Abstract
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and its fast variant, ultrafast DCE-MRI, are useful for the management of breast cancer. Segmentation of breast lesions is necessary for automatic clinical decision support. Despite the advantage of acquisition time, existing segmentation studies on ultrafast DCE-MRI are scarce, and they are mostly fully supervised studies with high annotation costs. Herein, we propose a semi-supervised segmentation approach that can be trained with small amounts of annotations for ultrafast DCE-MRI. A time difference map is proposed to incorporate the distinct time-varying enhancement pattern of the lesion. Furthermore, we present a novel loss function that efficiently distinguishes breast lesions from non-lesions based on triple loss. This loss reduces the potential false positives induced by the time difference map. Our approach is compared to that of five competing methods using the dice similarity coefficient and two boundary-based metrics. Compared to other models, our approach achieves better segmentation results using small amounts of annotations, especially for boundary-based metrics relevant to spatially continuous breast lesions. An ablation study demonstrates the incremental effects of our study. Our code is available on GitHub (https://github.com/yt-oh96/SSL-CTL).
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Breast cancer is the most frequently diagnosed cancer in women and the main cause of cancer-related deaths [1]. Early detection of breast cancer can significantly lower mortality rates [2]. The importance of early detection has been widely recognized; therefore, breast cancer screening has led to better patient care [3, 4]. Compared to commonly used mammography, dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is increasingly being adopted owing to its higher sensitivity in dense breasts [5,6,7].
Breast DCE-MRI has many phases, including precontrast, early, and delayed phases. After contrast agent (CA) injection, each phase is recorded with a different delay time, up to a few minutes, from the initial CA injection to measure the distinct time-varying enhancement [8]. Each phase takes approximately 60-120 s to acquire. High-resolution T2 weighted and diffusion-weighted sequences have been routinely added for the complete MRI sequence. This leads to an increased scan time, ranging from 20 to 40 min [9]. Because a long scan time is associated with high cost, it is urgent to shorten the scan protocol for the widespread adoption of breast DCE-MRI [10,11,12].
Ultrafast DCE-MRI records an early inflow of CA and can obtain whole-breast images at several time points within 1 min after CA injection [13]. Conventional DCE-MRI is typically performed immediately after ultrafast sequencing. Within the first minute of ultrafast DCE-MRI, malignant breast lesions show altered patterns compared to that of benign tissue in terms of shorter enhancement, steeper maximum slope, and higher initial contrast ratio [14,15,16,17]. This implies that there could be lesion-differentiating information in ultrafast DCE-MRI.
Manual segmentation of breast lesions in DCE-MRI is troublesome; therefore, many computer-aided detection systems have been developed to automatically segment breast lesions [18, 19]. These segmentation methods are increasingly adopting deep learning approaches [19, 20]. There is limited literature on the application of deep learning methods for ultrafast DCE-MRI, possibly moving toward a short scan time for breast MRI imaging [5, 19,20,21]. However, these methods are supervised learning approaches with high labeling costs.
In this study, we propose a semi-supervised segmentation method for breast lesions using ultrafast DCE-MRI with limited label data. In our method, we use a time difference map (TDM) to incorporate the distinct time-varying enhancement pattern of the lesion [21].Our TDM could locate enhanced regions, including the breast lesion, but could also enhance the blood vessel that receives the CA. To solve this problem, we introduce a distance-based learning approach of triplet loss to better contrast a lesion with a non-lesion area. Compared with various semi-supervised segmentation methods, our method can segment breast lesions well, even with a few labels. We obtained MRI data from 613 patients from Samsung Medical Center. Our method was evaluated using three metrics: dice similarity coefficient, average surface distance, and Hausdorff distance. The main contributions of our study are summarized as follows:
-
1.
As labeled medical image data are difficult to obtain, we propose a semi-supervised segmentation method based on pseudo-labels.
-
2.
We add TDM to explicitly model the distinct time-varying enhancement pattern of lesions.
