not find any significant differences in overall mortality or clinical outcome (Simonovich et al., 2021). However, in an uncontrolled case series of five critical COVID-19 patients with ARDS, the administration of convalescent plasma improved clinical outcomes, reduced fever, viral load, pulmonary lesions on CT, and the need for mechanical ventilation (Shen et al., 2020).
Moreover, a double-blind RCT on hospitalized non-intubated COVID-19 patients indicated that tocilizumab has no impact on intubation or death prevention, although it decreased the risk of developing serious secondary infections (Stone et al., 2020). In an ongoing phase 2 trial (BLAZE-1 trial), the efficacy of placebo and intravenous infusion of neutralizing mAb bamlanivimab (LY-CoV555) in either 700, 2800, or 7000 mg in treating mild or moderate COVID-19 was compared. It was demonstrated that compared to placebo, those treated with bamlanivimab had lower symptom severity and lower hospitalization rate, and the patients who received the highest dose had lower viral load at day 11. Since this viral load reduction was found in most patients, it was considered to be the result of natural disease course and not having a clinical significance (Chen et al., 2021). Three other clinical trials in more than 300 hospitals across 5 continents are investigating the benefit of therapeutic (high dose) and prophylactic (low dose) anticoagulants such as heparin in moderately ill-hospitalized patients who are not in the ICU and not on any organs support (e.g., mechanical ventilation). It demonstrated that high-dose heparin is safe and effective in preventing thromboembolism in these patients (National Heart, Lung, and Blood Institute (NHLBI), 2021).
In addition, some studies have found that vitamin D supplementation decreases mortality rate and viral load in COVID-19 patients; thus, currently, a nationwide RCT is being conducted in the USA and is to be completed by March 2021 (Sengupta et al., 2021). In fact, the deficiency in vitamin D (25-OH-D) has been reported to increase the risk of SARS-CoV-2 infection and hospitalization due to COVID-19 (Babaei et al., 2020), as it prevents CRS by inhibiting the proliferation of inflammatory cells, decreases the AT-II level by upregulating ACE2 level, and decreases the hypercoagulability by increasing antithrombotic factors (Sengupta et al., 2021). The effectiveness of NK cells, as well as stem cells (SC) such as umbilical cord (UC)/Wharton’s Jelly (WG) mesenchymal SC (MSCs), dental pulp SC, and human embryonic SC, are being investigated (Babaei et al., 2020). MSCs decrease inflammation by antagonizing the pro-inflammatory cytokines, such as IL-1α and TNF-α (Costela-Ruiz et al., 2020). It is reported that the injection of placental and UC/WG MSCs is well tolerated and can alleviate respiratory symptoms (Saleh et al., 2021).
It is noteworthy to mention that the stage at which these medications are administered plays a key role in the achieved outcome. For example, IFNs (that increase innate immune response), mAbs, and antivirals are most likely beneficial if given during the viral replication period at TDVR, while immunomodulatory drugs affecting innate immunity and anticoagulants are more effective if administered during the early inflammatory period at TEI (Griffin et al., 2021). Similarly, the unnecessary antibiotics given early in the course of the disease will not only be helpful but will increase antimicrobial resistance; hence, in order to be more effective and beneficial, they must be administered at the appropriate secondary infection phase at TSI (Griffin et al., 2021).
Isolation, Quarantine, and Social Distancing
One of the primary reactions against new contagious diseases is quarantine (Parmet & Sinha, 2020) that restricts and confines the movements of individuals who have been in contact with infectious agents in order to confirm whether or not they were infected (CDC, 2017). Two weeks of quarantine at home or in an assigned facility has been suggested for those who have come into contact with a confirmed or probable case of COVID-19, as well as travelers from an endemic area (WHO, 2021d). It is reported that quarantine is recommended in case of continuous or cumulative unprotected exposure of more than 15 min within a distance of 6 feet (≈1.83 m) or less (Griffin et al., 2021). Quarantine is distinct from isolation, which segregates infected individuals from healthy ones (CDC, 2017).
In China, the government used a “mass quarantine” strategy, imposing quarantines on almost 60 million people, which drastically reduced disease incidence (Roper, 2020). The speedy Chinese response to the pandemic was reported by Gregory Poland—the director of the Mayo Clinic’s Vaccine Research Group in Rochester, Minnesota, USA—as the critical factor that curtailed the spread of the virus, as compared to other countries, whose reactions were delayed despite a longer preparation time (Burki, 2020). It is worth mentioning that after the implementation of these preventative measures in Wuhan on January 23, 2020, R0 began to decline from a value of approximately four to below one by March 2020 (Rahman et al., 2020a). In other countries across the globe, such as the USA, quarantine measures have also been implemented; however, inadequate quarantines resulted in the USA, which has only 4% of the world’s population, having almost 26% of world cases and 24% of world deaths as of July 16, 2020 (Blumenthal et al., 2020).
Surprisingly, it was also reported that, compared to those under the age of 42, people with confirmed COVID-19 who are above the age of 42 experience longer incubation periods, suggesting that effective quarantine policies could be targeted at specific age groups as opposed to the existing “unified” policies (Pak et al., 2020). Girum et al. (2020) systematically reviewed studies on some COVID-19 prevention strategies, such as quarantine and isolation. They concluded that isolation combined with a 3-month self-quarantine could prevent 31% of COVID-19-related deaths, but that isolation by itself is not very effective since, without high levels of contact tracing and screening, this strategy will miss 75% of cases. In contrast, if SARS-CoV-2 completely adapts to humans, it would be difficult to contain the pandemic through quarantine or other public health measures (Ye et al., 2020).
In the pre-exposure period, vulnerable individuals should employ a range of steps, including social distancing and minimizing contact with suspected cases, in order to reduce their risk of SARS-CoV-2 infection (Griffin et al., 2021). Social distancing is defined as “keeping a safe space between yourself and other people who are not from your household” (CDC, 2020, para.1). It is worth noting that several weeks into the pandemic, many scholars have emphasized that the term “social distancing” is misleading and, in fact, counterproductive (Sørensen et al., 2021). It has been argued that “distant socialization” must be encouraged instead, and in order to prevent the spread of SARS-CoV-2, efforts should be made to establish social connections while also keeping physical distancing (Sørensen et al., 2021). Thus, the WHO Secretary-General, Dr. Tedros Adhanom Ghebreyesus, started using the term “physical distance” in his announcements and speeches (Sørensen et al., 2021).
The WHO (2021b) advises people to keep an interpersonal distance of at least 1 m in indoor spaces and when they talk, cough, and sneeze in order to decrease infection by SARS-CoV-2 droplets. However, as previously mentioned, the virus can be airborne in certain conditions, and computational fluid-particle dynamics (CFPD) models show that wind and relative humidity can lead to further movement of virus-laden droplets in the air, which would make the implemented social distancing rule inadequate (Feng et al., 2020). It is, however, important to note that without any specific pharmaceutical therapy or prophylaxis, strict social distancing (i.e., such as in China, where movement and contact of more than 500 million individuals across 80 cities were banned) is the primary means to control the pandemic (Du et al., 2020) and to prevent asymptomatic carriers—who are usually not identified and diagnosed—from transmitting the virus (Zhang et al., 2020c). It is reported that presymptomatic and asymptomatic individuals are responsible for at least 50% of community transmission (Subramanian et al., 2021). The median duration of virus shedding by asymptomatic cases has been varied significantly among different studies and ranged between 4.5 and 19 days, with one analysis reported positive RNA for up to 2 months in a few cases (Zhang et al., 2020c). Moreover, viral shedding occurs 2–3 days prior to the onset of symptoms (Peng et al., 2021).
A study among 149 countries concluded that using any of 5 different social and physical distancing techniques (closing schools, closing workplaces, limiting social gatherings, restricting movement, and implementing lockdowns) will result in a 13% reduction in disease incidence (Islam et al., 2020b), while the addition of 4 months of social distancing to isolation and quarantine interventions for those over 70 years old will decrease virus reproduction and decrease the number of deaths by almost half (Girum et al., 2020). It is reported that some countries, including France, Italy, Spain, Switzerland, and the UK, have been unable to contain the pandemic due to failure to implement and maintain adequate social distancing measures (Islam et al., 2020a). Moreover, with the exception of Switzerland and the UK, the other aforementioned countries failed to control the pandemic owing to a lack of prompt lockdown enforcement (Islam et al., 2020a).
In contrary to the aforementioned preventative measures, it has been argued that several other new and novel viruses have emerged throughout history (e.g., H2N2 influenza viruses (H2N2 in 1957, H3N2 in 1968, H5N1 in 2004, H1N1 in 2009, and seasonal influenza), SARS-CoV (2003), and MERS-CoV (2012)), yet, no social distancing, face masks, lockdown, and school closures or other extreme preventative measures were introduced (United Health Professionals, 2021), even though no vaccines were available at the time of outbreak onset. They further explain the unscientific logic behind the mandatory use of face masks by asymptomatic and healthy individuals by giving reasons such as 77% of influenza cases are asymptomatic (yet they are not being asked to no wear masks or practice social distancing), prolonged use of face masks increases mouth breathing, thus decreasing saliva and drying the mouth, leading to oral inflammation, cavities, and periodontal diseases. They also point out to the hygiene hypothesis, stating that strict hygiene measures can increase the risk of developing inflammatory, atopic, and autoimmune diseases, as well as some types of cancers (United Health Professionals, 2021). According to the 1989 hygiene hypothesis by an epidemiologist, Dr. David Strachan, lower incidence of unhygienic-induced childhood infections, such as viral respiratory infections (RSV, rhinoviruses, etc.) as a result of contact with other children (e.g., in daycare settings) can increase the risk of adulthood atopic diseases, such as asthma (Schaub et al., 2006), which is exacerbated in 85% of children by most viral respiratory tract infections (Morais-Almeida et al., 2020).
COVID-19 Risk Factors and Vulnerable Populations
People with Preexisting Conditions, the Elderly, and Others
According to CDC (2021b), regardless of age, any adults who have cancer, DM type-2, chronic kidney disease, immunocompromised status (e.g., solid organ transplant patients), obesity (i.e., body mass index [BMI] ≥ 40 kg/m2), or who are smokers are at increased risk of severe COVID-19. Adults with asthma, HTN, liver disease, a BMI of 25–30 kg/m2, pulmonary fibrosis, DM type-1, or immunocompromised status (e.g., blood or bone marrow transplants, HIV, or corticosteroid medication), among others, might be at increased risk (CDC, 2021b). It is reported that approximately 0.9% of all deaths were not linked with any preexisting medical conditions (Islam et al., 2020a). A systematic review and meta-analysis (Cao et al., 2020) reported 35.6% of COVID-19 patients had comorbidities. Another systematic review documented the most common comorbidities and conditions to be HTN (20.7%), CVD (9.6%), DM (9.55%), respiratory diseases (7%), and smoking (9%) (Hatmi, 2021). Abdi et al. (2020) reported DM as a risk factor for COVID-19, with diabetes being 14.5% prevalent in patients. While the signs and symptoms of COVID-19 are the same for both diabetics and nondiabetics, the former experience higher severity and mortality (Abdi et al., 2020). On the other hand, COVID-19 not only can exacerbate and complicate preexisting diabetes but can also result in the development of new-onset DM (due to endocrine pancreas involvement) (Rubino et al., 2020). Furthermore, the high levels of glucose, free fatty acids, and AT-II in those diabetic, obese, and hypertensive patients, respectively, are found to create a chronic inflammatory state leading to the hyperactivation of the NF-κB pathway, resulting in severe COVID-19 with the worst prognosis (Hariharan et al., 2020). Additionally, it is reported that individuals with DM type-2, hypertension, chronic pulmonary disease, as well as elderly, have greater expression of ACE2-R, thus making them more susceptible to SARS-CoV-2 infection (Solis & Nunn, 2021).
Moreover, age is clearly correlated with poor COVID-19 outcomes, evidenced by high mortality rates among nursing home residents (Fang et al., 2020), which account for 42%, 54%, and 44.6% of the total deaths reported in the USA, Ireland, and France, respectively (Thompson et al., 2020). Similarly, the mean age of death in both the UK and Italy was around 80, and in the UK, those above 65 years of age accounted for 87% of all deaths (Sornette et al., 2020). It is reported that the percentage of men aged 70–84 who may die from COVID-19 rises from about 5% for the original SARS-CoV-2 strain to more than 6% for B.1.1.7 variant. Furthermore, for males 85 years old or older, the risk of death increases from 17% for the original virus to almost 22% for the Alpha variant (Mallapaty, 2021). Elderly men are at higher risk of COVID-19 and death than elderly women, as the immune system reacts less robustly in older men, besides aging by itself in males substantially decreases the total numbers of lymphocytes (Perrotta et al., 2020). In addition, elderly men and postmenopausal women have reduced level of testosterone and estradiol, respectively, leading to the disinhibition of NF-κB pathway and hence increased level of TNFα and IL-6 and subsequent cytokine storm and resultant lung injury, since estrogen attenuates NF-κB signaling cascade and lower cytokine (e.g., IL-6, IL-8, and TNF-α) production (Hariharan et al., 2020).
Moreover, younger COVID-19 males have a better outcome than older men due to their higher levels of testosterone, as well as the peripheral conversion of testosterone to estrogen, which will double the anti-inflammatory effects; testosterone deficiency is also associated with higher levels of the inflammatory marker, such as CRP (Al-Lami et al., 2020). In fact, the therapeutic role of hormone replacement therapy (HRT) has been supported by a reduction in IL-1, IL-6, and TNF-α levels in postmenopausal COVID-19 patients, and the exogenous estrogen and testosterone are believed to act similar to corticosteroid in reducing SARS-CoV-2-induced multi-organ inflammatory damage without hindering the host antiviral immune response, while the anticatabolic effects of testosterone on respiratory muscles are believed to decrease the need for mechanical ventilation (Al-Lami et al., 2020). Furthermore, the lower COVID-19 incidence and mortality in young and middle-aged females compared to the male counterparts are due to their extra X chromosome which carries the majority of the genes responsible for controlling and regulating immune responses, thus providing women with a more effective innate immune response against viral infections while preventing cytokine storm development (Hariharan et al., 2020).
Preexisting mental distress can also have an impact on the likelihood of having COVID-19. For instance, in a cohort study performed in the USA, Nemani et al. (2021) reported the probability of having positive COVID-19 test for schizophrenia spectrum, mood, and anxiety disorders was reported to be 22.3%, 25.4%, and 24.1%, respectively, which could be attributed to SES, as well as high viral exposure due to environmental factors, including crowded living space, institutional settings, and lack of personal protective equipments. Additionally, schizophrenia is reported to be associated with abnormalities in cytokine signaling pathways, which result in severe COVID-19 and increased mortality (Nemani et al., 2021). Moreover, lower SES, and low social integration, will increase the risk of the development of chronic diseases, including heart, liver, and kidney disease, as well as DM, asthma, and stroke, resulting in higher susceptibility to SARS-CoV-2 infection (Solis & Nunn, 2021). Similarly, air pollution which affects 90% of people living in urban area globally increases the risk of chronic obstructive pulmonary disease (COPD), lung cancer, asthma, and respiratory infections. In fact, according to recent findings, air pollution is responsible for approximately 15% of worldwide COVID-19 mortality (Solis & Nunn, 2021).
Besides these non-modifiable factors, women are less likely to smoke and have other bad lifestyle habits and chronic conditions (e.g., HTN, diabetes, etc.), which lower their risk of infection (Zheng et al., 2020b). Moreover, even though men were shown to be more susceptible to SARS-CoV-2 infection, women account for a disproportionate percentage of healthcare workers, thus having a higher risk of hospital-acquired infection (Jensen et al., 2021). The prevalence of being a current smoker was reported to be significantly higher in critical or fatal COVID-19 patients compared to noncritical individuals (Zheng et al., 2020b). Furthermore, the percentage of blood type “A” was considerably greater in COVID-19 cases than that of healthy individuals (37.75% vs. 32.16%), whereas this value was significantly lower for type “O” (Zhao et al., 2020). This is explained by the presence of anti-A Abs (inhibitors of virus-ACE2-R interactions) in people with blood type “O,” which prevents viral binding to ACE2-R and makes them less susceptible to COVID-19 (Zhao et al., 2020). The aforementioned risk factors still do not explain the development of critical COVID-19, need for ICU admission, and eventually death in healthy young individuals; although a recent report suggests that previously formed undetected auto-Abs in some people may be the reason behind this (New Findings on the Pathophysiology of Severe COVID-19 Infections, 2021).
Pregnant and Breastfeeding Women and Neonates
The global incidence of COVID-19 in pregnant women is not known; however, a screening of all pregnant women admitted for delivery at a New York hospital revealed that 15.4% tested positive for SARS-CoV-2, and 87.9% of these cases were asymptomatic (Rodrigues et al., 2020). In general, pregnancy-induced immunosuppression is a risk factor for symptomatic infection which increases the risk of maternal (i.e., endotracheal intubation, MOF and ICU admission, DIC, etc.) and neonatal complications (i.e., vertical transmission, intrauterine growth retardation, spontaneous abortion), and thus prenatal screening for COVID-19 must be performed (Lopes de Sousa et al., 2020). A systematic review (Rodrigues et al., 2020) reported that pregnant women do not seem to be at higher risk of severe COVID-19 compared to nonpregnant women. Another systematic review and meta-analysis of pregnant women with COVID-19 reported that 50% needed C-sections (indications not reported) and 13% were admitted to the ICU, while 45% experienced complications such as placenta previa, premature rupture of membrane, or non-reassuring fetal status (Capobianco et al., 2020). According to Capobianco et al. (2020), 6% of neonates were infected—with unknown infection time (i.e., intrauterine, during vaginal delivery, or postnatal period)—and 39% of cases presented with neonatal complications, including fever, pneumonia, respiratory distress syndrome, and preterm birth (weight < 3.0 kg); however, no SARS-CoV-2-induced congenital malformations are yet reported (Rodrigues et al., 2020). In another systematic review, 68% of neonatal COVID-19 cases presented with symptoms, such as fever, GI symptoms, hypoxia, or cough, but even so, 75% breathed spontaneously (no intubation needed), and all patients were discharged after 10 days (Trevisanuto et al., 2020). Interestingly, vaginal delivery is not a contraindication in infected women due to negative vaginal screenings for the virus (Qiu et al., 2020) and of rare and mild cases of neonatal infection; thus, mother-infant separation is also not recommended by the WHO and UK guidelines (Gale et al., 2021), though a few countries (viz., China, Singapore, and South Korea) still recommended separation immediately after birth (Yeo et al., 2020). Except for Singapore and South Korea, most countries recommended that COVID-19-positive mothers breastfeed their neonates, while China recommends using pasteurized expressed breastmilk (Yeo et al., 2020).
Children and Adolescents
Children can still experience the same typical symptoms of fever, dry cough, fatigue, and so forth; however, they usually present with mild or asymptomatic disease, and unlike adults, age and gender are not risk factors (Castagnoli et al., 2020). The mild clinical symptoms in children are attributed to the lower expression of ACE2-R in their nasal epithelial compared to that of adults (Lee et al., 2020). However, childhood COVID-19 cases with complications were first observed and reported in the UK, where eight previously healthy children presented with symptoms of systemic hyper-inflammatory shock, called multisystem inflammatory syndrome in children (MIS-C) (Feldstein et al., 2020). MIS-C presented with CV (in 80%), hematologic (in 76%), mucocutaneous (in 74%), and respiratory (in 70%) symptoms, which were similar to those of KD—a rare pediatric vasculitis of unknown origin or possible abnormal immunological response to an infectious agent—yet, unlike KD, MIS-C predominantly affects children over the age of 5 and adolescents (Shaigany et al., 2020; Feldstein et al., 2020). This can result in coronary artery aneurysm in 25% of cases if left untreated (Gupta et al., 2020). Interestingly, cardiovascular manifestations have been reported in adults, including Kawasaki-like multisystem inflammatory syndrome, similar to the findings of complicated SARS-CoV-2 infection in children. For example, a 45-year-old male with no past medical history presented to the emergency department (ED) with fever, sore throat, diarrhea, bilateral pain in lower limbs, bilateral non-purulent conjunctivitis, periorbital erythema and edema, unilateral neck lymphadenopathy, diffuse skin rash, etc., meeting criteria for Kawasaki disease (KD) (Shaigany et al., 2020).
Severe COVID-19 GI complaints (e.g., diarrhea, vomit, or abdominal pain) were predominantly seen in children with COVID-19-related cardiac impairment (Giacomet et al., 2020). Among those hospitalized with MIS-C, 84.1% presented with GI symptoms (Miller et al., 2020). MIS-C was designated a reportable disease on May 14, 2020, by the CDC, which advises clinicians to report any cases meeting the criteria (Kest et al., 2020). However, according to Griffin et al. (2021), the MIS-C in children is similar to adult multisystem inflammatory syndrome (MIS-A).
