Abstract
Language–brain encoding experiments evaluate the ability of language models to predict brain responses elicited by language stimuli. The evaluation scenarios for this task have not yet been standardized which makes it difficult to compare and interpret results. We perform a series of evaluation experiments with a consistent encoding setup and compute the results for multiple fMRI datasets. In addition, we test the sensitivity of the evaluation measures to randomized data and analyze the effect of voxel selection methods. Our experimental framework is publicly available to make modelling decisions more transparent and support reproducibility for future comparisons.
The experiments were conducted in 2018 when all three authors were employed at the Institute of Logic, Language and Computation at the University of Amsterdam. The paper was presented in 2019. Since then, language modeling has progressed immensely. Experimental standards for robust, comparable, and reproducible evaluation for interpreting language–brain encoding experiments with respect to reasonable random permutation baselines need to be further developed and more widely adopted.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Representing language in a computationally usable format has been a research goal since the beginning of computational linguistics. In the last decade, distributional representations which interpret words, phrases, sentences, and even full stories as a high-dimensional vector in semantic space have become the most common standard. These representations are obtained by training language models on large corpora to optimally encode contextual information.
The quality of language representations is commonly evaluated on a set of downstream tasks. These tasks are either driven by engineering adequacy (e.g. the effect of the language representations on the performance of systems such as machine translation) or by the ability to reproduce human decisions (e.g. the performance of the representations on semantic similarity or entailment tasks). Many language researchers, however, are driven by the urge to better understand the underlying principles of human language processing.
With the increasing availability of brain imaging data, it has become popular to evaluate computational models by their ability to simulate brain signals related to human language processing [18, 19, 27]. If we can develop models that encode linguistic information in a way that is comparable to the activity in human brains, we will get one step closer to cognitively plausible models of human language understanding. While experimenting with human brains is evidently strictly constrained and regulated due to ethical reasons, we can easily query, adapt, constrain, degrade, and manipulate the computational model and analyze the effect on its language processing capabilities.
Although working with brain imaging data is highly promising from a cognitive perspective, it comes with many practical limitations. Brain datasets are usually too small for powerful machine learning models, the imaging technology produces noisy output that needs to be adjusted by statistical correction methods, and most importantly, only very few datasets are publicly available. Experiments in previous work are usually performed on a single dataset, so that it is unclear whether the observed effects are generalizable. In addition, the applied evaluation procedures have not yet been standardized. Understanding the subtle differences in the experimental setup to interpret the results can be particularly difficult because it has not yet become a common practice to publish the experimental code along with the results.
To the best of our knowledge, this paper provides the first analysis of language–brain encoding experiments which applies a consistent evaluation scenario across multiple fMRI datasets. We examine whether different evaluation measures provide different interpretations of the predictive power of the encoding model. Our experimental framework is publicly available to make modelling decisions more transparent and facilitate reproducibility for future comparisons. Due to its modular architecture, the pipeline can easily be extended to experiment with other datasets and language models.Footnote 1
2 Human-Centered Evaluation of Computational Models
As computational language models are trained on human-generated text, their performance is inherently optimized to simulate human behavior. Although novel architectural solutions attract notable interest in the research community, the ultimate benchmark for a model is the ability to approximate human language processing abilities. Models are supposed to reach a gold standard of human annotation decisions [29] and the difficulty of a task is often estimated by the inter-annotator agreement [5] or by error rates of human participants [8]. While these product-oriented evaluations focus on a final outcome, procedural measures of response times [25] or eye movements [7] are analyzed to provide deeper insights on sequential phenomena like attention or processing complexity. As neural network models are inspired by neuronal activities in the human brain, it is particularly interesting to analyze similarities and differences between distributed computational representations and low-level brain responses.