-
3.
We propose a local cross-triplet loss to discover the similarities and differences between breast lesions and non-lesion areas. This allows our model to focus on breast lesions with limited labeling data.
2 Related Work
Supervised Learning in DCE-MRI. Automatic segmentation technologies help with diagnosis and treatment planning tasks by reducing the time resources for manual annotation of breast cancer. In particular, deep learning algorithms show considerable potential and are gaining ground in breast imaging [22]. Several studies have proposed segmentation methods using conventional DCE-MRI. For example, Piantadosi et al. [23] proposed a fully automated breast lesion segmentation approach for breast DCE-MRI using 2D U-Net. Maicas et al. [24] used reinforcement learning to automatically detect breast lesions. Zhang et al. [25] proposed a breast mask to exclude confounding nearby structures and adopted two fully convolutional cascaded networks to detect breast lesions using the mask as a guideline. The aforementioned approaches worked well compared to those of the conventional machine learning approaches, they adopted conventional DCE-MRI with a long scan time.
Recently, the effectiveness of ultrafast DCE-MRI has been demonstrated [12,13,14,15], and studies using deep learning approaches in ultrafast DCE-MRI have been actively pursued. Ayatollahi et al. [19] detected breast lesions using spatial and temporal information obtained during the early stages of dynamic acquisition. Oh et al. [21] showed that ultrafast DCE-MRI could be used to generate conventional DCE-MRI, confirming the possibility of replacing conventional DCE-MRI with ultrafast DCE-MRI. These studies had shorter scan times for data acquisition than that of conventional DCE-MRI. However, because they adopted supervised learning, the labeling cost remained significant. Therefore, in this study, we propose a semi-supervised segmentation method using only a small amount of labeling data.
Semi-supervised Learning. Semi-supervised learning is widely used to reduce time-consuming and expensive manual pixel-level annotation [26, 27].
Consistency regularization imposes a constraint on consistency between predictions with and without perturbations applied to inputs, features, and networks [28]. “Mean Teacher” [29] updates the model parameter values of the teacher model by the exponential moving average of the model parameter values of the student model. Based on “Mean Teacher” [29], “Uncertainty Aware Mean Teacher” [30] proposed a consistency loss such that learning proceeds only with reliable predictions using the uncertainty information of the teacher model.
Entropy minimization forces the classifier to make predictions with low entropy for an unlabeled input. This method assumes that the classifier’s decision boundary does not traverse the dense area of the marginal data distribution [31]. “ADVENT” [32] proposed an entropy-based adversarial training strategy.
“Pseudo-label” [33] implicitly performs entropy minimization because the intermediate prediction probability undergoes one-hot encoding [31]. The recently proposed “cross-pseudo supervision” [28] is a consistency regularization approach with perturbations of the network that provides input after different initializations for two networks of the same structure. For unannotated inputs, the pseudo-segmentation map of one network was utilized to supervise the other network. This can increase the number of annotated training data, resulting in more stable and accurate prediction results.
Our proposed model is based on “cross-pseudo supervision” [28] and introduces TDM and local cross-triplet loss to incorporate the time-varying enhancement pattern of ultrafast DCE-MRI and contrast lesions from non-lesions. Our model performs an input perturbation to enforce the consistency of the intermediate features. The proposed method achieves reliable segmentation with fewer annotations.