Lower Socioeconomic Status (SES) and Ethnic Minorities
The pandemic has been found to disproportionately affect marginalized populations, people with lower SES, and racial and ethnic minorities (Fang et al., 2020). For instance, Black people (13% of the US population) account for 20% of COVID-19 cases and 22% of deaths, while Hispanics (18% of the US population) account for 33% of COVID-19 new cases (Blumenthal et al., 2020). A similar situation was reported in the UK, where COVID-19 mortality has been high among Black and Asian populations, as well as other ethnic minorities and those with lower SES (Han et al., 2020). In addition, compared to non-Hispanics, COVID-19 cases were more prevalent in American Indian/Native Alaskan and Hispanic/Latino people (CDC, 2021c). Furthermore, compared to white individuals, the American Indian/Native Alaskan group was 3.4 times more likely to be hospitalized, while this value was 2.8 times for Hispanic/Latino and Black/African American people. Moreover, the risk of COVID-19 death in these ethnicities was increased and ranged from 2.0 to 2.4 times (CDC, 2021c).
Moreover, exposure to SARS-CoV-2 is higher in those unable to social distance, including some of the world’s most vulnerable groups (e.g., prisoners, homeless, and refugees), and those who cannot work remotely from home (e.g., healthcare workers, cashiers, food preparation and waitresses/waiters, retail associates, and janitors) (Solis & Nunn, 2021). Moreover, low SES and unemployment create food insecurity, which force affected individuals to rely on food banks and/or work in hazardous settings, thus being at higher risk of SARS-CoV-2 exposure (Solis & Nunn, 2021). According to the US Bureau of Labor Statistics, in 2019, 29% of the population was able to work from home, while 25% could only occasionally do so; however, this value was lower in Hispanic (13%) and Black (18%) wage workers (Solis & Nunn, 2021). In addition, many of those able to work remotely had higher education level (at least a bachelor’s degree) and greater income (above the 75th percentile) (Solis & Nunn, 2021).
Another factor influencing access to health care is proximity to metropolitan areas, which have bigger hospitals with more advanced technology. For instance, people living in rural areas of the USA or sub-Saharan Africa have lower access to healthcare facilities and physicians for the treatment of severe COVID-19. Moreover, in the USA, other obstacles limiting equal healthcare access include race and gender discrimination, lack of health insurance, as well as disability status and SES (Solis & Nunn, 2021). Additionally, the imposed social distancing measures worsened healthcare access in some communities in the USA. A notable example is the Hualapai tribe that relies heavily on the revenue generated by tourists visiting the region; however, the social distancing measures compelled them to close down their tourist attraction site, which caused a scarcity of funds to sustain the healthcare facilities and clinics in the community (Solis & Nunn, 2021).
Hou et al. (2020) observed that the prevalence of ACE2 polymorphism was 54% and 39% among Non-Finnish European and African/African-American, respectively. This may explain the higher susceptibility to SARS-CoV-2 and even disease outcome in certain ethnic groups (Hou et al., 2020). In addition, the acquired genetic variantsFootnote 20 such as chromosome 3 (SLC6A20) and chromosome 9 (9q34), which interact with ACE2-R and ABO blood group locus, respectively, are also more prevalent in European ancestry and increase their risk of COVID-19 (Solis & Nunn, 2021). However mortality is still higher among Black, Native American, and Latinx communities, implying social and environmental parameters are particularly significant in determining COVID-19 outcome (Solis & Nunn, 2021).
In addition, comorbidities, such as obesity, DM, and vitamin D deficiency, show health disparities affecting COVID-19 outcomes. For example, in 2011–2012 in the USA, the rate of DM type-2 was higher among Black and Latinx communities, and American Indian/Alaskan native, Latinx, and Black communities (Solis & Nunn, 2021). Similarly, vitamin D deficiency is found disproportionately in 84.2% of non-Hispanic Black communities, compared to Hispanics (56.3%) and of non-Hispanic White American (34.8%) (Solis & Nunn, 2021).
Medical Insurance
“In an insurance-based health system, it’s insurers who foot the bill for pandemic care […] The pandemic is further complicating an already complicated system” (Roehr, 2020, p. 1). The US healthcare delivery and funding are based on a private for-profit insurance system (Ridic et al., 2012), and unlike most developed countries, where health insurance is provided to everyone regardless of their employment status (Santhanam, 2020), in the USA, health insurance is predominantly provided by the employer-sponsored health insurance (ESI) (Fronstin & Woodbury, 2020). Prior to the COVID-19 pandemic, more than 160 million Americans relied on ESI, and 30 million Americans had no health insurance (Santhanam, 2020). Analysis has already shown that if the unemployment rate reaches 20%, a maximum of 43 million people could lose their health insurance, which is fatal for those with serious diseases (The Lancet Oncology, 2020). The US pandemic-related unemployment reached its peak of 14.7% in April 2020 (Fronstin & Woodbury, 2020), and from March 2020 to September–October 2020, 60 million unemployment insurance (UI) claims have been filed, compared to the previous highest rate of 695,000 per week, which occurred the week of October 2, 1982, and for 20 weeks (from late March 2020), new unemployment claims surpassed 1 million per week (Cutler & Summers, 2020). Furthermore, minorities such as Black and Hispanic Americans, who are already affected by the pandemic as well as poverty and riskier jobs, disproportionately lack health insurance coverage, which further affects their health (Blumenthal et al., 2020). The US 2017 census has reported that the insurance coverage rate was higher for those above 400% of poverty (95.7%), as opposed to those under the 100% of poverty (83%); the uninsured rate of the US population was 8.7%, and of children (<19 years) was 5.4%, with the pediatric population (<19 years) living in poverty was to have the uninsured rate of 7.8%, compared to the uninsured rate of 4.9% for those children not in poverty (Berchick et al., 2018). Moreover, for those who are still able to work during the pandemic, employers have decreased insurance coverage, leaving many employees underinsured (Blumenthal et al., 2020). The 1985 Consolidated Omnibus Budget Reconciliation Act (COBRA) gives laid-off workers the option to still have health insurance even after unemployment; however, if not qualified for the government subsidy, the employees must pay $600 (for individual coverage) to $1800 (for family coverage) at maximum. In other words, the employee must pay 102% of the total premium (i.e., both the employee’s and the employer’s share), as well as a 2% administrative fee, leading to low COBRA demand (Agarwal & Sommers, 2020; Roehr, 2020).
Moreover, the 2014 Affordable Care Act (ACA) forbids private insurers to exclude, deny coverage, or charge a higher premium based on individual preexisting medical conditions (i.e., “declinable” medical conditions, such as pulmonary or heart diseases), jobs (i.e., “ineligible occupations” with a higher risk of infection), or serious psychological conditions (depression after losing a loved one); however, in June 2020, the Trump administration has asked the US Supreme Court to invalidate the ACA. If this happens, it will be catastrophic for patients, physicians, hospitals, etc., as insurers can even discriminate against individuals with acute or long-lasting COVID-19 (Pollitz & Michaud, 2020).
The need for health insurance has grown due to the need for the COVID-19 diagnosis and treatment (Agarwal & Sommers, 2020). The cost of COVID-19 diagnostic tests among the top two largest hospitals in each state was reported to range from $20 to $850 in the USA (Wapner, 2020), and the cost of severe COVID-19 treatments is up to $20,000 for hospitalization (or as high as $90,000 if ventilator support is indicated) and $3000 for 5-day remdesivir (Pollitz & Michaud, 2020). As a result, many Americans are not willing to be tested or to go to hospitals due to high costs and lack of insurance (Wapner, 2020). Even though The Families First Coronavirus Response Act (FFCRA) that was passed on March 18, 2020, ensures all persons, whether insured or not, have access to free testing, and The Coronavirus Aid, Relief, and Economic Security Act (CARES) also requires insurers to cover “out-network” testing claim, yet, due to legislation loopholes, many individuals still have to pay substantial amounts out of their pocket when visiting the emergency room (ER) or private healthcare facilities. For instance, if a healthcare facility is in the network of the patient insurer, but the ER physician is employed by another agency (i.e., out of the network of patient insurance plan), the patient may not be covered for the cost; or when asymptomatic or mild COVID-19 patients are asked to return for a follow-up (if their status worsens), they may be charged in case no testing is required; or in case a doctor requests for influenza (not COVID-19) to be ruled, CARES and FFCRA don’t mandate insurers to cover such costs (Wapner, 2020). Furthermore, as previously mentioned, COVID-19 affects older individuals more frequently, yet, several of the large insurance companies, such as Prudential, Lincoln National, and Protective Life, are imposing restrictions on new life insurance policies sold to those older than 80 by either suspending or delaying policy applications, and Securian does not take any applications from those who are 76 or older (Markowitz, 2021).
The failure of these health policies highlights the need to switch to a nonprofit social insurance model (Himmelstein & Woolhandler, 2020). For example, in the US neighbor, Canada, all the provincial healthcare systems must meet each and every principle of the Canada Health Act (CHA), including public administration, comprehensiveness, universality, and accessibility. The public administration refers to the provincial healthcare insurance being publicly administered and on a nonprofit basis; the comprehensiveness means that it is mandatory for provincial healthcare insurance to cover all the required and necessary healthcare services; by universality and accessibility, all Canadians must be provided with public healthcare insurance, and at no cost, respectively (Martin et al., 2018). Similarly, in order to promote a better healthcare system, the payments of health insurance benefits, off-site settlement, and financial compensation were all been established in China during the third phase of the pandemic (Islam et al., 2020a). Moreover, in China, everyone is covered for the cost of complex medical procedures through the established universal health coverage; hospitals are provided with additional funds and emergency equipment; and pharmaceutical companies are granted reimbursements to cover the cost of medications (Shadmi et al., 2020).
Death Tolls Among Different Nations
COVID-19 Infection Fatality Rate (IFR = COVID-19 deaths/total number of cases) varies significantly across different countries but is estimated to be 0.68% (Meyerowitz-Katz & Merone, 2020), while the mortality rate (MR) is reported to be 6.76% (Lu et al., 2020). Currently, the mortality rate associated with COVID-19 pandemics is markedly lower than that of SARS (9.6%) and notably lower than the mortality rate of MERS (34%) (Islam et al., 2020a). Sornette et al. (2020) have compared the number of deaths per million inhabitants among Western and Eastern block, developed Southeast Asian, Northern Hemisphere developing, and Southern Hemisphere countries (Fig. 7). Interestingly, the mortality patterns revealed that COVID-19 is responsible for more deaths in Western nations, which are most likely due to their higher proportion of elderly people (Sornette et al., 2020). For instance, high MR in Italy, where almost 60% of people are over the age of 40, is thought to be caused by population age (Triggle et al., 2020), and the high MR among developed countries (except Norway and Japan) that have “lavish healthcare systems” compared to developed Southeast Asian countries is also attributed to the higher age of their populations (Sornette et al., 2020). Even with the same public health measures, those countries with an older population still experienced higher IFR (e.g., Italy vs. Israel with a median age of 45.4 and 30 years, respectively) (Meyerowitz-Katz & Merone, 2020). It is estimated that IFR exponentially increases with age, from 0.005% for pediatric age groups to 0.2% for 50, 0.75% for 60, and 27% for those 85 years of age or older (Meyerowitz-Katz & Merone, 2020). It must be noted that depending on countries’ testing and screening strategies, as well as an official report, IFR can be overestimated or underestimated (Subramanian et al., 2021).
Population-normalized deaths per million among 55 countries. Notes: Logarithmic (top) and linear (bottom) population-normalized deaths per million among 55 countries as of July 14, 2020. Countries with no lockdown policy (Sweden, Brazil, and Belarus) are in bold. (Source: Sornette et al. (2020))
Moreover, the difference in higher infection and MR of various geographic regions can be attributed to disparities in SARS-CoV-2 mutations distribution. For instance, ORF3a mutation is found to be associated with higher MR, and it is reported to be more prevalent in countries with higher rates of infection and mortality (e.g., Brazil, Mexico, Spain, and the UK) (Majumdar & Niyogi, 2020). Furthermore, a preliminary study conducted in Italy on COVID-19 patients demonstrated a higher rate of SARS-CoV-2 infection and death in individuals carrying HLA-DRB1*08 since this allele and its subtypes are not capable of viral peptides recognition. Interestingly, the higher frequency of HLA-DRB1*08 allele was detected in most of Northern Italy, where a more cumulative incidence of COVID-19 was reported. On the other hand, the two provinces with low incidence showed a low level of this allele in their population (Amoroso et al., 2020).
In an evidence-based review, healthcare policies, access to resources, demographics, population characteristics (comorbidity, genetics [e.g., blood group], etc., discussed earlier), and viral variants, among other factors, were found to influence MR (Abu Hammad et al., 2020). As the COVID-19 death toll is on the rise globally, it is being recognized that disease mortality is distributed disproportionately among vulnerable groups, including the elderly, those with lower SES, residents of crowded places (e.g., prisons), migrants, refugees, and other minorities; the pandemic has also increased all-cause mortality, as a result of people losing their jobs, homes, and health insurance (Shadmi et al., 2020). For example, in Iran, the lack of medical supplies due to US-imposed sanctions and the government’s failure to implement COVID-19 measures is thought to have attributed to high MR (Triggle et al., 2020), while in other countries, the lack of ventilators could have increased the death rate among the elderly and other vulnerable groups, since medical staffs have been forced to choose some patients over others (Abu Hammad et al., 2020). Moreover, in India, a low-middle-income country with a high proportion of the low-income population and limited healthcare access, the implemented preventative measures (e.g., limiting gatherings and events, hand hygiene, etc.) by The Indian Ministry of Health and Welfare were most effective among the high-income population; this, combined with other sociocultural factors, such as congested living, religious gatherings, public transportation use, and so on, exacerbated the health disparity (Solis & Nunn, 2021).
The higher death rates in the USA and the UK compared to China and South Korea were due to the lack of screening of those with travel histories or contacts with suspected or confirmed cases (Yoo et al., 2020). Early detection can reduce MR (Das et al., 2020), and according to the chief executive of WHO:
“The most effective way to prevent infections and save lives is breaking the chains of transmission. You cannot fight a fire blindfolded, and we cannot stop this pandemic if we don’t know who is infected. We have a simple message for all countries: test, test, test, test” (Yoo et al., 2020, p. 10). According to reports, one of the reasons for the inability to control the pandemic in countries such as Turkey, the UK, and the USA was their failure to implement nationwide testing and contact tracing (Islam et al., 2020a).
Between May 10, 2020, and September 19, 2020, the top 6 countries with the highest COVID-19 MR still had lower deaths per 100,000 population than the USA (e.g., Italy 9.1/100000 vs. US’s 36.9/100000) (Bilinski & Emanuel, 2020). A lack of adequate measures by public health officials has made COVID-19 the leading cause of death in the USA (Woolf et al., 2020).
Biomedical Waste (BMW) Management
The composition of the generated BMW is 85% general nonhazardous and 15% hazardous infectious, and the sudden global surge in the volume of COVID-19-related hazardous infectious waste has created an additional burden for most governments, as they already did not own adequate capacities for the management of BMW generated even during regular times (United Nations Environment Programme, 2020). At the peak of the pandemic, 247 tons/day of BMW were produced in Wuhan, six times more than pre-pandemic levels (Singh et al., 2020). The daily amount of BMW (tons/day) during the COVID-19 pandemic has also increased in other countries, including the USA (8055.03), Brazil (2774.35), India (2160.34), Iran (81.31), and Italy (45.09) (Behera, 2021). According to the report, on average, 2.5 kg/bed/day of COVID-19-related BMW is produced in developed countries (United Nations Environment Programme, 2020). This pandemic-generated waste must be handled properly, as it will act as vectors not only for SARS-CoV-2 transmission and spread (Shammi et al., 2021) but will also contribute to the spread of other communicable diseases such as hepatitis, HIV, cholera, and typhoid (Singh et al., 2020). It is estimated that BMW-related diseases are responsible for killing 5.2 million people, including 4 million children, worldwide (Rahman et al., 2020b). It is still not known precisely how long SARS-CoV-2 survives on various fomites (WHO & UNICEF, 2020), but it is stated that it can remain viable on various fomites for 2–9 days, depending on the surface, temperature, relative humidity, and viral strain, as well as in serum, stool, and respiratory samples, for 11–21 days, 17–31 days, and 13–29 days, respectively (Behera, 2021). It is reported that when compared to aerosols, copper, and cardboard, the virus survives for a longer period of time (up to 72 h) on both stainless steel and plastic, and even for up to 96 h on glass surfaces (Islam et al., 2020a).
All the generated BMW during patient care, including those of COVID-19 patients, are considered infectious (Behera, 2021), and for proper handling, the 3Rs-based (reduce, reuse, and recycle) waste management hierarchy model must be followed to reduce waste generation and disposal in the first place and recover and recycle as many wastes as possible. This process starts from source segregation, storage, collection/transport, and treatment to the eventual disposal. The source segregation uses a double-layered bag, leak-proof and puncture-resistant (for sharp objects), color-coded and labeled bins, and separating reusable/recyclable items; on-site and off-site collection and transport of BMW must be done at regular times and routes, respectively, and utilize separate trollies for infectious/hazardous and general wastes and minimize contact with patients and other people; the storage site must be well-ventilated and secure to prevent human and animal pests access (United Nations Environment Programme, 2020). It is recommended that all trollies and transport vehicles be disinfected with 1% sodium hypochlorite, and the temporary storage of COVID-19 waste must not exceed 12 h (Behera, 2021). All BMW, especially COVID-19-generated waste, must be treated according to the local guidelines before their final disposal, and the most common ways for the treatment and dispose of these BMWs are chemical treatment, autoclaving, or incineration (Behera, 2021), with incineration being the most effective method (Peng et al., 2020). In China, to address this waste, an emergency incineration plant with a capacity of 30 tons/day was built, the national capacity for BMW management was also increased, and mobile incinerators were used to dispose the huge amount of BMW that accumulated during the lockdown and social distancing period (Singh et al., 2020). Even though incineration can be utilized for treatment and disposal, it produces toxic gases, such as furans and dioxins that increase the risk of developing cancer, diabetes, neurotoxicity, immunotoxicity, etc.; thus, many countries have used other alternative methods, including high-temperature pyrolysis and medium-temperature microwave (especially for on-site treatment), that completely eliminate digoxin (Behera, 2021). In situations where the number of incinerators does not meet the pandemic-generated waste volume, the final disposal can be done in a designated standard small landfill pit or in engineered sanitary landfills built far from residential and public areas (Behera, 2021).
The bodies of deceased suspected or confirmed COVID-19 individuals must be placed in fabric or cloth and sent to the mortuary as soon as possible (Hossain, 2020). However, before transferring them, all catheters and tubes must be removed, and their insertion sites must be disinfected with 1% hypochlorite, then sutured or covered with proper dressings; oral opening and nostrils must be packed completely; finally, the body must be put in a leak-proof plastic bag, and the outside must be disinfected using 1% hypochlorite and transported by a separate vehicle for cremation or burial (Behera, 2021).
In spite of all these rules and regulations, as a result of inadequate funding, shortage of equipment, poor knowledge of the dangers of hazardous wastes, and lack of qualified personnel, many countries (e.g., Bangladesh, India, etc.) still do not manage BMW properly (Shammi et al., 2021). According to reports, there are great disparities between rural and urban BMW handling, with rural regions lacking the practical knowledge of proper waste segregation and management. For instance, it is reported that in India, 82%, 60%, and 54% of primary, secondary, and tertiary healthcare institutes, respectively, do not own a proper BMW management facility (Shammi et al., 2021). Hence training healthcare staff and encouraging them to use PPE are recommended (United Nations Environment Programme, 2020). Table 6 summarizes COVID-19-related BMW types, with their segregation, treatment, and final disposal.