Electroencephalography (EEG) measures can be used to study specific semantic or syntactic phenomena [15, 18, 31] and compare the processing complexity of computational models to brain responses, for example, with respect to the N400 and P600 effects [14]. Signals with higher spatial resolution like magnetoencephalography (MEG) and functional magnetic resonance imaging (fMRI) are often used for experiments which are known as brain decoding and brain encoding. In the decoding setup, a computational model learns to identify differences in the signal and to discriminate between the responses for abstract and concrete words [4], for different syntactic classes [9, 22], for levels of syntactic complexity [10], and many other linguistic categories. Mitchell et al. [24] have shown that it is not only possible to distinguish between semantic categories but that a model can even learn to distinguish which word a participant is reading. The reverse direction of predicting the brain response that would most likely be observed for a novel linguistic stimulus is commonly called encoding. The encoding task requires a strong computational representation of the stimulus that reflects the shared properties of different stimuli and the relations between stimuli. For the remainder of this paper, we will focus on the language–brain encoding task and on fMRI datasets.
Many word representations have been tested on the Mitchell et al. [24] data including information from lexical resources, distributional, and multimodal representations [1, 4, 11, 34]. It has also been proposed to directly feed the brain signal into the language model as an additional source of information [6, 16]. Recently, new approaches for encoding and decoding of datasets using longer linguistic stimuli such as sentences [27] and even full stories [10, 13, 19, 32] are emerging. In some experiments, it has been shown that contextualized representations obtained from recurrent neural networks [19, 33] seem to represent the continuous stimuli slightly better than models that represent sentences as a conglomerate of context-independent word representations [13, 27]. However, these results are hard to generalize because they have been tested only on a single dataset. Gauthier and Ivanova [17] raise doubts about the informativeness of encoding results because differences between models are not reflected. Our robust evaluation experiments can serve as a comparative testbed for future analyses.
3 Datasets
We use four fMRI datasets that have been collected by different researchers (see Table 1). All datasets use English language stimuli and the participants are native speakers. Standard fMRI preprocessing methods such as motion correction, slice timing correction and co-registration to an MNI template had already been applied.
3.1 Isolated Stimuli
We use two datasets that work with isolated stimuli. The stimuli are not related and can be presented in varying order to the participants. Each stimulus is represented with only a single brain activation vector by averaging over several scans obtained during the presentation of the stimulus.
Words. For the Words dataset, 9 participants were shown a word paired with a line drawing of the object denoted by the word and were instructed to think about the properties of the object [24]. Six scans were taken during the presentation of each word. The scans were temporally detrended and smoothed. The activation values were normalized by computing the percent signal change relative to the fixation condition. Scans and stimuli were aligned with an offset of 4 s to account for the haemodynamic delay. The brain activation for each word is calculated by taking the mean over the six scans.
Stories. For the Stories dataset, 30 participants were reading 40 short personal stories that had been collected from weblogs [13]. The stories consisted of 11 sentences on average and were presented in three consecutive batches on a screen. The dataset also contains data for Farsi and Chinese stories but for the sake of comparison, we focus on the English subset here. The scans were preprocessed with detrending, temporal smoothing and spatial smoothing. The activation values were normalized by calculating z-scores with respect to the fixation condition. The authors then discretized the continuous story stimulus by calculating the mean over all story scans. We exclude subject 30 from the data because the voxel values are all zero.
3.2 Continuous Stimuli
Humans process language incrementally and in context. In order to simulate a more naturalistic language setting, recent approaches to brain encoding use continuous stimuli and analyze the fMRI scans as a sequence of responses.
Harry. For the Harry dataset by Wehbe et al. [32], 8 participants read chapter 9 of Harry Potter and the Sorcerer’s stone [30]. The story was split into four blocks and presented word by word on a screen. Each word was displayed for 0.5 s and an fMRI scan was taken every 2 s. We follow their protocol and apply detrending and temporal smoothing but do not smooth spatially because it did not have an effect on the results in pilot experiments.
Alice. For the Alice dataset by Brennan et al. [10], 27 participants were listening to an audio recording of the first chapter of Alice in Wonderland [12]. The published data contains the preprocessed signal averaged for 6 regions of interests defined using functional and anatomical criteria. The raw signal is not available.