An overview of the proposed method. The upstream model (purple) and downstream model (orange) receive both the original input (X) and transformed input T(X). P1 and P2 are segmentation confidence maps derived from the two models. Y1 and Y2 are the final segmentation confidence maps with TDM applied. Flows corresponding to the blue dotted arrow for one model lead to pseudo-labels for the other model. The product sign means element-wise multiplication. (Color figure online)
Deep Metric Learning. Deep metric learning maps an image to a feature vector in a manifold space through deep neural networks [34]. The mapped feature vector is trained using a distance function. Deep metric learning approaches typically optimize the loss functions defined on pairs or triplets of training examples [35]. The contrastive loss of pairs minimizes the distance from the feature vectors of the same class while ensuring the separation of the feature vectors of different classes [36]. Alternatively, triplet loss is defined based on three points: anchor point, positive point (i.e., a feature vector belonging to the same class as the anchor), and negative point (i.e., a feature vector belonging to a different class than the anchor). This loss forces the distance of the positive pair to be smaller than that of the negative pair [37]. “CUT” [38], a recently proposed patch-based approach, defines negatives within the input image itself. This leads to efficient discrimination between the target and nontarget areas. Inspired by this method, we propose a local cross-triplet loss to discriminate breast lesions from non-lesions in the input image. This is discussed in detail in Sect. 3.2.
3 Methodology
This study aims to accurately segment breast cancer lesions with small annotation data based on pseudo-labels. First, TDM is defined to incorporate time-varying enhancement patterns in ultrafast DCE-MRI. Next, we discuss the drawbacks of TDM in our task, as well as the proposed loss terms to overcome the shortcomings using metric learning. An overview of the proposed method is shown in Fig. 1.
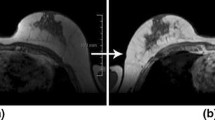
Effects of time difference map (TDM). (a) Representative 1st phase image of ultrafast DCE-MRI. (b) TDM is computed as the difference between the last-phase image of ultrafast DCE-MRI and (a). (c) Example of the training result of “cross-pseudo supervision” (5% of the labeled data is used for training). This shows the focused regions for segmentation (white, cancer; red, vessel). (d) Ground truth. (Color figure online)
Illustration of the proposed local cross-triplet loss. DS denotes downsampling, and RS denotes random spatial sampling. Anchor and positive points come from the positive group \(G_{pos}\) defined locally in the cancer region. Negative points come from the negative group \(G_{neg}\) defined locally in the non-cancer region. Cross-triplet comes from the loss defined across two streams. The loss is computed for each stream (\(M_{up}\), \(M_{down}\)) and the two losses(\(L^{up}_{lct}\),\(L^{down}_{lct}\)) are combined to obtain \(L_{lct}\).
3.1 TDM
Ultrafast DCE-MRI has up to 20 phasic images, each taking 2-3 s, within 1 min after CA. Our ultrafast DCE-MRI sequence has 17 phases. We introduce the TDM to incorporate time-varying information. Lesions appear brighter than those in the normal regions of the breast. In general, as we traverse the time steps in the ultrafast sequence, the slope of the intensity change in the lesion is positive, while that in the normal region remains flat, close to zero. TDMs computed from consecutive phases or the averaged TDM are certainly possible, but for computational efficiency and the linear trend of enhancement, TDM is defined as the difference between the first and last (17th) phases in our ultrafast data following Oh et al. [21]. Our ultrafast DCE-MRI(V) are of dimension \(H\times W\times D\times F\times \), where \(D=1\) and \(F=17\). Unlike color images, we have only one channel \(D=1\). TDM is defined as the difference between the last frame and the first frame. TDM can be obtained as follows:
The TDM generated in this manner is multiplied by the prediction of the model so that the model could focus only on the enhanced regions, as shown in the final segmentation results (Fig. 1). However, the results show that the blood vessel region is enhanced in addition to the lesion (Fig. 2). This could be especially detrimental because our approach is limited to a small amount of label data. To overcome this problem, we propose a local cross-triplet loss.
3.2 Local-Cross Triplet Loss
Starting from \(D^l\) with N labeled images and \(D^u\) with M unlabeled images. our local cross-triplet loss \(L_{lct}\) is designed to avoid false detection in situations where there is a limited amount of label data available (\(N<<< M\)). The input image \(X \in R^{H\times W}\), with height (H) and width (W), is mapped to anchor, positive, and negative points that are \(h\times w\) K-dimensional vectors \({\textbf {z}},{\textbf {z}}^+,{\textbf {z}}^- \in R^K\) through the encoder of U-net [39] used as the backbone, where h,w are the height and width of the feature volume. respectively, We use h=7, w=7, K=256 for our setup.