Artificial Intelligence and Other Technology Applications
Artificial intelligence (AI) techniques have the potential to be used for COVID-19 diagnosis, treatment, vaccine discovery, prognosis, epidemiology, and awareness-raising (Abd-Alrazaq et al., 2020) or reportedly even predicting pandemics before they erupt. The most common techniques are deep learning (DL) models (e.g., convolutional neural network [CNN], recurrent neural network [RNN]), machine learning (ML) models, and natural language processing (NLP) (Abd-Alrazaq et al., 2020). Having extracted data from various sources (e.g., global traveler movement, online papers, local healthcare workers reports, climate, animal data, etc.), the Toronto-based company, BlueDot, utilizes AI, ML, big data, and NLP to predict and track communicable disease outbreaks and claims to have warned officials in the private sectors of the novel coronavirus days before its detection in China (Bowles, 2020). CNN could be used as diagnostic and prognostic tools to detect and interpret changes seen in patients’ CXR and CT scans (Asraf et al., 2020). Furthermore, DL models (e.g., AlphaFold) can rapidly predict viral protein structures and help develop vaccines (Khemasuwan et al., 2020). Moreover, RNN and CNN algorithms are used in the laboratory for the rapid and accurate diagnosis of SARS-CoV-2 (Poongodi et al., 2021), and a hybrid of CNN and RNN (called pretrained molecule transformer-drug target interactions [MT-DTI]) can identify and predict the effect of existing antiviral drugs on SARS-CoV-2 life cycle (Beck et al., 2020). BenevolentAI was able to predict the potential efficacy of baricitinib, a drug used in rheumatoid arthritis patients, against COVID-19 (Zhou et al., 2020b). ML combined with bioinformatics and supercomputing can determine the Abs that are able to target RBD (Dey et al., 2020). Moreover, to better protect the public and to develop future preventative measures, big data technology is used to collect a huge amount of information from COVID-19 patients (Haleem et al., 2020), pretrained DL is used to classify cases according to all CXR and CT findings, and computer vision can be used to interpret and detect these findings (Ulhaq et al., 2020). Moreover, POCUS is an incredibly useful imaging option in remote locations since it is low in cost, can be connected to smartphones and tablets, and uses AI to assist in diagnosis (Yau et al., 2020).
Furthermore, IoT-based devices such as wearables, drones, robots, buttons, and smartphone applications are utilized for different purposes (Table 7) (Nasajpour et al., 2020). For example, Khan et al. (2020) reported that China and South Korea have successfully controlled the pandemic by using advanced technologies that reduced human interactions through various robots such as robot receptionists, doctors, and nurses; sampling robots; surgical robots for biopsies; CXR and LUS robots; sanitizer-dispensing robots; self-driving cars to transfer patients and collect lab samples; disinfectant robots (outdoor spraying robots and indoor UV robots); medicine-dispensing robots; and telemedicine robots, among others. Automated technology such as the Internet of Things (IoT) devices was also used in Wuhan to better control and dispose BMW (Singh et al., 2020).
In order to inform public health officials, several IoT technologies, such as “Worldometer,” monitor COVID-19 prevalence, incidence, and outcomes across countries (Ting et al., 2020), and Johns Hopkins University’s Center for Systems Science and Engineering used all the data from the US CDC, WHO, the European CDC, the Chinese CDC, and China’s National Health Commission to create a real-time tracking map that follows cases globally (Ting et al., 2020). Using the NLP algorithm, the Canadian Stallion company built Chatbots, a virtual healthcare assistant, to provide SARS-CoV-2-related information, answer any questions regarding coronavirus, monitor symptoms of infected cases, and give them appropriate recommendations whether they must take rest at home or visit hospitals for screening (Poongodi et al., 2021). Moreover, in order to monitor people’s maintenance of the 6-feet social distancing rule, Andrew Ng’s startup Landing AI developed a detector that transfers the result to a video screen in red and green color modes, indicating inadequate and adequate social distancing, respectively (Poongodi et al., 2021) Furthermore, once a likely infected person has been detected, Google Location History (GLH) can follow individuals’ movements and identify the places they have visited (Mohammed et al., 2020).
Research, Education, and Clinical Lessons Learned
The pandemic has had a dramatic long-term impact on scientific research, either by curtailing or closing in-progress clinical research or by redirecting it toward COVID-19 (Weiner et al., 2020). Almost a year into the pandemic, more than 64,000 related papers have been published (Fang et al., 2020), some without undergoing peer review (Weiner et al., 2020). More than 4500 COVID-19-related papers have been submitted to The Journal of Clinical Infectious Diseases, more than the usual number of yearly submissions, and browsing this tsunami of literature is very challenging for researchers and clinicians (Fang et al., 2020). In addition, the COVID-19 pandemic has drawn more attention to the already existing academic gender inequality by disproportionately impacting females more than males in regards to academic research productivity. For example, in the USA, since the implementation of the lockdown, women have approximately written 14% fewer social science research papers than men, and this decrease in research productivity was observed in many other countries, including Japan, China, Australia, Italy, the Netherlands, Switzerland, and the UK (Cui et al., 2020). This is attributed to the more and more countries having implemented social distancing and lockdowns of restaurants, schools, and daycares, resulting in a higher number of females, including women researchers have to unequally perform the majority of childcare and household works, which has been the case even for those gender-egalitarian northern European countries (Cui et al., 2020). Similarly, the pandemic lockdown has further increased the gap in academic gender inequality by preventing women from contributing less to COVID-19-related research papers, with women making up only 34%, 29%, and 26% of all authors, first authors, last authors, respectively (Pinho-Gomes et al., 2020). The percentage of women authors for 2020 pandemic-related studies has decreased by 16% relative to the percentage of women authors for all 37,531 papers published in 13 US medical journals in 2019; women have also registered fewer clinical trials and research projects in March–April 2020, compared to the same month in the previous year (Viglione, 2020).
The pandemic has also affected clinical trials. Weiner et al. (2020) reported the majority of ongoing and recruitment stage trials were either paused or switched to home administration, which highly impacted patients and researchers. Due to the rush to find an effective treatment for COVID-19, thousands of trials and studies have been published that contain misinformation and that do not meet clinical trial or FDA standards, endangering lives and causing resources to be diverted from more promising therapeutic agents (Weiner et al., 2020). Hopefully, the pandemic will lead to the emergence of better research models and sustained research infrastructure for public health emergencies and disasters (Weiner et al., 2020). The outbreak has also drawn attention to the shortage of critical care resources in low- and middle-income countries and to the need for preparation, education of healthcare staff, and modification of medical guidelines to better match and manage the needs of the local populations (The Lancet Respiratory Medicine, 2020).
The most important clinical lesson learned is the inaccuracy of the conventional airborne-droplet classification, as previously mentioned (Fang et al., 2020). The successful control of the pandemic in China, South Korea, Taiwan, and Vietnam due to aggressive and early action is also a valuable clinical lesson (Triggle et al., 2020), as is the value of increasing test availability in order to identify patients in the early stages of the disease since patients are contagious 1–2 days before becoming symptomatic (Fang et al., 2020). Further, the use of telemedicine for healthcare delivery and artificial intelligence for clinical decision-making could be beneficial (Gunasekeran et al., 2021), with the potential of expansion for future use (Kichloo et al., 2020). It is also worth mentioning other clinical lessons learned, such as IFN’s important role in the early phase of infection, remdesivir’s effect of shortening symptom duration, the usefulness of corticosteroids in critical cases on mechanical ventilation, and HCQ’s ineffectiveness (Fang et al., 2020). Moreover, the failure of many RCTs is likely due to the inappropriate timing of the administration of pharmaceutical agents. For example, the best time to administer prophylactic agents, active immunization (vaccines), or passive immunization (mAbs) is during the pre-exposure period (Griffin et al., 2021). Similarly, IFN-I therapy is beneficial in the preinfection or early infection period, while it is not effective or detrimental if administered at the late disease stage (Sa Ribero et al., 2020).
Following the COVID-19 pandemic, which has left governments and public health agencies in a state of shock and confusion, a call has been made for the adoption of “One Health” (OH) approaches in order to address the failure to predict and prevent the emergence of COVID-19 (de Garine-Wichatitsky et al., 2020). The WHO defines OH as “an approach to designing and implementing programs, policies, legislation and research in which multiple sectors communicate and work together to achieve better public health outcomes” (WHO, 2017). Adopted more than a decade ago and with the goal of creating a more widespread societal responsibility for human and environmental health (de Garine-Wichatitsky et al., 2020), OH approach acknowledges that the health of people, animals, and the environment are all intertwined and interdependent (Ruckert et al., 2020). This implies that those form a variety of sectors (e.g., human, animal, and plant health, as well as the environment) must collaborate to develop a response infrastructure that places an emphasis on information sharing and action coordination among various sectors (Ruckert et al., 2020). Solis and Nunn (2021) have coined the term OH disparities to argue that social environment is equally important to OH and may aid in illuminating disparities in the COVID-19 pandemic, such as viral origin, transmission, exposure, and interindividual susceptibility. Health disparities, according to The National Institute of Minority Health and Health Disparities, are the preventable diseases that emerge as a result of underlying systemic social issues, such as combination of lower SES, unemployment, education level, and social systemic racism (Solis & Nunn, 2021).
The term “spillover” refers to the transmission of a pathogen from nonhuman animals to human host, while the word “pillback” is commonly used to describe the transfer from humans to animals (Solis & Nunn, 2021). Therefore, identifying which animals may serve as new reservoirs is essential in the public health setting, while determining which human hosts will have the greatest interaction with those animals and become infected is necessary in the context of One Health Disparities (Solis & Nunn, 2021). For instance, the agricultural sector would be particularly vulnerable if SARS-CoV-2 were to establish itself in farm animals, leading to an increased demand for animal vaccinations and greater strain on financial and scientific resources, as well as the potential of emerging a new viral variant that would evade such vaccines (Solis & Nunn, 2021). In order for an animal species to be an effective reservoir host, SARS-CoV-2 must initially be transferred from humans to the specific species and further get established in the species population through intraspecies spread and eventually be reintroduced to humans (Sharun et al., 2021). Thus, it is reasonable to hypothesize about the potential presence of animal host reservoir, which as a result necessitates animal surveillance to monitor viral frequency in animal populations and the risk of spillover into human populations (Sharun et al., 2021).
Moreover, geographic information systems (GIS) and methods are increasingly being regarded by health professionals as important tools in monitoring and controlling infectious diseases, especially when a disease spread so rapidly; it is essential for knowledge and information to disseminate at an even faster pace (Kamel Boulos & Geraghty, 2020). In such circumstances, several SARS-CoV-2 map-based dashboards (e.g., WHO dashboard and Johns Hopkins University’s Center for Systems Science and Engineering (JHU CSSE) dashboard) become an essential tools in making information easily available, enhancing data transparency, while also assisting health official in the dissemination of information (Kamel Boulos & Geraghty, 2020). Developed by an epidemiologist (Lauren Gardner), JHU CSSE dashboard presents an interactive map that displays the number of confirmed cases, deaths, and recoveries, while graphs offer a visual representation of the progression of the virus over time; however, it lacks complete retrospective data visualization (Kamel Boulos & Geraghty, 2020). It relies on five reliable data sources, including WHO, the US-CDC, National Health Commission of the People’s Republic of China, European Centre for Disease Prevention and Control, and the Chinese online medical resource DXY.cn (Kamel Boulos & Geraghty, 2020). Moreover, the WHO dashboard only showed laboratory-confirmed cases, the JHU CSSE dashboard included cases that were diagnosed based on a combination of symptoms and chest imaging, resulting in approximately 18,000 additional reports. However, as of February 19, 2020, both dashboards display similar total case counts, and their numbers are consistent (Kamel Boulos & Geraghty, 2020). Even with GIS technology, it is difficult to track the pandemic, and according to Lauren Gardner, “it is especially challenging to collect good data at a fine spatial resolution, which is what most people want to know, and without having travel data in real-time that captures these altered mobility patterns, it is hard to assess what the geographic risk profile will look like moving forward” (Kamel Boulos & Geraghty, 2020).
Discussion and Conclusion
This chapter reviewed some of the most recent literature (from 2019 to 2021) on SARS-CoV-2 and the COVID-19 and various medical aspects of the virus and disease itself. There are still enormous unknown issues that are yet to be elucidated, and it would be difficult to reach a conclusion regarding SARS-CoV-2 evolutionary origin since the related viruses in bats are still poorly analyzed. The finding that pangolin SARS-CoV-2-related viruses and SARS-CoV-2 have nearly identical S-RBD does not make pangolin a definite intermediate host (Han, 2020). There are still several possible pathways for SARS-CoV-2 interspecies transmission, and hence future studies are warranted in order to find the exact origin (Ye et al., 2020). Moreover, it will not be known where and under what circumstances the recombination occurred that resulted in the emergence of SARS-CoV-2. Did the recombination take place between wild viral strains of bats, pangolins, or another species? Did it adapt to humans in a farm or laboratory animals and accidentally escaped from these places? Or was it the result of an intentional bioterrorism act of the viral genome modification by molecular engineering (Sallard et al., 2021)? In order to prevent other future outbreaks, regardless of the origin, it is essential to know how the virus breached the species boundary and acquired high transmissibility from human to human (Sallard et al., 2021). On the other hand, relying on R0 to measure transmissibility is not reliable, as the value of R0 and the transmission rate of the virus vary among various cohorts (e.g., low vs. high SES or refugees vs. non-refugees); hence, it is still challenging to measure SARS-CoV-2 transmissibility using R0 (Shaw & Kennedy, 2021). However, it has been reported that the reliability of R0 will increase as more information become available about this novel virus (Liu, Gayle, et al., 2020).
Moreover, the immune cells response to SARS-CoV-2 infections is still not well understood. Several studies attribute COVID-19 to the overactivity of the immune response, while others believe the T-cell dysfunction and exhaustion are responsible (Mathew et al., 2020). The underlying mechanisms behind the defective and dichotomous humoral and cellular responses in asymptomatic/mild or moderate/severe cases (Gao et al., 2021a), or the three different identified phenotypes among critically ill patients, remain unknown. As a result, various responses to pharmacotherapies, such as anti-inflammatory or immunomodulatory medications, are expected among different COVID-19 immunophenotypes (Dupont et al., 2020). The emerging mutations and variants of SARS-CoV-2 that allow the virus to evade host defense mechanisms, vaccines, or neutralizing Abs are the ones that raise public health concerns (Callaway, 2020). Thus, it is critical to understand SARS-CoV-2 genomic variants to better understand the pathogenesis and disease progression, implementation of therapeutic and preventative measures, and vaccine or drug developments (Laamarti et al., 2020).
Furthermore, Jaber et al. (2021) performed a study among 3167 participants from Jordan and Iraq to determine their level of knowledge and perception of COVID-19. They have reported that in both populations, the most common sources to obtain pandemic-related information in decreasing order were doctors/healthcare professionals, social media, and newspapers. On the other hand, citizens of Australia and other European countries (e.g., Italy, Germany, and the Netherlands) have used traditional media, including the television, to obtain such information, which might be due to the differences in the way their government officials reported pandemic news (Jaber et al., 2021). Moreover, their study explained that social media, which is flooded with misleading and inaccurate information, may explain why only 80% of the participants were aware of the already-established droplet route of transmission of SARS-CoV-2. Furthermore, those who are not aware of the route of transmission are less likely to adhere to the recommended preventative measures, which highlight the significance of healthcare authorities and other sectors using such platforms to raise public awareness to prevent the spread of the virus (Jaber et al., 2021).
In addition, in regards to the implemented preventative measures by governments, it should be noted that social distancing alone is not a “magic bullet,” and other factors, such as environmental setting, air ventilation, time spent indoors with others, viral loads, face mask use, and host factors play a role (Qureshi et al., 2020). Moreover, more stringent preventative measures by public health officials are warranted to reduce transmission of the VOCs and control the pandemic (Grubaugh et al., 2021). Furthermore, identifying the factors that contribute to the health disparities among various populations will enable the government, policymakers, and public health officials to distribute resources more effectively in order to achieve more equal health outcomes (Solis & Nunn, 2021). Additionally, modern-GIS technologies such as map-based dashboards have played a significant role in increasing awareness, as well as facilitating the surveillance, preparedness, and response to the COVID-19 outbreak (Kamel Boulos & Geraghty, 2020). Given the fact that viruses such as SARS-CoV-2 have little regard for national or continental borders, and the likelihood of similar pandemics occurring more frequently in the future, (i.e., it is not a matter of if but when and where the next outbreak will happen), it is important to consider the potential benefits of a such comprehensive GIS platforms in supporting the surveillance, preparedness, and response of another future outbreak (Kamel Boulos & Geraghty, 2020).
In addition, even though the use of technologies, such as AI, is fast compared to the conventional methods (e.g., an almost 30-min diagnosis with RT-PCR vs. few seconds with AI-inspected CT) (Abd-Alrazaq et al., 2020), choosing the appropriate technique to get the most accurate results could be challenging (Albahri et al., 2020). Additionally, even if accurate results are achieved, AI by itself is not the only solution to the pandemic; nevertheless, in the absence of AI, we will not be able to handle the next pandemic efficiently, and thus, in the future, AI-driven automation will play an important role (Poongodi et al., 2021). The COVID-19 pandemic has also resulted in unemployment and loss of medical insurance, further exacerbating the already complicated healthcare system. Thus more efficient health insurance policies are required to cover everyone, especially the underprivileged individuals of lower SES. Furthermore, the pandemic has resulted in an exponential increase in the amount of BMWs produced. For this reason, in order to better control future pandemics and decrease further spread of the virus, there is an urgent need to increase the capacity for BMW treatment and disposal facilities; to identify shortcomings in rural and urban places; to supply adequate equipment (e.g., bins, bags, transportation trollies, PPEs); and to train all the healthcare professionals involved in COVID-19-related issues (United Nations Environment Programme, 2020).
In conclusion, there are still a plethora of unanswered questions and concerns that must be addressed in regards to SARS-CoV-2 and COVID-19. More research is required to elucidate and fully understand the origin and pathogenesis of SARS-CoV-2, the types of immunologic response, risk factors, interindividual variabilities in clinical findings, and most promising treatments for each specific case, and the duration of vaccine-induced immunity. Finally, what could have been done differently, and more effectively in the first place, and when the pandemic will resolve we will return to the pre-pandemic state, are all crucial issues to consider.
Notes
- 1.
Upon microbe (antigen) entry into the host body, host immune cells are stimulated to produce antibodies (Abs), such as immunoglobulin M and/or G that are specific to the corresponding antigen (Clem, 2011).
- 2.
Detection of antigen-specific Abs in patient serum, using various serological immunoassays (Vainionpää & Leinikki, 2008).
- 3.
Serotherapy is a type of passive immunization against numerous infectious diseases, using purified serum of infected or vaccinated individuals that contain specific Abs against the disease in question (Hifumi et al., 2017).
- 4.
For example, the production of new strains of influenza virus due to genetic drift and the drastic genetic shift are responsible for the seasonal and pandemic influenza, respectively (Casanova & Abel, 2020).
- 5.
Antimicrobial substances can select for and result in the emergence of new strains of microbe that are resistance to antimicrobial agents (Casanova & Abel, 2020).
- 6.
In contrast to monozygotic twins, who are genetically identical, dizygotic twins are genetically distinct in that they only share approximately about half of their genetic material (Burgner et al., 2006).
- 7.
NK cells and CD8+ T cells, components of innate and adaptive immune systems, respectively, each have distinct mechanisms to recognize and kill infected cells (Rosenberg & Huang, 2018).
- 8.
Recombination refers to the transfer genetic materials, as well as harmful traits between same virus, but different strains, which allows for the emergence of a novel virus that the host has never encountered or acquired immunity against have not previously been encountered by the host population (Gibson et al., 2015).
- 9.
United Health Professionals has more than 1500 members, who are professors of medicine, intensive care unit doctors, and infectologists (United Health Professionals, 2021).
- 10.
Transplacental and intrapartum refers to viral transmission across placenta and direct contact of the baby with the genital tract during vaginal delivery, respectively (Konstantinidou et al., 2021).
- 11.
Opsonization is an immunological process that involves the attachment of opsonins, such as preformed Abs, to tag invading pathogens and then allowing them to be destroyed by phagocytes.
- 12.
Opsonization is an immunological process that involves the attachment of opsonins, such as preformed Abs, to tag invading pathogens and then allowing them to be destroyed by phagocytes.
- 13.
The term variant is misleading, as two viral variants may vary by a single mutation or by a large number of mutations (Lauring & Hodcroft, 2021).
- 14.
An antigen ability to elicit a cellular and humoral immune response is termed immunogenicity, whereas the ability to be recognized by antigen-specific antibodies is called antigenicity (Ilinskaya & Dobrovolskaia, 2016).
- 15.
In the substitution mutations, one amino acid (first letter) is replaced at a specific position in the protein sequence (middle number) with another amino acid (second letter). For example, D614G refers to a substitution of aspartic acid (D) to glycine (G) at amino acid position 614 of the spike glycoprotein (Khateeb et al., 2021).
- 16.
The ability of a screening test to accurately detect all individuals who have the disease (true positive) is referred to as its sensitivity (Trevethan, 2017).
- 17.
The ability of a screening test to accurately detect all individuals who do not have the disease (true negative) is referred to as its specificity (Trevethan, 2017).
- 18.
The positive predictive value (PPV) is the probability that those individuals who have tested positive for the disease in the screening test, truly have the disease in question (Trevethan, 2017).
- 19.
The negative predictive value (NPV) is the probability that those individuals who have tested negative for the disease in the screening test, truly do not have the disease in question (Trevethan, 2017).
- 20.
References
Abbas, A. K., Lichtman, A. H., & Pillai, S. (2015). Cellular and molecular immunology. Saunders/Elsevier.
Abd-Alrazaq, A., Alajlani, M., Alhuwail, D., Schneider, J., Al-Kuwari, S., Shah, Z., Hamdi, M., & Househ, M. (2020). Artificial intelligence in the fight against COVID-19: Scoping review. Journal of Medical Internet Research, 22(12), e20756. https://doi.org/10.2196/20756
Abdi, A., Jalilian, M., Sarbarzeh, P. A., & Vlaisavljevic, Z. (2020). Diabetes and COVID-19: A systematic review on the current evidences. Diabetes Research and Clinical Practice, 166, 108347. https://doi.org/10.1016/j.diabres.2020.108347
Abu Hammad, O., Alnazzawi, A., Borzangy, S. S., Abu-Hammad, A., Fayad, M., Saadaledin, S., Abu-Hammad, S., & Dar Odeh, N. (2020). Factors influencing global variations in COVID-19 cases and fatalities; A review [Review]. Healthcare, 8(3), 216. https://doi.org/10.3390/healthcare8030216
Adam, D. C., Wu, P., Wong, J. Y., Lau, E. H. Y., Tsang, T. K., Cauchemez, S., Leung, G. M., & Cowling, B. J. (2020). Clustering and superspreading potential of SARS-CoV-2 infections in Hong Kong. Nature Medicine, 26(11), 1714–1719. https://doi.org/10.1038/s41591-020-1092-0
Adams, H. J. A., Kwee, T. C., Yakar, D., Hope, M. D., & Kwee, R. M. (2020). Systematic review and meta-analysis on the value of chest CT in the diagnosis of coronavirus disease (COVID-19): Sol Scientiae, Illustra Nos. American Journal of Roentgenology, 215(6), 1342–1350. https://doi.org/10.2214/ajr.20.23391
Agarwal, S., & Sommers, B. D. (2020). Insurance Coverage after Job Loss — The Importance of the ACA during the Covid-Associated Recession. The New England Journal of Medicine, 383(17), 1603–1606. https://doi.org/10.1056/nejmp2023312
Agyeman, A. A., Chin, K. L., Landersdorfer, C. B., Liew, D., & Ofori-Asenso, R. (2020). Smell and taste dysfunction in patients with COVID-19: A systematic review and meta-analysis. Mayo Clinic Proceedings, 95(8), 1621–1631. https://doi.org/10.1016/j.mayocp.2020.05.030
Alanagreh, L., Alzoughool, F., & Atoum, M. (2020). The human coronavirus disease COVID-19: Its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens, 9(5), 331. https://doi.org/10.3390/pathogens9050331
Albahri, O. S., Zaidan, A. A., Albahri, A. S., Zaidan, B. B., Abdulkareem, K. H., Al-qaysi, Z. T., Alamoodi, A. H., Aleesa, A. M., Chyad, M. A., Alesa, R. M., Kem, L. C., Lakulu, M. M., Ibrahim, A. B., & Rashid, N. A. (2020). Systematic review of artificial intelligence techniques in the detection and classification of COVID-19 medical images in terms of evaluation and benchmarking: Taxonomy analysis, challenges, future solutions and methodological aspects. Journal of Infection and Public Health, 13(10), 1381–1396. https://doi.org/10.1016/j.jiph.2020.06.028
Al-Horani, R. A., Kar, S., & Aliter, K. F. (2020). Potential anti-COVID-19 therapeutics that block the early stage of the viral life cycle: Structures, mechanisms, and clinical trials. International Journal of Molecular Sciences, 21(15), 5224. https://doi.org/10.3390/ijms21155224
Al-Lami, R. A., Urban, R. J., Volpi, E., Algburi, A. M., & Baillargeon, J. (2020). Sex hormones and novel corona virus infectious disease (COVID-19). Mayo Clinic Proceedings, 95(8), 1710–1714. https://doi.org/10.1016/j.mayocp.2020.05.013
Al-Samkari, H., Karp Leaf, R. S., Dzik, W. H., Carlson, J. C., Fogerty, A. E., Waheed, A., ... & Rosovsky, R. P. (2020). COVID-19 and coagulation: bleeding and thrombotic manifestations of SARS-CoV-2 infection. Blood, 136(4), 489–500.
Alzahrani, A. S., Mukhtar, N., Aljomaiah, A., Aljamei, H., Bakhsh, A., Alsudani, N., Elsayed, T., Alrashidi, N., Fadel, R., Alqahtani, E., Raef, H., Butt, M. I., & Sulaiman, O. (2021). The impact of COVID-19 viral infection on the hypothalamic-pituitary-adrenal axis. Endocrine Practice, 27(2), 83–89. https://doi.org/10.1016/j.eprac.2020.10.014
Amoroso, A., Magistroni, P., Vespasiano, F., Bella, A., Bellino, S., Puoti, F., Alizzi, S., Vaisitti, T., Boros, S., Grossi, P. A., Trapani, S., Lombardini, L., Pezzotti, P., Deaglio, S., Brusaferro, S., & Cardillo, M. (2020). HLA and AB0 polymorphisms may influence SARS-CoV-2 infection and COVID-19 severity. Transplantation, 105(1), 193–200. https://doi.org/10.1097/tp.0000000000003507
Andersen, K. G., Rambaut, A., Lipkin, W. I., Holmes, E. C., & Garry, R. F. (2020). The proximal origin of SARS-CoV-2. Nature Medicine, 26(4), 450–452. https://doi.org/10.1038/s41591-020-0820-9
Arnold, M. L., & Martin, N. H. (2009). Adaptation by introgression. Journal of Biology, 8(9), 82. https://doi.org/10.1186/jbiol176
Asraf, A., Islam, M. Z., Haque, M. R., & Islam, M. M. (2020). Deep learning applications to combat novel coronavirus (COVID-19) pandemic. SN Computer Science, 1(6), 363. https://doi.org/10.1007/s42979-020-00383-w
Awulachew, E., Diriba, K., Anja, A., Getu, E., & Belayneh, F. (2020). Computed tomography (CT) imaging features of patients with COVID-19: Systematic review and meta-analysis. Radiology Research and Practice, 2020, 1–8. https://doi.org/10.1155/2020/1023506
Babaei, F., Mirzababaei, M., Nassiri-Asl, M., & Hosseinzadeh, H. (2020). Review of registered clinical trials for the treatment of COVID-19. Drug Development Research, 82, 474. https://doi.org/10.1002/ddr.21762
Bakheet, N., Fouad, R., Kassem, A. M., Hussin, W., & El-Shazly, M. (2020). Persistent hiccup: A rare presentation of COVID-19. Respiratory Investigation, 59(2), 263–265. Advance online publication. https://doi.org/10.1016/j.resinv.2020.11.003
Banerjee, A. K., Blanco, M. R., Bruce, E. A., Honson, D. D., Chen, L. M., Chow, A., Bhat, P., Ollikainen, N., Quinodoz, S. A., Loney, C., Thai, J., Miller, Z. D., Lin, A. E., Schmidt, M. M., Stewart, D. G., Goldfarb, D., De Lorenzo, G., Rihn, S. J., Voorhees, R. M., et al. (2020). SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell, 183(5), 1325–1339.e21. https://doi.org/10.1016/j.cell.2020.10.004
Bastian, B., Vauclair, C.-M., Loughnan, S., Bain, P., Ashokkumar, A., Becker, M., Bilewicz, M. ł., Collier-Baker, E., Crespo, C., Eastwick, P. W., Fischer, R., Friese, M., Gómez, Á., Guerra, V. M., Guevara, J. L. C., Hanke, K., Hooper, N., Huang, L.-L., Junqi, S., et al. (2019). Explaining illness with evil: Pathogen prevalence fosters moral vitalism. Proceedings of the Royal Society B: Biological Sciences, 286(1914), 20191576. https://doi.org/10.1098/rspb.2019.1576
Beck, B. R., Shin, B., Choi, Y., Park, S., & Kang, K. (2020). Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Computational and Structural Biotechnology Journal, 18, 784–790. https://doi.org/10.1016/j.csbj.2020.03.025
Behera, B. C. (2021). Challenges in handling COVID-19 waste and its management mechanism: A review. Environmental Nanotechnology, Monitoring and Management, 15, 100432. https://doi.org/10.1016/j.enmm.2021.100432
Beigel, J. H., Tomashek, K. M., Dodd, L. E., Mehta, A. K., Zingman, B. S., Kalil, A. C., Hohmann, E., Chu, H. Y., Luetkemeyer, A., Kline, S., Lopez de Castilla, D., Finberg, R. W., Dierberg, K., Tapson, V., Hsieh, L., Patterson, T. F., Paredes, R., Sweeney, D. A., Short, W. R., & Touloumi, G. (2020). Remdesivir for the treatment of Covid-19 – Final report. New England Journal of Medicine, 383(19), 1813–1826. https://doi.org/10.1056/NEJMoa2007764
Belizário, J. E. (2021). Immunity, virus evolution, and effectiveness of SARS-CoV-2 vaccines. Brazilian Journal of Medical and Biological Research, 54(5), e10725. https://doi.org/10.1590/1414-431x202010725
Berchick, E. R., Hood, E., & Barnett, J. C. (2018). Health insurance coverage in the United States: 2017. https://www.census.gov/content/dam/Census/library/publications/2018/demo/p60-264.pdf. This link is external to health.gov
Bhoi, S., Sahu, A. K., Mathew, R., & Sinha, T. P. (2020). Point-of-care ultrasound in COVID-19 pandemic. Postgraduate Medical Journal, 97(1143), 62–63. https://doi.org/10.1136/postgradmedj-2020-137853
Bilinski, A., & Emanuel, E. J. (2020). COVID-19 and excess all-cause mortality in the US and 18 comparison countries. JAMA, 324(20), 2100. https://doi.org/10.1001/jama.2020.20717
Blackwell, J. M., Jamieson, S. E., & Burgner, D. (2009). HLA and infectious diseases. Clinical Microbiology Reviews, 22(2), 370–385. https://doi.org/10.1128/cmr.00048-08
Blumenthal, D., Fowler, E. J., Abrams, M., & Collins, S. R. (2020). COVID-19—Implications for the health care system. New England Journal of Medicine, 383(15), 1483–1488. https://doi.org/10.1056/NEJMsb2021088
Bowles, J. (2020, March 10). How Canadian AI start-up BlueDot spotted coronavirus before anyone else had a clue. Diginomica. Accessed 18 Apr 2020.
Burgner, D., Jamieson, S. E., & Blackwell, J. M. (2006). Genetic susceptibility to infectious diseases: Big is beautiful, but will bigger be even better? The Lancet Infectious Diseases, 6(10), 653–663. https://doi.org/10.1016/s1473-3099(06)70601-6
Burki, T. (2020). China’s successful control of COVID-19. Lancet. Infectious Diseases, 20(11), 1240–1241. https://doi.org/10.1016/S1473-3099(20)30800-8
Callaway, E. (2020). The coronavirus is mutating—Does it matter? Nature, 585(7824), 174–177. https://doi.org/10.1038/d41586-020-02544-6
Callaway, E. (2021). Delta coronavirus variant: Scientists brace for impact. Nature, 595(7865), 17–18. https://doi.org/10.1038/d41586-021-01696-3
Cao, Y., Liu, X., Xiong, L., & Cai, K. (2020). Imaging and clinical features of patients with 2019 novel coronavirus SARS‐CoV‐2: a systematic review and meta‐analysis. Journal of Medical vi-rology, 92(9), 1449–1459.
Campagnano, S., Angelini, F., Fonsi, G. B., Novelli, S., & Drudi, F. M. (2021). Diagnostic imaging in COVID-19 pneumonia: A literature review. Journal of Ultrasound, 24, 383–395. https://doi.org/10.1007/s40477-021-00559-x
Cantuti-Castelvetri, L., Ojha, R., Pedro, L. D., Djannatian, M., Franz, J., Kuivanen, S., van der Meer, F., Kallio, K., Kaya, T. Ğ., Anastasina, M., Smura, T., Levanov, L., Szirovicza, L., Tobi, A., Kallio-Kokko, H., Österlund, P., Joensuu, M., Meunier, F. A., Butcher, S. J., et al. (2020). Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science, 370(6518), 856–860. https://doi.org/10.1126/science.abd2985
Capobianco, G., Saderi, L., Aliberti, S., Mondoni, M., Piana, A., Dessole, F., Dessole, M., Cherchi, P. L., Dessole, S., & Sotgiu, G. (2020). COVID-19 in pregnant women: A systematic review and meta-analysis. European Journal of Obstetrics, Gynecology, and Reproductive Biology, 252, 543–558. https://doi.org/10.1016/j.ejogrb.2020.07.006
Casanova, J. L., & Abel, L. (2007). Human genetics of infectious diseases: A unified theory. The EMBO Journal, 26(4), 915–922. https://doi.org/10.1038/sj.emboj.7601558
Casanova, J. L., & Abel, L. (2013). The genetic theory of infectious diseases: A brief history and selected illustrations. Annual Review of Genomics and Human Genetics, 14(1), 215–243. https://doi.org/10.1146/annurev-genom-091212-153448
Casanova, J. L., & Abel, L. (2018). Human genetics of infectious diseases: Unique insights into immunological redundancy. Seminars in Immunology, 36, 1–12. https://doi.org/10.1016/j.smim.2017.12.008
Casanova, J. L., & Abel, L. (2020). The human genetic determinism of life-threatening infectious diseases: Genetic heterogeneity and physiological homogeneity? Human Genetics, 139(6–7), 681–694. https://doi.org/10.1007/s00439-020-02184-w
Castagnoli, R., Votto, M., Licari, A., Brambilla, I., Bruno, R., Perlini, S., Rovida, F., Baldanti, F., & Marseglia, G. L. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents. JAMA Pediatrics, 174(9), 882. https://doi.org/10.1001/jamapediatrics.2020.1467
Centers for Disease Control and Prevention. (2017, September 27). Quarantine and isolation. https://www.cdc.gov/quarantine/index.html
Centers for Disease Control and Prevention. (2020, February 11). COVID-19 and your health: Social distancing. https://www.cdc.gov/coronavirus/2019-ncov/prevent-getting-sick/social-distancing.html
Centers for Disease Control and Prevention. (2021a, July 27). Coronavirus disease 2019 (COVID-19). https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html#Consequence
Centers for Disease Control and Prevention. (2021b, May 13). COVID-19 and your health. (2020, February 11). https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html?CDC_AA_refVal=https%3A%2F%2Fwww.cdc.gov%2Fcoronavirus%2F2019-ncov%2Fneed-extra-precautions%2Fgroups-at-higher-risk.html
Centers for Disease Control and Prevention. (2021c, July 16). Risk for COVID-19 infection, hospitalization, and death by race/ethnicity. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-discovery/hospitalization-death-by-race-ethnicity.html
Cevik, M., Marcus, J. L., Buckee, C., & Smith, T. C. (2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) transmission dynamics should inform policy. Clinical Infectious Diseases, 73, S170. https://doi.org/10.1093/cid/ciaa1442
Chen, P., Nirula, A., Heller, B., Gottlieb, R. L., Boscia, J., Morris, J., Huhn, G., Cardona, J., Mocherla, B., Stosor, V., Shawa, I., Adams, A. C., Van Naarden, J., Custer, K. L., Shen, L., Durante, M., Oakley, G., Schade, A. E., Sabo, J., et al. (2021). SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. New England Journal of Medicine, 384(3), 229–237. https://doi.org/10.1056/nejmoa2029849
Choo, S. Y. (2007). The HLA system: Genetics, immunology, clinical testing, and clinical implications. Yonsei Medical Journal, 48(1), 11. https://doi.org/10.3349/ymj.2007.48.1.11
Chu, D., & Wei, L. (2019). Nonsynonymous, synonymous and nonsense mutations in human cancer-related genes undergo stronger purifying selections than expectation. BMC Cancer, 19(1). https://doi.org/10.1186/s12885-019-5572-x
Clem, A. (2011). Fundamentals of vaccine immunology. Journal of Global Infectious Diseases, 3(1), 73. https://doi.org/10.4103/0974-777x.77299
Colson, P., Finaud, M., Levy, N., Lagier, J.-C., & Raoult, D. (2020). Evidence of SARS-CoV-2 re-infection with a different genotype. Journal of Infection, 82, 84. https://doi.org/10.1016/j.jinf.2020.11.011
Coppo, A., Bellani, G., Winterton, D., Di Pierro, M., Soria, A., Faverio, P., Cairo, M., Mori, S., Messinesi, G., Contro, E., Bonfanti, P., Benini, A., Valsecchi, M. G., Antolini, L., & Foti, G. (2020). Feasibility and physiological effects of prone positioning in non-intubated patients with acute respiratory failure due to COVID-19 (PRON-COVID): A prospective cohort study. The Lancet Respiratory Medicine, 8(8), 765–774. https://doi.org/10.1016/s2213-2600(20)30268-x
Costela-Ruiz, V. J., Illescas-Montes, R., Puerta-Puerta, J. M., Ruiz, C., & Melguizo-Rodríguez, L. (2020). SARS-CoV-2 infection: The role of cytokines in COVID-19 disease. Cytokine & Growth Factor Reviews, 54, 62–75. https://doi.org/10.1016/j.cytogfr.2020.06.001
Cox, R. J., & Brokstad, K. A. (2020). Not just antibodies: B cells and T cells mediate immunity to COVID-19. Nature Reviews Immunology, 20(10), 581–582. https://doi.org/10.1038/s41577-020-00436-4
Cui, R., Ding, H., & Zhu, F. (2020). Gender inequality in research productivity during the COVID-19 pandemic. SSRN Electronic Journal, 1–30. https://doi.org/10.2139/ssrn.3623492
Cutler, D. M., & Summers, L. H. (2020). The COVID-19 pandemic and the $16 trillion virus. JAMA, 324(15), 1495–1496. https://doi.org/10.1001/jama.2020.19759
Danwang, C., Endomba, F. T., Nkeck, J. R., Wouna, D. L. A., Robert, A., & Noubiap, J. J. (2020). A meta-analysis of potential biomarkers associated with severity of coronavirus disease 2019 (COVID-19). Biomarker Research, 8(1), 37. https://doi.org/10.1186/s40364-020-00217-0
Das, R., Hossain, A., Mandal, S., Pariari, D., Rohj, R. K., Shukla, S., Varma, R. M., & Sarma, D. D. (2020). A comparative study of infection and mortality in COVID-19 across countries: A scaling analysis. medRxiv. https://doi.org/10.1101/2020.10.13.20212175
Davies, N. G., Abbott, S., Barnard, R. C., Jarvis, C. I., Kucharski, A. J., Munday, J. D., Pearson, C. A. B., Russell, T. W., Tully, D. C., Washburne, A. D., Wenseleers, T., Gimma, A., Waites, W., Wong, K. L. M., van Zandvoort, K., Silverman, J. D., Diaz-Ordaz, K., Keogh, R., Eggo, R. M., et al. (2021). Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science, 372(6538), eabg3055. https://doi.org/10.1126/science.abg3055
Davis, J. S., Ferreira, D., Denholm, J. T., & Tong, S. Y. C. (2020). Clinical trials for the prevention and treatment of COVID-19: Current state of play. Medical Journal of Australia, 213(2), 86–93. https://doi.org/10.5694/mja2.50673
de Garine-Wichatitsky, M., Binot, A., Morand, S., Kock, R., Roger, F., Wilcox, B. A., & Caron, A. (2020). Will the COVID-19 crisis trigger a One Health coming-of-age? The Lancet Planetary Health, 4(9), e377–e378. https://doi.org/10.1016/s2542-5196(20)30179-0
de Rivero Vaccari, J. C., Dietrich, W. D., Keane, R. W., & de Rivero Vaccari, J. P. (2020). The inflammasome in times of COVID-19. Frontiers in Immunology, 11, 583373. https://doi.org/10.3389/fimmu.2020.583373
Deshmukh, V., Motwani, R., Kumar, A., Kumari, C., & Raza, K. (2021). Histopathological observations in COVID-19: A systematic review. Journal of Clinical Pathology, 74(2), 76–83. https://doi.org/10.1136/jclinpath-2020-206995
Dey, L., Chakraborty, S., & Mukhopadhyay, A. (2020). Machine learning techniques for sequence-based prediction of viral–host interactions between SARS-CoV-2 and human proteins. Biomedical Journal, 43(5), 438–450. https://doi.org/10.1016/j.bj.2020.08.003
Dhama, K., Khan, S., Tiwari, R., Sircar, S., Bhat, S., Malik, Y. S., Singh, K. P., Chaicumpa, W., Bonilla-Aldana, D. K., & Rodriguez-Morales, A. J. (2020). Coronavirus disease 2019-COVID-19. Clinical Microbiology Reviews, 33(4), e00028–e00020. https://doi.org/10.1128/CMR.00028-20
Dhangadamajhi, G., & Rout, R. (2021). Association of TLR3 functional variant (rs3775291) with COVID-19 susceptibility and death: A population-scale study. Human Cell, 34(3), 1025–1027. https://doi.org/10.1007/s13577-021-00510-6
Du, Z., Xu, X., Wang, L., Fox, S. J., Cowling, B. J., Galvani, A. P., & Meyers, L. A. (2020). Effects of proactive social distancing on COVID-19 outbreaks in 58 cities, China. Emerging Infectious Diseases, 26(9), 2267–2269. https://doi.org/10.3201/eid2609.201932