The positive candidate group \(G_{pos}\) corresponding to the cancer region and the negative candidate group \(G_{neg} \)corresponding to the non-cancer region, are defined in the upstream model \(M_{up}\) and downstream model \(M_{down}\) of Fig. 3. \(G_{pos}\) is obtained by downsampling the given binary label mask to the size of the feature map. Similarly, \(G_{neg}\) can be obtained by downsampling the input image that contains the entire breast and others. However, background regions outside the breast lack the necessary information. Therefore, we set \(G_{neg}\) to exclude background and cancer regions (Fig. 3). Both groups are obtained from one given image, and thus, our design allows for effective local discrimination of breast lesions from non-lesions.
We adopt the local cross-triplet loss approach for both streams (\(M_{up}\) and \(M_{down}\)), The loss of \(M_{up}\) is as follows. “Locality” comes from how the triplet points are defined. Anchor and positive points come from the positive group \(G_{pos}\) defined locally in the cancer region. Negative points come from the negative group \(G_{neg}\) defined locally in the non-cancer region. “Cross-triplet” comes from the loss defined across two streams, where anchor points of one stream are compared with positive/negative points of the other stream. A K-dimensional vector \({\textbf {z}}\) is chosen at random from \(G_{pos}\) of \(M_{up}\) to work as an anchor point and the K-dimensional vectors \({\textbf {z}}^+\) and \({\textbf {z}}^-\) are chosen at random from \(G_{pos}\) and \(G_{neg}\) of \(M_{down}\) to work as positive and negative points, respectively. Then, \(L^{up}_{lct}\) can be written as :
where \(\tau \) is an important tuning parameter for supervised feature learning [38, 40]. We set this value at 0.07, as in previous studies [38]. The loss for \(M_{down}\) can be calculated in the same manner as the loss for \(L^{up}_{lct}\). We use the final \(L_{lct}\) obtained by combining \(L^{up}_{lct}\) and \(L^{down}_{lct}\) in the following manner.
3.3 Cross Pseudo Supervision Loss
In both labeled and unlabeled data, the loss for semantic segmentation consists of supervision loss \(L_s\) and cross-pseudo supervision loss \(L_{cps}\) [28]. Our models \(M_{up}\) and \(M_{down}\) for \(L_s\) use an original input image and a transformed input image as input, respectively. The segmentation confidence maps created in this manner are P1(X) and P2(T(X)). The final segmentation confidence maps Y1,Y2 are created by applying the TDM and the transformed TDM introduced in Sect. 3.1. Standard pixel-wise cross-entropy for the confidence vectors \({\textbf {y}}_{1i}\),\({\textbf {y}}_{2i}\) at each i position is as follows:
where T is the geometric transformation for the input perturbation, the circle dot is the element-wise multiplication, and \({\textbf {l}}_{1i}\) and \({\textbf {l}}_{2i}\) are one-hot vector corresponding to each pixel i of the ground truth Label, T(Label).
The cross-pseudo supervision loss proposed in “cross-pseudo supervision” [28] is similar to the supervision loss above, but uses a pixel-wise one-hot label map created by the one-hot encoding of segmentation confidence maps from different stream models as the ground truth. The loss for the unlabeled data is as follows:
where \({\textbf {pl}}_{1i}\), \({\textbf {pl}}_{2i}\) are the hot vectors for each pixel i in a pixel-wise one-hot label map created using different stream models. Because pseudo supervision cannot access ground truth information, the addition of TDM might lead to focusing on unimportant regions, such as vessels, making the learning unstable. Therefore, we do not use the TDM when generating pseudo-supervision.