Duan, L., Zheng, Q., Zhang, H., Niu, Y., Lou, Y., & Wang, H. (2020). The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: Implications for the design of spike-based vaccine immunogens. Frontiers in Immunology, 11, 576622. https://doi.org/10.3389/fimmu.2020.576622
Dupont, T., Caillat-Zucman, S., Fremeaux-Bacchi, V., Morin, F., Lengliné, E., Darmon, M., Peffault de Latour, R., Zafrani, L., Azoulay, E., & Dumas, G. (2020). Identification of distinct immunophenotypes in critically ill coronavirus disease 2019 patients. Chest, 159, 1884. https://doi.org/10.1016/j.chest.2020.11.049
Ebrahimi, M., Malehi, A. S., & Rahim, F. (2020). COVID-19 patients: A systematic review and meta-analysis of laboratory findings, comorbidities, and clinical outcomes comparing medical staff versus the general population. Osong Public Health and Research Perspectives, 11(5), 269–279. https://doi.org/10.24171/j.phrp.2020.11.5.02
Falahi, S., & Kenarkoohi, A. (2020). COVID-19 reinfection: Prolonged shedding or true reinfection? New Microbes and New Infections, 38, 100812. https://doi.org/10.1016/j.nmni.2020.100812
Fang, F. C., Benson, C. A., del Rio, C., Edwards, K. M., Fowler, V. G., Fredricks, D. N., Limaye, A. P., Murray, B. E., Naggie, S., Pappas, P. G., Patel, R., Paterson, D. L., Pegues, D. A., Petri, W. A., & Schooley, R. T. (2020). COVID-19—Lessons learned and questions remaining. Clinical Infectious Diseases, 72, 2225. https://doi.org/10.1093/cid/ciaa1654
Faria, N. R., Claro, I. M., Candido, D., Franco, L. A. M., Andrade, P. S., Coletti, T. M., Silva, C. A. M., Sales, F. C., Manuli, E. R., Aguiar, R. S., Gaburo, N., Camilo, C. d. C., Fraiji, N. A., Crispim, M. A. E., Carvalho, M. D. P. S. S., Rambaut, A., Loman, N., Pybus, O. G., Sabino, E. C., & CADDE Genomic Network. (2021). Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: Preliminary findings. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manauspreliminary-findings/586
Farinholt, T., Doddapaneni, H., Qin, X., Menon, V., Meng, Q., Metcalf, G., Chao, H., Gingras, M. C., Farinholt, P., Agrawal, C., Muzny, D. M., Piedra, P. A., Gibbs, R. A., & Petrosino, J. (2021). Transmission event of SARS-CoV-2 Delta variant reveals multiple vaccine breakthrough infections. MedRvix. https://doi.org/10.1101/2021.06.28.21258780
Feldstein, L. R., Rose, E. B., Horwitz, S. M., Collins, J. P., Newhams, M. M., Son, M. B. F., Newburger, J. W., Kleinman, L. C., Heidemann, S. M., Martin, A. A., Singh, A. R., Li, S., Tarquinio, K. M., Jaggi, P., Oster, M. E., Zackai, S. P., Gillen, J., Ratner, A. J., Walsh, R. F., Fitzgerald, J. C., et al. (2020). Multisystem inflammatory syndrome in U.S. children and adolescents. New England Journal of Medicine, 383(4), 334–346. https://doi.org/10.1056/NEJMoa2021680
Feng, Y., Marchal, T., Sperry, T., & Yi, H. (2020). Influence of wind and relative humidity on the social distancing effectiveness to prevent COVID-19 airborne transmission: A numerical study. Journal of Aerosol Science, 147, 105585. https://doi.org/10.1016/j.jaerosci.2020.105585
Fennelly, K. P. (2020). Particle sizes of infectious aerosols: Implications for infection control. The Lancet Respiratory Medicine, 8(9), 914–924. https://doi.org/10.1016/S2213-2600(20)30323-4
Fernandez, C. E., Franz, C. K., Ko, J. H., Walter, J. M., Koralnik, I. J., Ahlawat, S., & Deshmukh, S. (2021). Imaging review of peripheral nerve injuries in patients with COVID-19. Radiology, 298(3), E117–E130. https://doi.org/10.1148/radiol.2020203116
Fox, G. J., Orlova, M., & Schurr, E. (2016). Tuberculosis in newborns: The lessons of the “Lübeck disaster” (1929–1933). PLoS Pathogens, 12(1), e1005271. https://doi.org/10.1371/journal.ppat.1005271
Fronstin, P., & Woodbury, S. A. (2020, October 7). How many Americans have lost jobs with employer health coverage during the pandemic? The Commonwealth Fund. https://www.commonwealthfund.org/publications/issue-briefs/2020/oct/how-many-lost-jobs-employer-coverage-pandemic#1
Gale, C., Quigley, M. A., Placzek, A., Knight, M., Ladhani, S., Draper, E. S., Sharkey, D., Doherty, C., Mactier, H., & Kurinczuk, J. J. (2021). Characteristics and outcomes of neonatal SARS-CoV-2 infection in the UK: A prospective national cohort study using active surveillance. The Lancet Child and Adolescent Health, 5(2), 113–121. https://doi.org/10.1016/S2352-4642(20)30342-4
Gan, J. M., Kho, J., Akhunbay-Fudge, M., Choo, H. M., Wright, M., Batt, F., Mandal, A. K. J., Chauhan, R., & Missouris, C. G. (2020). Atypical presentation of COVID-19 in hospitalised older adults. Irish Journal of Medical Science, 190, 469. https://doi.org/10.1007/s11845-020-02372-7
Gao, J., Tian, Z., & Yang, X. (2020). Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Bioscience Trends, 14(1), 72–73. https://doi.org/10.5582/bst.2020.01047
Gao, L., Zhou, J., Yang, S., Wang, L., Chen, X., Yang, Y., Li, R., Pan, Z., Zhao, J., Li, Z., Huang, Q., Tang, J., Hu, L., Liu, P., Zhang, G., Chen, Y., & Ye, L. (2021a). The dichotomous and incomplete adaptive immunity in COVID-19 patients with different disease severity. Signal Transduction and Targeted Therapy, 6(1), 113. https://doi.org/10.1038/s41392-021-00525-3
Gao, Z., Xu, Y., Sun, C., Wang, X., Guo, Y., Qiu, S., & Ma, K. (2021b). A systematic review of asymptomatic infections with COVID-19. Journal of Microbiology, Immunology and Infection, 54(1), 12–16. https://doi.org/10.1016/j.jmii.2020.05.001
Garg, M., Gupta, P., Maralakunte, M., Kumar, M. P., Sinha, A., Kang, M., Agarwal, R., & Sandhu, M. S. (2021). Diagnostic accuracy of CT and radiographic findings for novel coronavirus 2019 pneumonia: Systematic review and meta-analysis. Clinical Imaging, 72, 75–82. https://doi.org/10.1016/j.clinimag.2020.11.021
Garry, R. F. (2021). Mutations arising in SARS-CoV-2 spike on sustained human-to-human transmission and human-to-animal passage. https://virological.org/t/mutations-arising-in-sars-cov-2-spike-on-sustained-human-to-human-transmission-and-human-to-animal-passage/578
Gavriatopoulou, M., Korompoki, E., Fotiou, D., Ntanasis-Stathopoulos, I., Psaltopoulou, T., Kastritis, E., Terpos, E., & Dimopoulos, M. A. (2020). Organ-specific manifestations of COVID-19 infection. Clinical and Experimental Medicine, 20(4), 493–506. https://doi.org/10.1007/s10238-020-00648-x
Giacomet, V., Barcellini, L., Stracuzzi, M., Longoni, E., Folgori, L., Leone, A., Zuccotti, G. V., & COVID-19 Pediatric Network. (2020). Gastrointestinal symptoms in severe COVID-19 children. Pediatric Infectious Disease Journal, 39(10), e317–e320. https://doi.org/10.1097/INF.0000000000002843
Gibson, W., Peacock, L., Ferris, V., Fischer, K., Livingstone, J., Thomas, J., & Bailey, M. (2015). Genetic recombination between human and animal parasites creates novel strains of human pathogen. PLoS Neglected Tropical Diseases, 9(3), e0003665. https://doi.org/10.1371/journal.pntd.0003665
Girum, T., Lentiro, K., Geremew, M., Migora, B., & Shewamare, S. (2020). Global strategies and effectiveness for COVID-19 prevention through contact tracing, screening, quarantine, and isolation: A systematic review. Tropical Medicine and Health, 48(1), 91. https://doi.org/10.1186/s41182-020-00285-w
Gómez, C. E., Perdiguero, B., & Esteban, M. (2021). Emerging SARS-CoV-2 variants and impact in global vaccination programs against SARS-CoV-2/COVID-19. Vaccine, 9(3), 243. https://doi.org/10.3390/vaccines9030243
Goodman, L. B., & Whittaker, G. R. (2021). Public health surveillance of infectious diseases: Beyond point mutations. The Lancet Microbe, 2(2), e53–e54. https://doi.org/10.1016/S2666-5247(21)00003-3
Government of Canada. (2021, July 21). SARS-CoV-2 variants: National definitions, classifications and public health actions. https://www.canada.ca/en/public-health/services/diseases/2019-novel-coronavirus-infection/health-professionals/testing-diagnosing-case-reporting/sars-cov-2-variants-national-definitions-classifications-public-health-actions.html
Grant, M. C., Geoghegan, L., Arbyn, M., Mohammed, Z., McGuinness, L., Clarke, E. L., & Wade, R. G. (2020). The prevalence of symptoms in 24,410 adults infected by the novel coronavirus (SARS-CoV-2; COVID-19): A systematic review and meta-analysis of 148 studies from 9 countries. PLoS One, 15(6), e0234765. https://doi.org/10.1371/journal.pone.0234765
Griffin, D. O., Brennan-Rieder, D., Ngo, B., Kory, P., Confalonieri, M., Shapiro, L., Iglesias, J., Dube, M., Nanda, N., In, G. K., Arkfeld, D., Chaudhary, P., Campese, V. M., Hanna, D. L., Sawcer, D., Ehresmann, G., Peng, D., Smorgorzewski, M., Amstrong, A., et al. (2021). The importance of understanding the stages of COVID-19 in treatment and trials. Aids Reviews, 23(1), 40–47. https://doi.org/10.24875/aidsrev.200001261
Grubaugh, N. D., Hodcroft, E. B., Fauver, J. R., Phelan, A. L., & Cevik, M. (2021). Public health actions to control new SARS-CoV-2 variants. Cell, 184(5), 1127–1132. https://doi.org/10.1016/j.cell.2021.01.044
Gubernatorova, E. O., Gorshkova, E. A., Polinova, A. I., & Drutskaya, M. S. (2020). IL-6: Relevance for immunopathology of SARS-CoV-2. Cytokine & Growth Factor Reviews, 53, 13–24. https://doi.org/10.1016/j.cytogfr.2020.05.009
Gunasekeran, D. V., Tham, Y. C., Ting, D. S. W., Tan, G. S. W., & Wong, T. Y. (2021). Digital health during COVID-19: Lessons from operationalising new models of care in ophthalmology. The Lancet. Digital Health, 3(2), e124–e134. https://doi.org/10.1016/S2589-7500(20)30287-9
Guo, T., Fan, Y., Chen, M., Wu, X., Zhang, L., He, T., Wang, H., Wan, J., Wang, X., & Lu, Z. (2020). Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19). JAMA Cardiology, 5(7), 811–818. https://doi.org/10.1001/jamacardio.2020.1017
Gupta, A., Madhavan, M. V., Sehgal, K., Nair, N., Mahajan, S., Sehrawat, T. S., Bikdeli, B., Ahluwalia, N., Ausiello, J. C., Wan, E. Y., Freedberg, D. E., Kirtane, A. J., Parikh, S. A., Maurer, M. S., Nordvig, A. S., Accili, D., Bathon, J. M., Mohan, S., Bauer, K. A., Leon, M. B., et al. (2020). Extrapulmonary manifestations of COVID-19. Nature Medicine, 26(7), 1017–1032. https://doi.org/10.1038/s41591-020-0968-3
Gurumurthy, B., Das, S. K., Hiremath, R., Shetty, S., Hiremath, A., & Gowda, T. (2021). Spectrum of atypical pulmonary manifestations of COVID-19 on computed tomography. Egyptian Journal of Radiology and Nuclear Medicine, 52(1), 72. https://doi.org/10.1186/s43055-021-00448-7
Hajar, R. (2013). The air of history (part V): Ibn Sina (Avicenna): The great physician and philosopher. Heart Views, 14(4), 196. https://doi.org/10.4103/1995-705x.126893
Haleem, A., Javaid, M., Khan, I. H., & Vaishya, R. (2020). Significant applications of big data in COVID-19 pandemic. Indian Journal of Orthopaedics, 54(4), 1–3. https://doi.org/10.1007/s43465-020-00129-z
Hall, L. (2020, March 12). Coronavirus conspiracy theory that Covid-19 originated in US spreading in China. The Independent. https://www.independent.co.uk/news/world/americas/coronavirus-start-originate-conspiracy-china-us-wuhan-cdc-robert-redfield-a9398711.html
Halpert, G., & Shoenfeld, Y. (2020). SARS-CoV-2, the autoimmune virus. Autoimmunity Reviews, 19(12), 102695. https://doi.org/10.1016/j.autrev.2020.102695
Han, G.-Z. (2020). Pangolins Harbor SARS-CoV-2-related coronaviruses. Trends in Microbiology, 28(7), 515–517. https://doi.org/10.1016/j.tim.2020.04.001
Han, E., Tan, M. M. J., Turk, E., Sridhar, D., Leung, G. M., Shibuya, K., Asgari, N., Oh, J., García-Basteiro, A. L., Hanefeld, J., Cook, A. R., Hsu, L. Y., Teo, Y. Y., Heymann, D., Clark, H., McKee, M., & Legido-Quigley, H. (2020). Lessons learnt from easing COVID-19 restrictions: An analysis of countries and regions in Asia Pacific and Europe. Lancet, 396(10261), 1525–1534. https://doi.org/10.1016/S0140-6736(20)32007-9
Hariharan, A., Hakeem, A. R., Radhakrishnan, S., Reddy, M. S., & Rela, M. (2020). The role and therapeutic potential of NF-kappa-B pathway in severe COVID-19 patients. Inflammopharmacology, 29(1), 91–100. https://doi.org/10.1007/s10787-020-00773-9
Hatmi, Z. N. (2021). A systematic review of systematic reviews on the COVID-19 pandemic. SN Comprehensive Clinical Medicine, 3, 419. https://doi.org/10.1007/s42399-021-00749-y
Hayashi, T., Yaegashi, N., & Konishi, I. (2021). Effect of RBD, Y453f mutation in spike glycoprotein of SARS-CoV-2 on neutralizing IgG affinity. medRxiv. https://doi.org/10.1101/2021.01.28.21250577
He, B., Zhong, A., Wu, Q., Liu, X., Lin, J., Chen, C., He, Y., Guo, Y., Zhang, M., Zhu, P., Wu, J., Wang, C., Wang, S., & Xia, X. (2020). Tumor biomarkers predict clinical outcome of COVID-19 patients. Journal of Infection, 81(3), 452–482. https://doi.org/10.1016/j.jinf.2020.05.069
Hifumi, T., Yamamoto, A., Ato, M., Sawabe, K., Morokuma, K., Morine, N., Kondo, Y., Noda, E., Sakai, A., Takahashi, J., & Umezawa, K. (2017). Clinical serum therapy: Benefits, cautions, and potential applications. The Keio Journal of Medicine, 66(4), 57–64. https://doi.org/10.2302/kjm.2016-0017-ir
Himmelstein, D. U., & Woolhandler, S. (2020). The U.S. health care system on the eve of the Covid-19 epidemic: A summary of recent evidence on its impaired performance. International Journal of Health Services: Planning, Administration, Evaluation, 50(4), 408–414. https://doi.org/10.1177/0020731420937631
Hossain, D. I. (2020). Pandemic COVID-19 and biomedical waste handling: A review study. Journal of Medical Science and Clinical Research, 8(5), 497–502. https://doi.org/10.18535/jmscr/v8i5.88
Hossain, K., Hassanzadeganroudsari, M., & Apostolopoulos, V. (2021). The emergence of new strains of SARS-CoV-2. What does it mean for COVID-19 vaccines? Expert Review of Vaccines, 20(6), 635–638. https://doi.org/10.1080/14760584.2021.1915140
Hou, Y., Zhao, J., Martin, W., Kallianpur, A., Chung, M. K., Jehi, L., Sharifi, N., Erzurum, S., Eng, C., & Cheng, F. (2020). New insights into genetic susceptibility of COVID-19: An ACE2 and TMPRSS2 polymorphism analysis. BMC Medicine, 18(1), 216. https://doi.org/10.1186/s12916-020-01673-z
Hu, Y., Wang, T., Hu, Z., Wang, X., Zhang, Z., Li, L., & Peng, P. (2020). Clinical efficacy of glucocorticoid on the treatment of patients with COVID-19 pneumonia: A single-center experience. Biomedicine & Pharmacotherapy, 130, 110529. https://doi.org/10.1016/j.biopha.2020.110529
Hussain, A., Via, G., Melniker, L., Goffi, A., Tavazzi, G., Neri, L., Villen, T., Hoppmann, R., Mojoli, F., No-ble, V., Zieleskiewicz, L., Blanco, P., Ma, I. W. Y., Wahab, M. A., Alsaawi, A., Al Salamah, M., Balik, M., Barca, D., Bendjelid, K., … & Arabi, Y. (2020). Multi-organ point-of-care ultrasound for COVID-19 (PoCUS4COVID): international expert consensus. Critical Care, 24(1). https://doi.org/10.1186/s13054-020-03369-5
Ibekwe, T. S., Fasunla, A. J., & Orimadegun, A. E. (2020). Systematic review and meta-analysis of smell and taste disorders in COVID-19. OTO Open, 4(3), X2095797. https://doi.org/10.1177/2473974X20957975
Ilinskaya, A. N., & Dobrovolskaia, M. A. (2016). Understanding the immunogenicity and antigenicity of nanomaterials: Past, present and future. Toxicology and Applied Pharmacology, 299, 70–77. https://doi.org/10.1016/j.taap.2016.01.005
Inui, S., Kurokawa, R., Nakai, Y., Watanabe, Y., Kurokawa, M., Sakurai, K., Fujikawa, A., Sugiura, H., Kawahara, T., Yoon, S. H., Uwabe, Y., Uchida, Y., Gonoi, W., & Abe, O. (2020). Comparison of chest CT grading systems in coronavirus disease 2019 (COVID-19) pneumonia. Radiology: Cardiothoracic Imaging, 2(6), e200492. https://doi.org/10.1148/ryct.2020200492
Iranzadasl, M., Karimi, Y., Moadeli, F., & Pasalar, M. (2021). Persian medicine recommendations for the prevention of pandemics related to the respiratory system: A narrative literature review. Integrative Medicine Research, 10(1), 100483. https://doi.org/10.1016/j.imr.2020.100483
Islam, M., Sobur, M., Akter, M., Nazir, K., Toniolo, A., & Rahman, M. (2020a). Coronavirus disease 2019 (COVID-19) pandemic, lessons to be learned! Journal of Advanced Veterinary and Animal Research, 7(2), 260. https://doi.org/10.5455/javar.2020.g418
Islam, N., Sharp, S. J., Chowell, G., Shabnam, S., Kawachi, I., Lacey, B., Massaro, J. M., D’Agostino, R. B., & White, M. (2020b). Physical distancing interventions and incidence of coronavirus disease 2019: Natural experiment in 149 countries. BMJ, 370, m2743. https://doi.org/10.1136/bmj.m2743
Jaber, R. M., Mafrachi, B., Al-Ani, A., & Shkara, M. (2021). Awareness and perception of COVID-19 among the general population: A Middle Eastern survey. PLoS One, 16(4), e0250461. https://doi.org/10.1371/journal.pone.0250461
Java, A., Apicelli, A. J., Liszewski, M. K., Coler-Reilly, A., Atkinson, J. P., Kim, A. H. J., & Kulkarni, H. S. (2020). The complement system in COVID-19: Friend and foe? JCI Insight, 5(15), e140711. https://doi.org/10.1172/jci.insight.140711
Jayaweera, M., Perera, H., Gunawardana, B., & Manatunge, J. (2020). Transmission of COVID-19 virus by droplets and aerosols: A critical review on the unresolved dichotomy. Environmental Research, 188, 109819. https://doi.org/10.1016/j.envres.2020.109819
Jensen, N., Kelly, A. H., & Avendano, M. (2021). The COVID-19 pandemic underscores the need for an equity-focused global health agenda. Humanities and Social Sciences Communications, 8(1), 15. https://doi.org/10.1057/s41599-020-00700-x
Kaina, B. (2021). On the origin of SARS-CoV-2: Did cell culture experiments lead to increased virulence of the progenitor virus for humans? In Vivo, 35(3), 1313–1326. https://doi.org/10.21873/invivo.12384
Kamel Boulos, M. N., & Geraghty, E. M. (2020). Geographical tracking and mapping of coronavirus disease COVID-19/severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic and associated events around the world: How 21st century GIS technologies are supporting the global fight against outbreaks and epidemics. International Journal of Health Geographics, 19(1), 8. https://doi.org/10.1186/s12942-020-00202-8
Kanduc, D., & Shoenfeld, Y. (2020). Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: Implications for the vaccine. Immunologic Research, 68(5), 310–313. https://doi.org/10.1007/s12026-020-09152-6
Karamanou, M., Panayiotakopoulos, G., Tsoucalas, G., Kousoulis, A. A., & Androutsos, G. (2012). From miasmas to germs: A historical approach to theories of infectious disease transmission. Infezioni in Medicina, 20(1), 58–62.
Kaufer, A. M., Theis, T., Lau, K. A., Gray, J. L., & Rawlinson, W. D. (2020). Laboratory biosafety measures involving SARS-CoV-2 and the classification as a Risk Group 3 biological agent. Pathology, 52(7), 790–795. https://doi.org/10.1016/j.pathol.2020.09.006
Kazemi, E., Soldoozi Nejat, R., Ashkan, F., & Sheibani, H. (2021). The laboratory findings and different COVID-19 severities: A systematic review and meta-analysis. Annals of Clinical Microbiology and Antimicrobials, 20(1), 17. https://doi.org/10.1186/s12941-021-00420-3
Kest, H., Kaushik, A., DeBruin, W., Colletti, M., & Goldberg, D. (2020). Multisystem inflammatory syndrome in children (MIS-C) associated with 2019 novel coronavirus (SARS-CoV-2) infection. Case Reports in Pediatrics, 2020, 8875987. https://doi.org/10.1155/2020/8875987
Khafaie, M. A., & Rahim, F. (2020). Cross-country comparison of case fatality rates of COVID-19/SARS-COV-2. Osong Public Health and Research Perspectives, 11(2), 74–80. https://doi.org/10.24171/j.phrp.2020.11.2.03
Khan, Z. H., Siddique, A., & Lee, C. W. (2020). Robotics utilization for healthcare digitization in global COVID-19 management. International Journal of Environmental Research and Public Health, 17(11), 3819. https://doi.org/10.3390/ijerph17113819
Khanmohammadi, S., & Rezaei, N. (2021). Role of toll-like receptors in the pathogenesis of COVID-19. Journal of Medical Virology, 93(5), 2735–2739. https://doi.org/10.1002/jmv.26826
Khateeb, J., Li, Y., & Zhang, H. (2021). Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Critical Care, 25(1), 244. https://doi.org/10.1186/s13054-021-03662-x
Khemasuwan, D., Sorensen, J. S., & Colt, H. G. (2020). Artificial intelligence in pulmonary medicine: Computer vision, predictive model and COVID-19. European Respiratory Review, 29(157), 200181. https://doi.org/10.1183/16000617.0181-2020
Kichloo, A., Albosta, M., Dettloff, K., Wani, F., El-Amir, Z., Singh, J., Aljadah, M., Chakinala, R. C., Kanugula, A. K., Solanki, S., & Chugh, S. (2020). Telemedicine, the current COVID-19 pandemic and the future: A narrative review and perspectives moving forward in the USA. Family Medicine and Community Health, 8(3), e000530. https://doi.org/10.1136/fmch-2020-000530
Kircheis, R., Haasbach, E., Lueftenegger, D., Heyken, W. T., Ocker, M., & Planz, O. (2020). NF-κB pathway as a potential target for treatment of critical stage COVID-19 patients. Frontiers in Immunology, 11, 598444. https://doi.org/10.3389/fimmu.2020.598444
Konstantinidou, A.-E., Skaltsounis, P., Eleftheriades, M., Antsaklis, P., Charitou, A., Anatolitou, F., Moutafi, A., Petropoulos, P., & Daskalakis, G. (2021). Pharyngeal sampling for PCR-testing in the investigation of SARS-COV-2 vertical transmission in pregnancy. European Journal of Obstetrics & Gynecology and Reproductive Biology, 260, 18. https://doi.org/10.1016/j.ejogrb.2021.02.026
Laamarti, M., Alouane, T., Kartti, S., Chemao-Elfihri, M. W., Hakmi, M., Essabbar, A., Laamarti, M., Hlali, H., Bendani, H., Boumajdi, N., Benhrif, O., Allam, L., El Hafidi, N., El Jaoudi, R., Allali, I., Marchoudi, N., Fekkak, J., Benrahma, H., Nejjari, C., Amzazi, S., et al. (2020). Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. PLoS One, 15(11), e0240345. https://doi.org/10.1371/journal.pone.0240345
Lapointe, C. P., Grosely, R., Johnson, A. G., Wang, J., Fernández, I. S., & Puglisi, J. D. (2021). Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proceedings of the National Academy of Sciences, 118(6), e2017715118. https://doi.org/10.1073/pnas.2017715118
Lauring, A. S., & Hodcroft, E. B. (2021). Genetic variants of SARS-CoV-2—What do they mean? JAMA, 325(6), 529–531. https://doi.org/10.1001/jama.2020.27124
Lee, S. J., Channappanavar, R., & Kanneganti, T.-D. (2020). Coronaviruses: Innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends in Immunology, 41(12), 1083–1099. https://doi.org/10.1016/j.it.2020.10.005
Lei, X., Dong, X., Ma, R., Wang, W., Xiao, X., Tian, Z., Wang, C., Wang, Y., Li, L., Ren, L., Guo, F., Zhao, Z., Zhou, Z., Xiang, Z., & Wang, J. (2020). Activation and evasion of type I interferon responses by SARS-CoV-2. Nature Communications, 11(1), 3810. https://doi.org/10.1038/s41467-020-17665-9
LePan, N. (2020, March). A visual history of pandemics. Visual Capitalist. https://www.visualcapitalist.com/history-of-pandemics-deadliest/
Lester, S. N., & Li, K. (2014). Toll-like receptors in antiviral innate immunity. Journal of Molecular Biology, 426(6), 1246–1264. https://doi.org/10.1016/j.jmb.2013.11.024
Leung, N. H. L. (2021). Transmissibility and transmission of respiratory viruses. Nature Reviews Microbiology, 19(8), 528–545. https://doi.org/10.1038/s41579-021-00535-6
Lew, L. (2020, December 2). American study finds signs of coronavirus in US before China outbreak. Global Research. https://www.globalresearch.ca/american-study-finds-signs-coronavirus-us-before-china-outbreak/5731055
Li, L. Q., Huang, T., Wang, Y. Q., Wang, Z. P., Liang, Y., Huang, T. B., Zhang, H. Y., Sun, W., & Wang, Y. (2020a). COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. Journal of Medical Virology, 92(6), 577–583. https://doi.org/10.1002/jmv.25757
Li, M. Y., Li, L., Zhang, Y., & Wang, X. S. (2020b). Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infectious Diseases of Poverty, 9(1), 45. https://doi.org/10.1186/s40249-020-00662-x
Li, Q., Wu, J., Nie, J., Zhang, L., Hao, H., Liu, S., Zhao, C., Zhang, Q., Liu, H., Nie, L., Qin, H., Wang, M., Lu, Q., Li, X., Sun, Q., Liu, J., Zhang, L., Li, X., Huang, W., & Wang, Y. (2020c). The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell, 182(5), 1284–1294.e9. https://doi.org/10.1016/j.cell.2020.07.012
Liu, Y., Gayle, A. A., Wilder-Smith, A., & Rocklöv, J. (2020). The reproductive number of COVID-19 is higher compared to SARS coronavirus. Journal of Travel Medicine, 27(2). https://doi.org/10.1093/jtm/taaa021
Liu, J., Xie, W., Wang, Y., Xiong, Y., Chen, S., Han, J., & Wu, Q. (2020a). A comparative overview of COVID-19, MERS and SARS: Review article. International Journal of Surgery (London, England), 81(1), 8. https://doi.org/10.1016/j.ijsu.2020.07.032
Liu, X., Zhou, H., Zhou, Y., Wu, X., Zhao, Y., Lu, Y., Tan, W., Yuan, M., Ding, X., Zou, J., Li, R., Liu, H., Ewing, R. M., Hu, Y., Nie, H., & Wang, Y. (2020b). Temporal radiographic changes in COVID-19 patients: Relationship to disease severity and viral clearance. Scientific Reports, 10(1), 10263. https://doi.org/10.1038/s41598-020-66895-w
Liu, Y., Zhang, C., Huang, F., Yang, Y., Wang, F., Yuan, J., Zhang, Z., Qin, Y., Li, X., Zhao, D., Li, S., Tan, S., Wang, Z., Li, J., Shen, C., Li, J., Peng, L., Wu, W., Cao, M., Xing, L., Xu, Z., Chen, L., Zhou, C., Liu, W. J., Liu, L., & Jiang, C. (2020c). 2019-novel coronavirus (2019-nCoV) infections trigger an exaggerated cytokine response aggravating lung injury. http://www.chinaxiv.org/abs/202002.00018
Long, Q.-X., Tang, X.-J., Shi, Q.-L., Li, Q., Deng, H.-J., Yuan, J., Hu, J.-L., Xu, W., Zhang, Y., Lv, F.-J., Su, K., Zhang, F., Gong, J., Wu, B., Liu, X.-M., Li, J.-J., Qiu, J.-F., Chen, J., & Huang, A.-L. (2020). Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nature Medicine, 26(8), 1200–1204. https://doi.org/10.1038/s41591-020-0965-6
Lopes de Sousa, Á. F., Carvalho, H. E. F., Oliveira, L. B., Schneider, G., Camargo, E. L. S., Watanabe, E., de Andrade, D., Fernandes, A. F. C., Mendes, I. A. C., & Fronteira, I. (2020). Effects of COVID-19 infection during pregnancy and neonatal prognosis: What is the evidence? International Journal of Environmental Research and Public Health, 17(11), 4176. https://doi.org/10.3390/ijerph17114176
Lopez-Leon, S., Wegman-Ostrosky, T., Perelman, C., Sepulveda, R., Rebolledo, P. A., Cuapio, A., & Villapol, S. (2021). More than 50 long-term effects of COVID-19: A systematic review and meta-analysis. medRxiv. https://doi.org/10.1101/2021.01.27.21250617
Lorente, L., Martín, M., Franco, A., Barrios, Y., Cáceres, J., Solé-Violán, J., Perez, A., Marcos, Y., Ramos, J., Ramos-Gómez, L., Ojeda, N., Jiménez, A., Lorente, L., Franco, A., Barrios, Y., Perez, A., Jiménez, A., Pérez-Cejas, A., Pérez-Llombet, A., Uribe, L., et al. (2021). HLA genetic polymorphisms and prognosis of patients with COVID-19. Medicina Intensiva, 45(2), 96–103. https://doi.org/10.1016/j.medin.2020.08.004
Lu, L., Zhong, W., Bian, Z., Li, Z., Zhang, K., Liang, B., Zhong, Y., Hu, M., Lin, L., Liu, J., Lin, X., Huang, Y., Jiang, J., Yang, X., Zhang, X., & Huang, Z. (2020). A comparison of mortality-related risk factors of COVID-19, SARS, and MERS: A systematic review and meta-analysis. The Journal of Infection, 81(4), e18–e25. https://doi.org/10.1016/j.jinf.2020.07.002
Maison, D. P., Ching, L. L., Shikuma, C. M., & Nerurkar, V. R. (2021). Genetic characteristics and phylogeny of 969-bp S gene sequence of SARS-CoV-2 from Hawaii reveals the worldwide emerging P681H mutation. bioRxiv. https://doi.org/10.1101/2021.01.06.425497
Majumdar, P., & Niyogi, S. (2020). ORF3a mutation associated with higher mortality rate in SARS-CoV-2 infection. Epidemiology and Infection, 148, e262. https://doi.org/10.1017/S0950268820002599
Mak, T. W., Saunders, M. E., & Jett, B. D. (2014). Primer to the immune response (2nd ed.). Academic Cell.
Malik, G. R., Wolfe, A. R., Soriano, R., Rydberg, L., Wolfe, L. F., Deshmukh, S., Ko, J. H., Nussbaum, R. P., Dreyer, S. D., Jayabalan, P., Walter, J. M., & Franz, C. K. (2020). Injury-prone: Peripheral nerve injuries associated with prone positioning for COVID-19-related acute respiratory distress syndrome. British Journal of Anaesthesia, 125(6), e478–e480. https://doi.org/10.1016/j.bja.2020.08.045
Mallapaty, S. (2021). What’s the risk of dying from a fast-spreading COVID-19 variant? Nature, 590(7845), 191–192. https://doi.org/10.1038/d41586-021-00299-2
Marjot, T., Webb, G. J., Barritt, A. S., Moon, A. M., Stamataki, Z., Wong, V. W., & Barnes, E. (2021). COVID-19 and liver disease: Mechanistic and clinical perspectives. Nature Reviews Gastroenterology & Hepatology, 18(5), 348–364. https://doi.org/10.1038/s41575-021-00426-4
Markowitz, B. A. (2021, March 5). AARP answers: Your insurance coverage and the coronavirus. AARP. https://www.aarp.org/money/budgeting-saving/info-2020/insurance-policies-during-coronavirus-faq.html
Martin, D., Miller, A. P., Quesnel-Vallée, A., Caron, N. R., Vissandjée, B., & Marchildon, G. P. (2018). Canada’s universal health-care system: Achieving its potential. The Lancet, 391(10131), 1718–1735. https://doi.org/10.1016/s0140-6736(18)30181-8
Martin Webb, L., Matzinger, S., Grano, C., Kawasaki, B., Stringer, G., Bankers, L., & Herlihy, R. (2021). Identification of and surveillance for the SARS-CoV-2 variants B.1.427 and B.1.429—Colorado, January–March 2021. MMWR. Morbidity and Mortality Weekly Report, 70(19), 717–718. https://doi.org/10.15585/mmwr.mm7019e2
Mateos González, M., García de Casasola Sánchez, G., Muñoz, F. J. T., Proud, K., Lourdo, D., Sander, J.-V., Jaimes, G. E. O., Mader, M., Canora Lebrato, J., Restrepo, M. I., & Soni, N. J. (2021). Comparison of lung ultrasound versus chest X-ray for detection of pulmonary infiltrates in COVID-19. Diagnostics, 11(2), 373. https://doi.org/10.3390/diagnostics11020373
Mathew, D., Giles, J. R., Baxter, A. E., Oldridge, D. A., Greenplate, A. R., Wu, J. E., Alanio, C., Kuri-Cervantes, L., Pampena, M. B., D’Andrea, K., Manne, S., Chen, Z., Huang, Y. J., Reilly, J. P., Weisman, A. R., Ittner, C. A. G., Kuthuru, O., Dougherty, J., Nzingha, K., et al. (2020). Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science, 369(6508), eabc8511. https://doi.org/10.1126/science.abc8511
Matschke, J., Lütgehetmann, M., Hagel, C., Sperhake, J. P., Schröder, A. S., Edler, C., Mushumba, H., Fitzek, A., Allweiss, L., Dandri, M., Dottermusch, M., Heinemann, A., Pfefferle, S., Schwabenland, M., Sumner Magruder, D., Bonn, S., Prinz, M., Gerloff, C., Püschel, K., et al. (2020). Neuropathology of patients with COVID-19 in Germany: A post-mortem case series. The Lancet Neurology, 19(11), 919–929. https://doi.org/10.1016/s1474-4422(20)30308-2
McCallum, M., Bassi, J., Marco, A. D., Chen, A., Walls, A. C., Iulio, J. D., Tortorici, M. A., Navarro, M. J., Silacci-Fregni, C., Saliba, C., Agostini, M., Pinto, D., Culap, K., Bianchi, S., Jaconi, S., Cameroni, E., Bowen, J. E., Tilles, S. W., Pizzuto, M. S., et al. (2021). SARS-CoV-2 immune evasion by variant B.1.427/B.1.429. bioRxiv. https://doi.org/10.1101/2021.03.31.437925
McGill, A. R., Kahlil, R., Dutta, R., Green, R., Howell, M., Mohapatra, S., & Mohapatra, S. S. (2021). SARS–CoV-2 immuno-pathogenesis and potential for diverse vaccines and therapies: Opportunities and challenges. Infectious Disease Reports, 13(1), 102–125. https://doi.org/10.3390/idr13010013
Mehlotra, R. K. (2020). Chemokine receptor gene polymorphisms and COVID-19: Could knowledge gained from HIV/AIDS be important? Infection, Genetics and Evolution, 85, 104512. https://doi.org/10.1016/j.meegid.2020.104512
Menter, T., Haslbauer, J. D., Nienhold, R., Savic, S., Hopfer, H., Deigendesch, N., Frank, S., Turek, D., Willi, N., Pargger, H., Bassetti, S., Leuppi, J. D., Cathomas, G., Tolnay, M., Mertz, K. D., & Tzankov, A. (2020). Postmortem examination of COVID-19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology, 77(2), 198–209. https://doi.org/10.1111/his.14134
Meyerowitz-Katz, G., & Merone, L. (2020). A systematic review and meta-analysis of published research data on COVID-19 infection fatality rates. International Journal of Infectious Diseases, 101, 138–148. https://doi.org/10.1016/j.ijid.2020.09.1464
Miao, G., Zhao, H., Li, Y., Ji, M., Chen, Y., Shi, Y., Bi, Y., Wang, P., & Zhang, H. (2021). ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Developmental Cell, 56(4), 427–442.e5. https://doi.org/10.1016/j.devcel.2020.12.010
Miller, J., Cantor, A., Zachariah, P., Ahn, D., Martinez, M., & Margolis, K. G. (2020). Gastrointestinal symptoms as a major presentation component of a novel multisystem inflammatory syndrome in children that is related to coronavirus disease 2019: A single center experience of 44 cases. Gastroenterology, 159(4), 1571–1574.e2. https://doi.org/10.1053/j.gastro.2020.05.079