The cross-pseudo supervision loss \(L_{cps}^{l}\) for labeled data can be defined in the same manner. The cross-pseudo supervision loss for both labeled and unlabeled data is \(L_{cps}\)=\(L_{cps}^{u}\)+\(L_{cps}^{l}\). The final objective function to which the trade-off weight \(\lambda \) is applied is as follows:
4 Experiments
4.1 Dataset
The institutional review board of Samsung Medical Center authorized this study, and the requirement for informed consent was waived. The total number of patients was 613, 500 as the training set, 50 as the validation set, and the remaining 63 as the test set. An expert manually segmented the breast lesions in the entire dataset. The following MRI protocol was used to acquire the imaging data. First, images of the precontrast phase were captured before CA injection. Following CA administration, ultrafast DCE-MRI images were collected for approximately 1 min, followed by conventional DCE-MRI images of the three phases recorded at 70 s intervals. There were three imaging acquisition settings available for ultrafast DCE-MRI, and the typical settings were as follows. The imaging data were collected using a Philips 3T Ingenia CX scanner. Echo time, repetition time, and field-of-view were 2.1 ms, 4.1 ms, and 330 \(\times \) 330 mm\(^{2}\), respectively. In-plane resolution, slice thickness, and temporal resolution per phase were 1.0 \(\times \) 1.0 mm, 1.0 mm, and 3.4 s, respectively. Because three settings were employed in the data collection process, 1.0 \(\times \) 1.0 \(\times \) 1.0 resampling was used to unify them. Only images containing lesions were selected and further divided into left- and right-breast images by splitting them in half in the horizontal direction.
4.2 Implementation Details
To evaluate the effectiveness of the proposed approach, we compare ours with five models: 1) U-net [39]; 2) cross-pseudo supervision [28]; 3) mean teacher [29]; 4) uncertainty aware mean teacher [30]; and 5) entropy minimization [32]. The U-net is adopted to evaluate the fully supervised learning scenario, and the associated results using 100% labeled data represent the upper bound of our semi-supervised approach. All models, including ours, use a Vanilla U-net as their backbone. U-net-related codes are implemented using those proposed by Luo and Xiangde [41]. Our model is based on cross-pseudo supervision [28], which requires the application of geometric transforms to the input. Random rotation, flipping, and cropping are performed.
We train various models on four NVIDIA TITAN XP GPUs. For all experiments, the batch size is set to 24, and the maximum number of iterations is 15000. All codes are implemented using Pytorch1.8.0 and are available on GitHub [42].
4.3 Evaluation Metrics
We measure segmentation performance using three metrics: 1) dice similarity coefficient (DSC); 2) average surface distance (ASD); 3) Hausdorff distance (HD). DSC is the most representative metric based on the spatial overlap between the predictive segmentation and ground truth. However, it is insensitive to the spatial continuity of the segmentation results [43]. As shown in Fig. 4, one result is spatially continuous, while the other is spatially disparate. Both cases have the same DSCs, but the spatially continuous result is relevant to the context of the breast lesion. Therefore, we further adopt the ASD and HD metrics based on the boundary distances. The ASD is the average of all distances from the point of the boundary of the predictive segmentation to that of the ground truth. HD is the maximum distance between a point in one of the two segmentation results and its nearest point in the other. Due to the spatially continuous nature of the breast lesion, it is important to have low ASD and HD, which penalize spatially disparate segmentation maps that are likely to be false positives.
4.4 Results
Figure 5 shows the performance metrics related to the increasing proportion of labeled data used for training. Because this study aims at an environment with limited labeled data, we show the results using up to 50% of the labeled data. As the proportion of labeled data approaches 100%, all models perform equally well, making the comparison pointless. DSCs are comparable among the methods; however, our model performs better than that of other methods for ASD and HD, which are more relevant for spatially continuous breast lesions.