Mohammed, M. N., Syamsudin, G., Al-Zubaidi, S., Abdulkarim, S., Ramli, R., & Yusuf, E. (2020). Novel COVID-19 detection and diagnosis system using IOT based smart helmet. International Journal of Psychosocial Rehabilitation, 24(7), 2296–2303. https://doi.org/10.37200/IJPR/V24I7/PR270221
Morais-Almeida, M., Pité, H., Aguiar, R., Ansotegui, I., & Bousquet, J. (2020). Asthma and the coronavirus disease 2019 pandemic: A literature review. International Archives of Allergy and Immunology, 181(9), 680–688. https://doi.org/10.1159/000509057
Moutchia, J., Pokharel, P., Kerri, A., McGaw, K., Uchai, S., Nji, M., & Goodman, M. (2020). Clinical laboratory parameters associated with severe or critical novel coronavirus disease 2019 (COVID-19): A systematic review and meta-analysis. PLoS One, 15(10), e0239802. https://doi.org/10.1371/journal.pone.0239802
Mullol, J., Alobid, I., Mariño-Sánchez, F., Izquierdo-Domínguez, A., Marin, C., Klimek, L., Wang, D. Y., & Liu, Z. (2020). The loss of smell and taste in the COVID-19 outbreak: A tale of many countries. Current Allergy and Asthma Reports, 20(10), 61. https://doi.org/10.1007/s11882-020-00961-1
Naemi, F. M. A., Al-adwani, S., Al-khatabi, H., & Al-nazawi, A. (2021). Association between the HLA genotype and the severity of COVID-19 infection among south Asians. Journal of Medical Virology, 93(7), 4430–4437. https://doi.org/10.1002/jmv.27003
Nardacci, R., Colavita, F., Castilletti, C., Lapa, D., Matusali, G., Meschi, S., Del Nonno, F., Colombo, D., Capobianchi, M. R., Zumla, A., Ippolito, G., Piacentini, M., & Falasca, L. (2021). Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death & Disease, 12(3), 263. https://doi.org/10.1038/s41419-021-03527-9
Nasajpour, M., Pouriyeh, S., Parizi, R. M., Dorodchi, M., Valero, M., & Arabnia, H. R. (2020). Internet of things for current COVID-19 and future pandemics: An exploratory study. Journal of Healthcare Informatics Research, 4(4), 1–40. https://doi.org/10.1007/s41666-020-00080-6
National Heart, Lung, and Blood Institute (NHLBI). (2021, January 22). Full-dose blood thinners decreased need for life support and improved outcome in hospitalized COVID-19 patients. https://www.nih.gov/news-events/news-releases/full-dose-blood-thinners-decreased-need-life-support-improved-outcome-hospitalized-covid-19-patients
Nemani, K., Li, C., Olfson, M., Blessing, E. M., Razavian, N., Chen, J., Petkova, E., & Goff, D. C. (2021). Association of psychiatric disorders with mortality among patients with COVID-19. JAMA Psychiatry, 78(4), 380. https://doi.org/10.1001/jamapsychiatry.2020.4442
Neumann, J., Prezzemolo, T., Vanderbeke, L., Roca, C. P., Gerbaux, M., Janssens, S., Willemsen, M., Burton, O., van Mol, P., van Herck, Y., Wauters, J., Wauters, E., Liston, A., Humblet-Baron, S., Bosisio, F., de Smet, F., Dooms, C., Garg, A., Gunst, J., et al. (2020). Increased IL-10-producing regulatory T cells are characteristic of severe cases of COVID-19. Clinical & Translational Immunology, 9(11), e1204. https://doi.org/10.1002/cti2.1204
New Findings on the Pathophysiology of Severe COVID-19 Infections. (2021). American Journal of Nursing, 121(2), 15. https://doi.org/10.1097/01.naj.0000734060.74732.9d
Nie, J. B. (2020). In the shadow of biological warfare: Conspiracy theories on the origins of COVID-19 and enhancing global governance of biosafety as a matter of urgency. Journal of Bioethical Inquiry, 17(4), 567–574. https://doi.org/10.1007/s11673-020-10025-8
Nikolich-Zugich, J., Knox, K. S., Rios, C. T., Natt, B., Bhattacharya, D., & Fain, M. J. (2020). SARS-CoV-2 and COVID-19 in older adults: What we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience, 42(2), 505–514. https://doi.org/10.1007/s11357-020-00186-0
Olbei, M., Hautefort, I., Modos, D., Treveil, A., Poletti, M., Gul, L., Shannon-Lowe, C. D., & Korcsmaros, T. (2021). SARS-CoV-2 causes a different cytokine response compared to other cytokine storm-causing respiratory viruses in severely ill patients. Frontiers in Immunology, 12, 629193. https://doi.org/10.3389/fimmu.2021.629193
Ortega, M., Fraile-Martínez, O., García-Montero, C., García-Gallego, S., Sánchez-Trujillo, L., Torres-Carranza, D., Álvarez-Mon, M., Pekarek, L., García-Honduvilla, N., Bujan, J., Álvarez-Mon, M., Asúnsolo, Á., & De la Torre, B. (2020). An integrative look at SARS-CoV2 [Review]. International Journal of Molecular Medicine, 47(2), 415–434. https://doi.org/10.3892/ijmm.2020.4828
Özel, M., Aslan, A., & Araç, S. (2021). Use of the COVID-19 Reporting and Data System (CO-RADS) classification and chest computed tomography involvement score (CT-IS) in COVID-19 pneumonia. La Radiologia Medica, 126, 679. https://doi.org/10.1007/s11547-021-01335-x
Pak, D., Langohr, K., Ning, J., Cortés Martínez, J., Gómez Melis, G., & Shen, Y. (2020). Modeling the coronavirus disease 2019 incubation period: Impact on quarantine policy. Mathematics, 8(9), 1631. https://doi.org/10.3390/math8091631
Parmet, W. E., & Sinha, M. S. (2020). Covid-19—The law and limits of quarantine. New England Journal of Medicine, 382(15), e28. https://doi.org/10.1056/NEJMp2004211
Patel, K. P., Vunnam, S. R., Patel, P. A., Krill, K. L., Korbitz, P. M., Gallagher, J. P., Suh, J. E., & Vunnam, R. R. (2020). Transmission of SARS-CoV-2: An update of current literature. European Journal of Clinical Microbiology and Infectious Diseases, 39(11), 2005–2011. https://doi.org/10.1007/s10096-020-03961-1
Patocka, J., Kuca, K., Oleksak, P., Nepovimova, E., Valis, M., Novotny, M., & Klimova, B. (2021). Rapamycin: Drug repurposing in SARS-CoV-2 infection. Pharmaceuticals, 14(3), 217. https://doi.org/10.3390/ph14030217
Peiró, Ó. M., Carrasquer, A., Sánchez-Gimenez, R., Lal-Trehan, N., Del-Moral-Ronda, V., Bonet, G., Fort-Gallifa, I., Picó-Plana, E., Bastón-Paz, N., Gutiérrez, C., & Bardaji, A. (2021). Biomarkers and short-term prognosis in COVID-19. Biomarkers: Biochemical Indicators of Exposure, Response, and Susceptibility to Chemicals, 26, 1–8. https://doi.org/10.1080/1354750X.2021.1874052
Peng, J., Wu, X., Wang, R., Li, C., Zhang, Q., & Wei, D. (2020). Medical waste management practice during the 2019–2020 novel coronavirus pandemic: Experience in a general hospital. American Journal of Infection Control, 48(8), 918–921. https://doi.org/10.1016/j.ajic.2020.05.035
Peng, Y., Tao, H., Satyanarayanan, S. K., Jin, K., & Su, H. (2021). A comprehensive summary of the knowledge on COVID-19 treatment. Aging and Disease, 12(1), 155. https://doi.org/10.14336/ad.2020.1124
Pereda, R., González, D., Rivero, H. B., Rivero, J. C., Pérez, A., López, L. D. R., Mezquia, N., Venegas, R., Betancourt, J. R., & Domínguez, R. E. (2020). Therapeutic effectiveness of interferon-α2b against COVID-19: The Cuban experience. Journal of Interferon & Cytokine Research, 40(9), 438–442. https://doi.org/10.1089/jir.2020.0124
Perrotta, F., Corbi, G., Mazzeo, G., Boccia, M., Aronne, L., D’Agnano, V., Komici, K., Mazzarella, G., Parrella, R., & Bianco, A. (2020). COVID-19 and the elderly: Insights into pathogenesis and clinical decision-making. Aging Clinical and Experimental Research, 32(8), 1599–1608. https://doi.org/10.1007/s40520-020-01631-y
Phan, T. G. (2020). Genetic diversity and evolution of SARS-CoV-2. Infection, Genetics and Evolution, 81, 104260. https://doi.org/10.1016/j.meegid.2020.104260
Philips, C. A., Mohan, N., Ahamed, R., Kumbar, S., Rajesh, S., George, T., Mohanan, M., & Augustine, P. (2020). One disease, many faces-typical and atypical presentations of SARS-CoV-2 infection-related COVID-19 disease. World Journal of Clinical Cases, 8(18), 3956–3970. https://doi.org/10.12998/wjcc.v8.i18.3956
Picard, C., Casanova, J. L., & Abel, L. (2006). Mendelian traits that confer predisposition or resistance to specific infections in humans. Current Opinion in Immunology, 18(4), 383–390. https://doi.org/10.1016/j.coi.2006.05.005
Pinho-Gomes, A.-C., Peters, S., Thompson, K., Hockham, C., Ripullone, K., Woodward, M., & Carcel, C. (2020). Where are the women? Gender inequalities in COVID-19 research authorship. BMJ Global Health, 5(7), e002922. https://doi.org/10.1136/bmjgh-2020-002922
Pollitz, K., & Michaud, K. J. J. (2020, September 30). Is COVID-19 a pre-existing condition? What could happen if the ACA is overturned. Kettering Family Foundation. https://www.kff.org/policy-watch/is-covid-19-a-pre-existing-condition-what-could-happen-if-the-aca-is-overturned/
Poongodi, M., Hamdi, M., Malviya, M., Sharma, A., Dhiman, G., & Vimal, S. (2021). Diagnosis and combating COVID-19 using wearable Oura smart ring with deep learning methods. Personal and Ubiquitous Computing, 26, 25. https://doi.org/10.1007/s00779-021-01541-4
Prince, G., & Sergel, M. (2020). Persistent hiccups as an atypical presenting complaint of COVID-19. The American Journal of Emergency Medicine, 38(7), 1546.e5–1546.e6. https://doi.org/10.1016/j.ajem.2020.04.045
Qiu, L., Liu, X., Xiao, M., Xie, J., Cao, W., Liu, Z., Morse, A., Xie, Y., Li, T., & Zhu, L. (2020). SARS-CoV-2 is not detectable in the vaginal fluid of women with severe COVID-19 infection. Clinical Infectious Diseases, 71(15), 813–817. https://doi.org/10.1093/cid/ciaa375
Qu, J.-M., Cao, B., & Chen, R.-C. (2021). Chapter 3-Clinical features of COVID-19. In B. Cao, R.-C. Chen, C. Chi, Q. Guo, Y. Guo, M. Hu, Y. Huang, R. Jiang, J. Li, Q. Li, L. Liu, J.-M. Qu, Y. Shi, Y.-L. Song, A. Wu, C. Wang, Y.-M. Wang, G. Zeng, J. Zhang, X. Zhang, et al. (Eds.), COVID-19: The essentials of prevention and treatment (pp. 13–39). Elsevier. https://doi.org/10.1016/b978-0-12-824003-8.00003-6
Qureshi, Z., Jones, N., Temple, R., Larwood, J. P. J., Greenhalgh, T., & Bourouiba, L. (2020). What is the evidence to support the 2-metre social distancing rule to reduce COVID-19 transmission? The Centre for Evidence-Based Medicine, University of Oxford. https://www.cebm.net/covid-19/what-is-the-evidence-to-support-the-2-metre-social-distancing-rule-to-reduce-covid-19-transmission/
Rahman, B., Sadraddin, E., & Porreca, A. (2020a). The basic reproduction number of SARS-CoV-2 in Wuhan is about to die out, how about the rest of the world? Reviews in Medical Virology, 30(4), e2111. https://doi.org/10.1002/rmv.2111
Rahman, M. M., Bodrud-Doza, M., Griffiths, M. D., & Mamun, M. A. (2020b). Biomedical waste amid COVID-19: Perspectives from Bangladesh. The Lancet Global Health, 8(10), e1262. https://doi.org/10.1016/s2214-109x(20)30349-1
Ramani, S. L., Samet, J., Franz, C. K., Hsieh, C., Nguyen, C. V., Horbinski, C., & Deshmukh, S. (2021). Musculoskeletal involvement of COVID-19: Review of imaging. Skeletal Radiology, 50, 1763. https://doi.org/10.1007/s00256-021-03734-7
Rambaut, A., Loman, N., Pybus, O., Barclay, W., Barrett, J., Carabelli, A., Connor, T., Peacock, T., Robertson, D., & Volz, E. (2020). Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563
Rastogi, M., Pandey, N., Shukla, A., & Singh, S. K. (2020). SARS coronavirus 2: From genome to infectome. Respiratory Research, 21(1), 318. https://doi.org/10.1186/s12931-020-01581-z
Resende, P. C., Bezerra, J. F., Vasconcelos, R. H. T., Arantes, I., Appolinario, L., Mendonça, A. C., Paixao, A. C., Rodrigues, A. C., Silva, T., Rocha, A. S., Pauvolid-Corrêa, A., Motta, F. C., Teixeira, D. L. F., de Oliveira Carneiro, T. F., Neto, F. P. F., Herbster, I. D., Leite, A. B., Riediger, I. N., Debur, M. C., Naveca, F. G., et al. (2020). Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil. Virological.org. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584
Ridic, G., Gleason, S., & Ridic, O. (2012). Comparisons of health care systems in the United States, Germany and Canada. Materia Socio-Medica, 24(2), 112–120. https://doi.org/10.5455/msm.2012.24.112-120
Rodrigues, C., Baía, I., Domingues, R., & Barros, H. (2020). Pregnancy and breastfeeding during COVID-19 pandemic: A systematic review of published pregnancy cases. Frontiers in Public Health, 8, 558144. https://doi.org/10.3389/fpubh.2020.558144
Rodriguez-Morales, A. J., Cardona-Ospina, J. A., Gutiérrez-Ocampo, E., Villamizar-Peña, R., Holguin-Rivera, Y., Escalera-Antezana, J. P., Alvarado-Arnez, L. E., Bonilla-Aldana, D. K., Franco-Paredes, C., Henao-Martinez, A. F., Paniz-Mondolfi, A., Lagos-Grisales, G. J., Ramírez-Vallejo, E., Suárez, J. A., Zambrano, L. I., Villamil-Gómez, W. E., Balbin-Ramon, G. J., Rabaan, A. A., Harapan, H., et al. (2020). Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Medicine and Infectious Disease, 34, 101623. https://doi.org/10.1016/j.tmaid.2020.101623
Roehr, B. (2020). The health of private insurance in the US during covid-19. BMJ, 370, m2606. https://doi.org/10.1136/bmj.m2606
Romano, M., Ruggiero, A., Squeglia, F., Maga, G., & Berisio, R. (2020). A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cell, 9(5), 1267. https://doi.org/10.3390/cells9051267
Roper, W. (2020). Mass quarantine effective against coronavirus in China. Statista. https://www.statista.com/chart/21076/china-quarantine-drops-coronavirus-cases/
Rosenberg, J., & Huang, J. (2018). CD8+ T cells and NK cells: Parallel and complementary soldiers of immunotherapy. Current Opinion in Chemical Engineering, 19, 9–20. https://doi.org/10.1016/j.coche.2017.11.006
Rubino, F., Amiel, S. A., Zimmet, P., Alberti, G., Bornstein, S., Eckel, R. H., Mingrone, G., Boehm, B., Cooper, M. E., Chai, Z., Del Prato, S., Ji, L., Hopkins, D., Herman, W. H., Khunti, K., Mbanya, J.-C., & Renard, E. (2020). New-onset diabetes in Covid-19. New England Journal of Medicine, 383(8), 789–790. https://doi.org/10.1056/nejmc2018688
Ruckert, A., Zinszer, K., Zarowsky, C., Labonté, R., & Carabin, H. (2020). What role for One Health in the COVID-19 pandemic? Canadian Journal of Public Health, 111(5), 641–644. https://doi.org/10.17269/s41997-020-00409-z
Ryan, K. J. (2004). Overview. In K. J. Ryan & C. G. Ray (Eds.), Sherris medical microbiology (pp. 1–2). McGraw-Hill.
Sa Ribero, M., Jouvenet, N., Dreux, M., & Nisole, S. (2020). Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathogens, 16(7), e1008737. https://doi.org/10.1371/journal.ppat.1008737
Sahu, A. K., Sreepadmanabh, M., Rai, M., & Chande, A. (2021). SARS-CoV-2: Phylogenetic origins, pathogenesis, modes of transmission, and the potential role of nanotechnology. Virusdisease, 32, 1. https://doi.org/10.1007/s13337-021-00653-y
Saleh, M., Vaezi, A. A., Aliannejad, R., Sohrabpour, A. A., Kiaei, S. Z. F., Shadnoush, M., Siavashi, V., Aghaghazvini, L., Khoundabi, B., Abdoli, S., Chahardouli, B., Seyhoun, I., Alijani, N., & Verdi, J. (2021). Cell therapy in patients with COVID-19 using Wharton’s jelly mesenchymal stem cells: A phase 1 clinical trial. Stem Cell Research & Therapy, 12(1), 410. https://doi.org/10.1186/s13287-021-02483-7
Sallard, E., Halloy, J., Casane, D., Decroly, E., & van Helden, J. (2021). Tracing the origins of SARS-COV-2 in coronavirus phylogenies: A review. Environmental Chemistry Letters, 19, 769. https://doi.org/10.1007/s10311-020-01151-1
Salleh, M. Z., Derrick, J. P., & Deris, Z. Z. (2021). Structural evaluation of the spike glycoprotein variants on SARS-CoV-2 transmission and immune evasion. International Journal of Molecular Sciences, 22(14), 7425. https://doi.org/10.3390/ijms22147425
Santhanam, L. (2020, September 1). How Canada got universal health care and what the U.S. could learn. PBS NewsHour. https://www.pbs.org/newshour/health/how-canada-got-universal-health-care-and-what-the-u-s-could-learn
Sasisekharan, V., Pentakota, N., Jayaraman, A., Tharakaraman, K., Wogan, G. N., & Narayanasami, U. (2021). Orthogonal immunoassays for IgG antibodies to SARS-CoV-2 antigens reveal that immune response lasts beyond 4 mo post illness onset. Proceedings of the National Academy of Sciences, 118(5), e2021615118. https://doi.org/10.1073/pnas.2021615118
Schaub, B., Lauener, R., & von Mutius, E. (2006). The many faces of the hygiene hypothesis. Journal of Allergy and Clinical Immunology, 117(5), 969–977. https://doi.org/10.1016/j.jaci.2006.03.003
Schmid, B., Feuerstein, D., Lang, C. N., Fink, K., Steger, R., Rieder, M., Duerschmied, D., Busch, H. J., & Damjanovic, D. (2020). Lung ultrasound in the emergency department—A valuable tool in the management of patients presenting with respiratory symptoms during the SARS-CoV-2 pandemic. BMC Emergency Medicine, 20(1), 96. https://doi.org/10.1186/s12873-020-00389-w
Sengupta, T., Majumder, R., & Majumder, S. (2021). Role of vitamin D in treating COVID-19-associated coagulopathy: Problems and perspectives. Molecular and Cellular Biochemistry, 476, 2421. https://doi.org/10.1007/s11010-021-04093-6
Sette, A., & Crotty, S. (2021). Adaptive immunity to SARS-CoV-2 and COVID-19. Cell, 184(4), 861–880. https://doi.org/10.1016/j.cell.2021.01.007
Shadmi, E., Chen, Y., Dourado, I., Faran-Perach, I., Furler, J., Hangoma, P., Hanvoravongchai, P., Obando, C., Petrosyan, V., Rao, K. D., Ruano, A. L., Shi, L., de Souza, L. E., Spitzer-Shohat, S., Sturgiss, E., Suphanchaimat, R., Uribe, M. V., & Willems, S. (2020). Health equity and COVID-19: Global perspectives. International Journal for Equity in Health, 19(1), 104. https://doi.org/10.1186/s12939-020-01218-z
Shaigany, S., Gnirke, M., Guttmann, A., Chong, H., Meehan, S., Raabe, V., Louie, E., Solitar, B., & Femia, A. (2020). An adult with Kawasaki-like multisystem inflammatory syndrome associated with COVID-19. The Lancet, 396(10246), e8–e10. https://doi.org/10.1016/s0140-6736(20)31526-9
Shammi, M., Behal, A., & Tareq, S. M. (2021). The escalating biomedical waste management to control the environmental transmission of COVID-19 pandemic: A perspective from two south Asian countries. Environmental Science & Technology, 55, 4087. https://doi.org/10.1021/acs.est.0c05117
Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020). Cell entry mechanisms of SARS-CoV-2. Proceedings of the National Academy of Sciences of the United States of America, 117(21), 11727–11734. https://doi.org/10.1073/pnas.2003138117
Sharma, A. K., Dhasmana, N., Dubey, N., Kumar, N., Gangwal, A., Gupta, M., & Singh, Y. (2016). Bacterial virulence factors: Secreted for survival. Indian Journal of Microbiology, 57(1), 1–10. https://doi.org/10.1007/s12088-016-0625-1
Sharun, K., Dhama, K., Pawde, A. M., Gortázar, C., Tiwari, R., Bonilla-Aldana, D. K., Rodriguez-Morales, A. J., de la Fuente, J., Michalak, I., & Attia, Y. A. (2021). SARS-CoV-2 in animals: Potential for unknown reservoir hosts and public health implications. Veterinary Quarterly, 41(1), 181–201. https://doi.org/10.1080/01652176.2021.1921311
Shaw, C. L., & Kennedy, D. A. (2021). What the reproductive number R0 can and cannot tell us about COVID-19 dynamics. Theoretical Population Biology, 137, 2–9. https://doi.org/10.1016/j.tpb.2020.12.003
Shen, C., Wang, Z., Zhao, F., Yang, Y., Li, J., Yuan, J., Wang, F., Li, D., Yang, M., Xing, L., Wei, J., Xiao, H., Yang, Y., Qu, J., Qing, L., Chen, L., Xu, Z., Peng, L., Li, Y., et al. (2020). Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA, 323(16), 1582. https://doi.org/10.1001/jama.2020.4783
Shors, T. (2021). Coronavirus. In AccessScience. Retrieved 26 Feb 2021, from https://doi.org/10.1036/1097-8542.163220
Simonovich, V. A., Burgos Pratx, L. D., Scibona, P., Beruto, M. V., Vallone, M. G., Vázquez, C., Savoy, N., Giunta, D. H., Pérez, L. G., Sánchez, M. L., Gamarnik, A. V., Ojeda, D. S., Santoro, D. M., Camino, P. J., Antelo, S., Rainero, K., Vidiella, G. P., Miyazaki, E. A., Cornistein, W., et al. (2021). A randomized trial of convalescent plasma in Covid-19 severe pneumonia. New England Journal of Medicine, 384(7), 619–629. https://doi.org/10.1056/nejmoa2031304
Simpson, S., Kay, F. U., Abbara, S., Bhalla, S., Chung, J. H., Chung, M., Henry, T. S., Kanne, J. P., Kligerman, S., Ko, J. P., & Litt, H. (2020). Radiological Society of North America expert consensus document on reporting chest CT findings related to COVID-19: Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiology: Cardiothoracic Imaging, 2(2), e200152. https://doi.org/10.1148/ryct.2020200152
Singh, D., & Yi, S. V. (2021). On the origin and evolution of SARS-CoV-2. Experimental & Molecular Medicine, 53(4), 537–547. https://doi.org/10.1038/s12276-021-00604-z
Singh, N., Tang, Y., Zhang, Z., & Zheng, C. (2020). COVID-19 waste management: Effective and successful measures in Wuhan, China. Resources, Conservation, and Recycling, 163, 105071. https://doi.org/10.1016/j.resconrec.2020.105071
Solis, A., & Nunn, C. L. (2021). One health disparities and COVID-19. Evolution, Medicine, and Public Health, 9(1), 70–77. https://doi.org/10.1093/emph/eoab003
Sørensen, K., Okan, O., Kondilis, B., & Levin-Zamir, D. (2021). Rebranding social distancing to physical distancing: Calling for a change in the health promotion vocabulary to enhance clear communication during a pandemic. Global Health Promotion, 28(1), 5–14. https://doi.org/10.1177/1757975920986126
Sornette, D., Mearns, E., Schatz, M., Wu, K., & Darcet, D. (2020). Interpreting, analysing and modelling COVID-19 mortality data. Nonlinear Dynamics, 101(3), 1–26. https://doi.org/10.1007/s11071-020-05966-z
Stone, J. H., Frigault, M. J., Serling-Boyd, N. J., Fernandes, A. D., Harvey, L., Foulkes, A. S., Horick, N. K., Healy, B. C., Shah, R., Bensaci, A. M., Woolley, A. E., Nikiforow, S., Lin, N., Sagar, M., Schrager, H., Huckins, D. S., Axelrod, M., Pincus, M. D., Fleisher, J., et al. (2020). Efficacy of tocilizumab in patients hospitalized with Covid-19. New England Journal of Medicine, 383(24), 2333–2344. https://doi.org/10.1056/nejmoa2028836
Straif-Bourgeois, S., Ratard, R., & Kretzschmar, M. (2014). Infectious disease epidemiology. In Handbook of epidemiology (pp. 2041–2119). Springer. https://doi.org/10.1007/978-0-387-09834-0_34
Subbian, S. (2021). The abstruse side of type I interferon immunotherapy for COVID-19 cases with comorbidities. Journal of Respiration, 1(1), 49–59. https://doi.org/10.3390/jor1010005
Subramanian, R., He, Q., & Pascual, M. (2021). Quantifying asymptomatic infection and transmission of COVID-19 in New York City using observed cases, serology, and testing capacity. Proceedings of the National Academy of Sciences, 118(9), e2019716118. https://doi.org/10.1073/pnas.2019716118
Sun, Z., Zhang, N., Li, Y., & Xu, X. (2020). A systematic review of chest imaging findings in COVID-19. Quantitative Imaging in Medicine and Surgery, 10(5), 1058–1079. https://doi.org/10.21037/qims-20-564
Swaroop, S. (1957). Index of endemicity. Bulletin of the World Health Organization, 16(6), 1083–1101. https://apps.who.int/iris/handle/10665/265708
Tacquard, C., Mansour, A., Godon, A., Godet, J., Poissy, J., Garrigue, D., Kipnis, E., Rym Hamada, S., Mertes, P. M., Steib, A., Ulliel-Roche, M., Bouhemad, B., Nguyen, M., Reizine, F., Gouin-Thibault, I., Besse, M. C., Collercandy, N., Mankikian, S., Levy, J. H., et al. (2021). Impact of high-dose prophylactic anticoagulation in critically ill patients with COVID-19 pneumonia. Chest, 159(6), 2417–2427. https://doi.org/10.1016/j.chest.2021.01.017
Tellier, R., Li, Y., Cowling, B. J., & Tang, J. W. (2019). Recognition of aerosol transmission of infectious agents: A commentary. BMC Infectious Diseases, 19(1), 101. https://doi.org/10.1186/s12879-019-3707-y
The Lancet Oncology. (2020). COVID-19 and the US health insurance conundrum (Editorial). Lancet Oncology, 21(6), 733. https://doi.org/10.1016/s1470-2045(20)30286-2
The Lancet Respiratory Medicine. (2020). The future of critical care: Lessons from the COVID-19 crisis (Editorial). The Lancet Respiratory Medicine, 8(6), 527. https://doi.org/10.1016/S2213-2600(20)30240-X
Thompson, D. C., Barbu, M. G., Beiu, C., Popa, L. G., Mihai, M. M., Berteanu, M., & Popescu, M. N. (2020). The impact of COVID-19 pandemic on long-term care facilities worldwide: An overview on international issues. BioMed Research International, 2020, 8870249. https://doi.org/10.1155/2020/8870249
Ting, D. S. W., Carin, L., Dzau, V., & Wong, T. Y. (2020). Digital technology and COVID-19. Nature Medicine, 26(4), 459–461. https://doi.org/10.1038/s41591-020-0824-5
Trevethan, R. (2017). Sensitivity, specificity, and predictive values: Foundations, pliabilities, and pitfalls in research and practice. Frontiers in Public Health, 5, 307. https://doi.org/10.3389/fpubh.2017.00307
Trevisanuto, D., Cavallin, F., Cavicchiolo, M. E., Borellini, M., Calgaro, S., & Baraldi, E. (2020). Coronavirus infection in neonates: A systematic review. Archives of Disease in Childhood. Fetal and Neonatal Edition, 106(3), 330–335. https://doi.org/10.1136/archdischild-2020-319837
Triggle, C. R., Bansal, D., Farag, E. A. B. A., Ding, H., & Sultan, A. A. (2020). COVID-19: Learning from lessons to guide treatment and prevention interventions. mSphere, 5(3), e00317-20. https://doi.org/10.1128/mSphere.00317-20
Tulchinsky, T. H., & Varavikova, E. A. (2014a). A history of public health. The New Public Health, 1–42. https://doi.org/10.1016/b978-0-12-415766-8.00001-x
Tulchinsky, T. H., & Varavikova, E. A. (2014b). Communicable diseases. The New Public Health, 149–236. https://doi.org/10.1016/b978-0-12-415766-8.00004-5
Ujike, M., & Taguchi, F. (2015). Incorporation of Spike and Membrane Glycoproteins into Coronavirus Virions. Viruses, 7(4), 1700–1725. https://doi.org/10.3390/v7041700
Ulhaq, A., Khan, A., Gomes, D. P. S., & Paul, M. (2020, April 15). Computer vision for COVID-19 control: A survey. https://doi.org/10.31224/osf.io/yt9sx
United Health Professionals. (2021, February 26). The Covid outbreak: “Biggest health scam of the 21st century.” Report by 1500 Health Professionals. Global Research. https://www.globalresearch.ca/the-covid-outbreak-biggest-health-scam-of-the-21st-century-report-by-1500-health-professionals/5737838
United Nations Environment Programme. (2020). Waste management during the COVID-19 pandemic from response to recovery. https://wedocs.unep.org/bitstream/handle/20.500.11822/33416/WMC-19.pdf?sequence=1&isAllowed=y
V’kovski, P., Kratzel, A., Steiner, S., Stalder, H., & Thiel, V. (2020). Coronavirus biology and replication: Implications for SARS-CoV-2. Nature Reviews Microbiology, 19(3), 155–170. https://doi.org/10.1038/s41579-020-00468-6
Vainionpää, R., & Leinikki, P. (2008). Diagnostic techniques: Serological and molecular approaches. Encyclopedia of Virology, 29–37. https://doi.org/10.1016/b978-012374410-4.00585-9
van der Made, C. I., Simons, A., Schuurs-Hoeijmakers, J., van den Heuvel, G., Mantere, T., Kersten, S., van Deuren, R. C., Steehouwer, M., van Reijmersdal, S. V., Jaeger, M., Hofste, T., Astuti, G., Corominas Galbany, J., van der Schoot, V., van der Hoeven, H., Wanda Hagmolen of Ten Have, Klijn, E., van den Meer, C., Fiddelaers, J., et al. (2020). Presence of genetic variants among young men with severe COVID-19. JAMA, 324(7), 663. https://doi.org/10.1001/jama.2020.13719
Verdecchia, P., Cavallini, C., Spanevello, A., & Angeli, F. (2020). The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. European Journal of Internal Medicine, 76, 14–20. https://doi.org/10.1016/j.ejim.2020.04.037
Viglione, G. (2020). Are women publishing less during the pandemic? Here’s what the data say. Nature, 581(7809), 365–366. https://doi.org/10.1038/d41586-020-01294-9
Wang, C., Zheng, Y., Niu, Z., Jiang, X., & Sun, Q. (2021a). The virological impacts of SARS-CoV-2 D614G mutation. Journal of Molecular Cell Biology, 13, 712. https://doi.org/10.1093/jmcb/mjab045
Wang, P., Nair, M. S., Liu, L., Iketani, S., Luo, Y., Guo, Y., Wang, M., Yu, J., Zhang, B., Kwong, P. D., Graham, B. S., Mascola, J. R., Chang, J. Y., Yin, M. T., Sobieszczyk, M., Kyratsous, C. A., Shapiro, L., Sheng, Z., Huang, Y., & Ho, D. D. (2021b). Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. bioRvix. https://doi.org/10.1101/2021.01.25.428137
Wapner, J. (2020). Covid-19: Medical expenses leave many Americans deep in debt. BMJ, 370, m3097. https://doi.org/10.1136/bmj.m3097
Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S., & Crispin, M. (2020). Site-specific glycan analysis of the SARS-CoV-2 spike. Science, 369, 330. https://doi.org/10.1126/science.abb9983
Wei, C., Wan, L., Yan, Q., Wang, X., Zhang, J., Yang, X., Zhang, Y., Fan, C., Li, D., Deng, Y., Sun, J., Gong, J., Yang, X., Wang, Y., Wang, X., Li, J., Yang, H., Li, H., Zhang, Z., et al. (2020). HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nature Metabolism, 2(12), 1391–1400. https://doi.org/10.1038/s42255-020-00324-0
Weiner, D. L., Balasubramaniam, V., Shah, S. I., Javier, J. R., & Pediatric Policy Council. (2020). COVID-19 impact on research, lessons learned from COVID-19 research, implications for pediatric research. Pediatric Research, 88(2), 148–150. https://doi.org/10.1038/s41390-020-1006-3
Williams, T. C., & Burgers, W. A. (2021). SARS-CoV-2 evolution and vaccines: Cause for concern? The Lancet Respiratory Medicine, 9, 333. https://doi.org/10.1016/S2213-2600(21)00075-8
Wool, G. D., & Miller, J. L. (2020). The impact of COVID-19 disease on platelets and coagulation. Pathobiology, 88(1), 15–27. https://doi.org/10.1159/000512007
Woolf, S. H., Chapman, D. A., & Lee, J. H. (2020). COVID-19 as the leading cause of death in the United States. JAMA, 325, 123. https://doi.org/10.1001/jama.2020.24865
World Health Organization. (2017, September 21). One Health. https://www.who.int/news-room/q-a-detail/one-health
World Health Organization. (2020, December 31). COVID-19 - Global. WHO. https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON305
World Health Organization. (2021a, September 29). Coronavirus disease (COVID-19) dashboard. https://covid19.who.int/
World Health Organization. (2021b, December 31). COVID-19 – Global. WHO. https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON305
World Health Organization. (2021c, October 12). https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/
World Health Organization. (2021d, June 25). Considerations for quarantine of contacts of COVID-19 cases. https://www.who.int/publications/i/item/considerations-for-quarantine-of-individuals-in-the-context-of-containment-for-coronavirus-disease-(covid-19)
World Health Organization & United Nations Children’s Fund (UNICEF). (2020). Water, sanitation, hygiene, and waste management for the COVID-19 virus: interim guidance. https://apps.who.int/iris/bitstream/handle/10665/331499/WHO-2019-nCoV-IPC_WASH-2020.2-eng.pdf?sequence=1&isAllowed=y
Xia, S., Zhu, Y., Liu, M., Lan, Q., Xu, W., Wu, Y., Ying, T., Liu, S., Shi, Z., Jiang, S., & Lu, L. (2020). Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cellular & Molecular Immunology, 17(7), 765–767. https://doi.org/10.1038/s41423-020-0374-2
Xu, Y., Li, X., Zhu, B., Liang, H., Fang, C., Gong, Y., Guo, Q., Sun, X., Zhao, D., Shen, J., Zhang, H., Liu, H., Xia, H., Tang, J., Zhang, K., & Gong, S. (2020). Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nature Medicine, 26(4), 502–505. https://doi.org/10.1038/s41591-020-0817-4
Yao, H., Lu, X., Chen, Q., Xu, K., Chen, Y., Cheng, M., Chen, K., Cheng, L., Weng, T., Shi, D., Liu, F., Wu, Z., Xie, M., Wu, H., Jin, C., Zheng, M., Wu, N., Jiang, C., & Li, L. (2020). Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discovery, 6(1), 76. https://doi.org/10.1038/s41421-020-00226-1
Yau, O., Gin, K., Luong, C., Jue, J., Abolmaesumi, P., Tsang, M., Nair, P., & Tsang, T. S. M. (2020). Point-of-care ultrasound in the COVID-19 era: A scoping review. Echocardiography, 38, 329. https://doi.org/10.1111/echo.14951
Ye, Z. W., Yuan, S., Yuen, K.-S., Fung, S.-Y., Chan, C.-P., & Jin, D.-Y. (2020). Zoonotic origins of human coronaviruses. International Journal of Biological Sciences, 16(10), 1686–1697. https://doi.org/10.7150/ijbs.45472
Yeo, K. T., Oei, J. L., De Luca, D., Schmölzer, G. M., Guaran, R., Palasanthiran, P., Kumar, K., Buonocore, G., Cheong, J., Owen, L. S., Kusuda, S., James, J., Lim, G., Sharma, A., Uthaya, S., Gale, C., Whittaker, E., Battersby, C., Modi, N., et al. (2020). Review of guidelines and recommendations from 17 countries highlights the challenges that clinicians face caring for neonates born to mothers with COVID-19. Acta Paediatrica, 109(11), 2192–2207. https://doi.org/10.1111/apa.15495
Yi, Y., Lagniton, P. N. P., Ye, S., Li, E., & Xu, R. H. (2020). COVID-19: What has been learned and to be learned about the novel coronavirus disease. International Journal of Biological Sciences, 16(10), 1753–1766. https://doi.org/10.7150/ijbs.45134
Yoo, J. Y., Dutra, S. V. O., Fanfan, D., Sniffen, S., Wang, H., Siddiqui, J., Song, H. S., Bang, S. H., Kim, D. E., Kim, S., & Groer, M. (2020). Comparative analysis of COVID-19 guidelines from six countries: A qualitative study on the US, China, South Korea, the UK, Brazil, and Haiti. BMC Public Health, 20(1), 1853. https://doi.org/10.1186/s12889-020-09924-7
Yoshimoto, F. K. (2021). A Biochemical Perspective of the Nonstructural Proteins (NSPs) and the Spike Protein of SARS CoV-2. Protein Journal, 40(3), 260–295. https://doi.org/10.1007/s10930-021-09967-8
Yu, J., Nie, L., Wu, D., Chen, J., Yang, Z., Zhang, L., Li, D., & Zhou, X. (2020). Prognostic value of a clinical biochemistry-based nomogram for coronavirus disease 2019. Frontiers in Medicine, 7, 597791. https://doi.org/10.3389/fmed.2020.597791
Yüce, M., Filiztekin, E., & Özkaya, K. G. (2021). COVID-19 diagnosis—A review of current methods. Biosensors and Bioelectronics, 172, 112752. https://doi.org/10.1016/j.bios.2020.112752
Zahan, M., Habibi, H., Pencil, A., Abdul-Ghafar, J., Ahmadi, S., Juyena, N., Rahman, M., & Parvej, M. (2021). Diagnosis of COVID-19 in symptomatic patients: An updated review. Vacunas, 23, 55. https://doi.org/10.1016/j.vacun.2021.06.002
Zhang, C., Wu, Z., Li, J.-W., Zhao, H., & Wang, G.-Q. (2020a). Cytokine release syndrome in severe COVID-19: Interleukin-6 receptor antagonist tocilizumab may be the key to reduce mortality. International Journal of Antimicrobial Agents, 55(5), 105954. https://doi.org/10.1016/j.ijantimicag.2020.105954
Zhang, L., Jackson, C. B., Mou, H., Ojha, A., Rangarajan, E. S., Izard, T., Farzan, M., & Choe, H. (2020b). The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv. https://doi.org/10.1101/2020.06.12.148726
Zhang, W., Wu, X., Zhou, H., & Xu, F. (2020c). Clinical characteristics and infectivity of asymptomatic carriers of SARS-CoV2 [Review]. Experimental and Therapeutic Medicine, 21(2), 115. https://doi.org/10.3892/etm.2020.9547
Zhang, Z.-L., Hou, Y.-L., Li, D.-T., & Li, F.-Z. (2020d). Laboratory findings of COVID-19: A systematic review and meta-analysis. Scandinavian Journal of Clinical and Laboratory Investigation, 80(6), 441–447. https://doi.org/10.1080/00365513.2020.1768587
Zhang, J., Xiao, T., Cai, Y., & Chen, B. (2021). Structure of SARS-CoV-2 spike protein. Current Opinion in Virology, 50, 173–182. https://doi.org/10.1016/j.coviro.2021.08.010
Zhao, J., Yang, Y., Huang, H., Li, D., Gu, D., Lu, X., Zhang, Z., Liu, L., Liu, T., Liu, Y., He, Y., Sun, B., Wei, M., Yang, G., Wang, X., Zhang, L., Zhou, X., Xing, M., & Wang, P. G. (2020). Relationship between the ABO blood group and the COVID-19 susceptibility. medRxiv. https://doi.org/10.1101/2020.03.11.20031096
Zheng, S., Fan, J., Yu, F., Feng, B., Lou, B., Zou, Q., Xie, G., Lin, S., Wang, R., Yang, X., Chen, W., Wang, Q., Zhang, D., Liu, Y., Gong, R., Ma, Z., Lu, S., Xiao, Y., Gu, Y., et al. (2020a). Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: Retrospective cohort study. BMJ, 369, m1443. https://doi.org/10.1136/bmj.m1443
Zheng, Z., Peng, F., Xu, B., Zhao, J., Liu, H., Peng, J., Li, Q., Jiang, C., Zhou, Y., Liu, S., Ye, C., Zhang, P., Xing, Y., Guo, H., & Tang, W. (2020b). Risk factors of critical and mortal COVID-19 cases: A systematic literature review and meta-analysis. Journal of Infection, 81(2), e16–e25. https://doi.org/10.1016/j.jinf.2020.04.021
Zhou, Q., Chen, V., Shannon, C. P., Wei, X. S., Xiang, X., Wang, X., Wang, Z. H., Tebbutt, S. J., Kollmann, T. R., & Fish, E. N. (2020a). Interferon-α2b treatment for COVID-19. Frontiers in Immunology, 11, 1061. https://doi.org/10.3389/fimmu.2020.01061
Zhou, Y., Wang, F., Tang, J., Nussinov, R., & Cheng, F. (2020b). Artificial intelligence in COVID-19 drug repurposing. The Lancet. Digital Health, 2(12), e667–e676. https://doi.org/10.1016/s2589-7500(20)30192-8
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Hosseini-Nezhad, P., Hosseini-Nezhad, S., Hosseini-Nezhad, A. (2023). Medical Perspective on COVID-19. In: Faghih, N., Forouharfar, A. (eds) Biopolitics and Shock Economy of COVID-19 . Contributions to Economics. Springer, Cham. https://doi.org/10.1007/978-3-031-27886-0_2
Download citation
DOI: https://doi.org/10.1007/978-3-031-27886-0_2
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-27885-3
Online ISBN: 978-3-031-27886-0
eBook Packages: Economics and FinanceEconomics and Finance (R0)