We performed a 5-fold cross validation. Table 1 shows a detailed comparison of the models using 5% or 50% of the labeled data. Our model using 50% labeled data achieves a performance similar to that of the upper bound obtained from supervised U-net results using 100% labeled data. More importantly, our model trained with only 5% of the labeling data shows similar performance in boundary-based metrics (ASD and HD) compared to the results of other models trained with 50% of the labeling data. This demonstrates that our proposed method does not detect false positives and segments spatially continuous breast lesions even with a small amount of labeling data.
Figure 6 shows representative segmentation results for various methods for different axial slices. All models use 5% of the labeled data. The U-net model uses 5% of the labeled data, while the others use 5% of the labeled data and 95% of the unlabeled data. Our method generally provides more accurate predictions than other comparative models. Furthermore, our model has fewer false positives, primarily due to enhanced blood vessels, and results in one spatially continuous cluster for the lesion. These results confirm that the loss terms of our approach are effective for the segmentation of breast lesions.
4.5 Ablation Study
An ablation study is conducted to demonstrate the incremental effects of the proposed contributions (Table 2). The cross-pseudo supervision model is used as the baseline model. 1) We apply TDM to incorporate the time-varying enhancement pattern of the lesion into the baseline model. DSC is improved because TDM can focus on enhanced regions (lesions and vessels), but the boundary distance-based metrics are not as favorable, possibly due to false positives owing to spatially disparate vessels. 2) This shortcoming of TDM is addressed with local cross-triplet loss, where spatially continuous lesions are explicitly encouraged. This results in improved performance for DSC, ASD, and HD. 3) In addition, we apply not only the network perturbation, but also the input perturbation by adopting geometric transformations, such as rotation, flipping, and cropping, to better enforce the consistency of the intermediate features. Finally, the model to which all proposed techniques are applied shows good performance for all three metrics.
5 Conclusion
As high-quality annotation is difficult to collect in the medical domain, an approach based on little or weak annotation is preferred. With only a small amount of annotation, we utilize the time-varying enhancement pattern of ultrafast DCE-MRI to segment breast lesions, and propose a loss to efficiently distinguish lesions from non-lesions. Our design allows the network to focus solely on breast lesions, reducing false positives using limited annotations. Compared to that of other models, our approach achieves significant qualitative and quantitative improvements. We plan to study a segmentation technique using a small number of weak annotations in the future. Rather than using TDM as self-attention, joint prediction across slices could be a more promising approach where the model can automatically figure out context by looking at consecutive slices.
References
Sung, H., et al.: Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clinic. 71, 209–249 (2021)
Maicas, G., Bradley, A.P., Nascimento, J.C., Reid, I., Carneiro, G.: Pre and post-hoc diagnosis and interpretation of malignancy from breast DCE-MRI. Med. Image Anal. 58, 101562 (2019)
Lauby-Secretan, B., et al.: Breast-cancer screening-viewpoint of the IARC working group. N. Engl. J. Med. 372, 2353–2358 (2015)
Morgan, M.B., Mates, J.L.: Applications of artificial intelligence in breast imaging. Radiol. Clinics 59, 139–148 (2021)
Ayatollahi, F., Shokouhi, S.B., Mann, R.M., Teuwen, J.: Automatic breast lesion detection in ultrafast DCE-MRI using deep learning. Med. Phys. 48, 5897–5907 (2021)
Gubern-Mérida, A., et al.: Automated localization of breast cancer in DCE-MRI. Med. Image Anal. 20, 265–274 (2015)
Pisano, E.D., et al.: Diagnostic accuracy of digital versus film mammography: exploratory analysis of selected population subgroups in DMIST. Radiology 246, 376 (2008)
Onishi, N., et al.: Ultrafast dynamic contrast-enhanced breast MRI may generate prognostic imaging markers of breast cancer. Breast Cancer Res. 22, 1–13 (2020)
Mus, R.D., et al.: Time to enhancement derived from ultrafast breast MRI as a novel parameter to discriminate benign from malignant breast lesions. Eur. J. Radiol. 89, 90–96 (2017)
Jing, X., Dorrius, M.D., Wielema, M., Sijens, P.E., Oudkerk, M., van Ooijen, P.: Breast tumor identification in ultrafast MRI using temporal and spatial information. Cancers 14, 2042 (2022)
Kuhl, C.K.: A call for improved breast cancer screening strategies, not only for women with dense breasts. JAMA Netw. Open 4, e2121492 (2021)
Mann, R.M., Kuhl, C.K., Moy, L.: Contrast-enhanced MRI for breast cancer screening. J. Magn. Reson. Imaging 50, 377–390 (2019)
Abe, H., et al.: Kinetic analysis of benign and malignant breast lesions with ultrafast dynamic contrast-enhanced MRI: comparison with standard kinetic assessment. AJR Am. J. Roentgenol. 207, 1159 (2016)
Kim, E.S., et al.: Added value of ultrafast sequence in abbreviated breast MRI surveillance in women with a personal history of breast cancer: a multireader study. Eur. J. Radiol. 151, 110322 (2022)
Onishi, N., et al.: Differentiation between subcentimeter carcinomas and benign lesions using kinetic parameters derived from ultrafast dynamic contrast-enhanced breast MRI. Eur. Radiol. 30, 756–766 (2020)
Honda, M., et al.: New parameters of ultrafast dynamic contrast-enhanced breast MRI using compressed sensing. J. Magn. Reson. Imaging 51, 164–174 (2020)
Goto, M., et al.: Diagnostic performance of initial enhancement analysis using ultra-fast dynamic contrast-enhanced MRI for breast lesions. Eur. Radiol. 29, 1164–1174 (2019)
Gubern-Mérida, A., et al.: Automated localization of breast cancer in DCE-MRI. Med. Image Anal. 20, 265–274 (2015)
Ayatollahi, F., Shokouhi, S.B., Mann, R.M., Teuwen, J.: Automatic breast lesion detection in ultrafast DCE-MRI using deep learning. Med. Phys. 48, 5897–5907 (2021)
Dalmis, M.U., et al.: Artificial intelligence-based classification of breast lesions imaged with a multiparametric breast MRI protocol with ultrafast DCE-MRI, t2, and DWI. Invest. Radiol. 54, 325–332 (2019)
Oh, Y.T., Ko, E., Park, H.: TDM-stargan: Stargan using time difference map to generate dynamic contrast-enhanced MRI from ultrafast dynamic contrast-enhanced MRI. In: 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), pp. 1–5. IEEE (2022)
Militello, C., et al.: Semi-automated and interactive segmentation of contrast-enhancing masses on breast DCE-MRI using spatial fuzzy clustering. Biomed. Signal Process. Control 71, 103113 (2022)
Piantadosi, G., Sansone, M., Sansone, C.: Breast segmentation in MRI via U-Net deep convolutional neural networks. In: 2018 24th International Conference on Pattern Recognition (ICPR), pp. 3917–3922. IEEE (2018)
Maicas, G., Carneiro, G., Bradley, A.P., Nascimento, J.C., Reid, I.: Deep reinforcement learning for active breast lesion detection from DCE-MRI. In: Descoteaux, M., Maier-Hein, L., Franz, A., Jannin, P., Collins, D.L., Duchesne, S. (eds.) MICCAI 2017. LNCS, vol. 10435, pp. 665–673. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-66179-7_76
Zhang, J., Saha, A., Zhu, Z., Mazurowski, M.A.: Hierarchical convolutional neural networks for segmentation of breast tumors in MRI with application to radiogenomics. IEEE Trans. Med. Imaging 38, 435–447 (2018)
Van Engelen, J.E., Hoos, H.H.: A survey on semi-supervised learning. Mach. Learn. 109, 373–440 (2020)
Cheplygina, V., de Bruijne, M., Pluim, J.P.: Not-so-supervised: a survey of semi-supervised, multi-instance, and transfer learning in medical image analysis. Med. Image Anal. 54, 280–296 (2019)
Chen, X., Yuan, Y., Zeng, G., Wang, J.: Semi-supervised semantic segmentation with cross pseudo supervision. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2613–2622 (2021)
Tarvainen, A., Valpola, H.: Mean teachers are better role models: weight-averaged consistency targets improve semi-supervised deep learning results. In: Advances in Neural Information Processing Systems 30 (2017)
Yu, L., Wang, S., Li, X., Fu, C.-W., Heng, P.-A.: Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11765, pp. 605–613. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32245-8_67
Berthelot, D., Carlini, N., Goodfellow, I., Papernot, N., Oliver, A., Raffel, C.A.: MixMatch: a holistic approach to semi-supervised learning. In: Advances in Neural Information Processing Systems 32 (2019)
Vu, T.H., Jain, H., Bucher, M., Cord, M., Pérez, P.: Advent: adversarial entropy minimization for domain adaptation in semantic segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2517–2526 (2019)
Lee, D.H., et al.: Pseudo-label: The simple and efficient semi-supervised learning method for deep neural networks. In: Workshop on Challenges in Representation Learning, vol. 3, pp. 896. ICML (2013)
Ge, W., Huang, W., Dong, D., Scott, M.R.: Deep metric learning with hierarchical triplet loss. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) ECCV 2018. LNCS, vol. 11210, pp. 272–288. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-01231-1_17
Cakir, F., He, K., Xia, X., Kulis, B., Sclaroff, S.: Deep metric learning to rank. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2019)
Chopra, S., Hadsell, R., LeCun, Y.: Learning a similarity metric discriminatively, with application to face verification. In: 2005 IEEE Computer Society Conference on Computer Vision and Pattern Recognition (CVPR2005), vol. 1, pp. 539–546 (2005)
Do, T.T., Tran, T., Reid, I., Kumar, V., Hoang, T., Carneiro, G.: A theoretically sound upper bound on the triplet loss for improving the efficiency of deep distance metric learning. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2019)
Park, T., Efros, A.A., Zhang, R., Zhu, J.-Y.: Contrastive learning for unpaired image-to-image translation. In: Vedaldi, A., Bischof, H., Brox, T., Frahm, J.-M. (eds.) ECCV 2020. LNCS, vol. 12354, pp. 319–345. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-58545-7_19
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Wu, Z., Xiong, Y., Yu, S.X., Lin, D.: Unsupervised feature learning via non-parametric instance discrimination. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2018)
Luo, X.: SSL4MIS. https://github.com/HiLab-git/SSL4MIS (2020)
Oh, Y.T., Ko, E., Park, H.: SSL-CTL. https://github.com/yt-oh96/SSL-CTL (2022)
Yeghiazaryan, V., Voiculescu, I.D.: Family of boundary overlap metrics for the evaluation of medical image segmentation. J. Med. Imaging 5, 1–19 (2018)
Acknowledgments
This research was supported by the National Research Foundation (NRF-2020M3E5D2A01084892), Institute for Basic Science (IBSR015- D1), Ministry of Science and ICT (IITP-2020-2018-0-01798), AI Graduate School Support Program (2019-0-00421), ICT Creative Consilience program (IITP-2020-0-01821), and Artificial Intelligence Innovation Hub (2021-0-02068).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Oh, Yt., Ko, E., Park, H. (2023). Semi-supervised Breast Lesion Segmentation Using Local Cross Triplet Loss for Ultrafast Dynamic Contrast-Enhanced MRI. In: Wang, L., Gall, J., Chin, TJ., Sato, I., Chellappa, R. (eds) Computer Vision – ACCV 2022. ACCV 2022. Lecture Notes in Computer Science, vol 13846. Springer, Cham. https://doi.org/10.1007/978-3-031-26351-4_13
Download citation
DOI: https://doi.org/10.1007/978-3-031-26351-4_13
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-26350-7
Online ISBN: 978-3-031-26351-4
eBook Packages: Computer ScienceComputer Science (R0)