4 Encoding Model
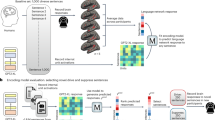
The fMRI data is obtained by measuring the so-called blood-oxygenation level dependent (BOLD) response. This signal indicates the level of oxygen in the blood (approximated by its magnetic susceptibility) and an increased BOLD response in an area of the brain is interpreted as increased neuronal activity in this region. In order to analyze the response, the brain is fragmented into stacked voxels which are cubes of constant size (e.g. 3\(\,\times \,3\,\times \,\)3 mm). The response thus consists of a three-dimensional matrix with activation values for each voxel. This matrix is flattened into a one-dimensional vector \(\textbf{v}\). In the brain encoding approach, the goal is to predict \(\textbf{v}\) given the stimulus \(\textbf{s}\) that was presented when measuring the response.
Mapping Model. A multiple linear ridge regression model is usually applied as encoding model to learn the response pattern \(\mathbf {v_n} {\in } \mathbb {R}^m\) for stimulus \(\mathbf {s_n} {\in } \mathbb {R}^d\) on a training set \(V {\in } \mathbb {R}^{m{\times }n}\) of responses to n other stimuli.Footnote 2 It requires a strong computational representation of the stimulus that reflects the relations between stimuli. The predictive power of this mapping model is evaluated on a set of held-out stimuli \(S {\in } \mathbb {R}^{d{\times }n}\). The mapping model learns a separate regression equation for every voxel \(v_i\) which is fitted by learning a weight \(w_{d}\) for each dimension \(\mathbf {s_d}\) of the stimulus representations and the weights are regularized by the L2 norm. The cost function f for learning the weight vector \(\textbf{w}\) for a voxel vector \(\mathbf {v_i}\) is:
4.1 Language Model
The linguistic stimuli are represented using vectors obtained from a language model. Previous work has compared the performance of different language models for brain encoding tasks showing that contextual models like long short-term memory networks perform better than standard word-based representations [19]. For a more robust comparison, we keep the language model constant for all datasets. We choose the language model ELMO because it produces contextualized representations on the sentence level and performs very well on semantic tasks [28]. ELMO is based on a bi-directional long short-term memory network and it uses character-based representations of the input which makes it perform very well on out-of-vocabulary words. This is an important property for modeling fictional texts. We use a pre-trained pytorch version of ELMO available on github.Footnote 3
For Words, we use the representations from the token layer. For all other datasets, we obtain contextualized representations from the first layer. We restrict the representation to the forward language model to simulate incremental processing and obtain a 512-dimensional vector. We take the representation of the last token of each sentence and average over all sentences for each story in Stories. For the continuous stimuli, we feed the language model the whole chapter and extract the representation of the last token of the sequence which had been presented between the previous and the current scan.
Haemodynamic Delay. The fMRI signal measures a brain response to a stimulus with a delay of up to ten seconds [23]. This delay needs to be considered when aligning stimuli with responses. Similarly to Mitchell et al. [24], we align scans to stimuli with a fixed offset of 4 s. The haemodynamic response decays slowly over a duration of several seconds. For continuous stimuli, this means that the response to previous stimuli will have an influence on the current signal. Wehbe et al. [32] use a feature-based representation and learn different weights for stimuli occurring at previous time steps. In this approach, the number of features increases linearly with the number of time steps considered. In contextual language models, a representation is build up incrementally using recurrent connections. The representation of a word thus implicitly contains information from the previous context. As ELMO processes language sentence by sentence, our context window comprises the current sentence up to the current word but the number of dimensions remains constant.
4.2 Voxel Selection
The number of voxels in a brain varies with respect to the voxel size and the shape of the subject’s brain. In the datasets used here, the number of voxels ranges from 20,000 to more than 40,000. The activity measured in many of these voxels is most likely not related to language processing but might change due to physical processes like the noise perception in the scanner. In these cases, learning a mapping model from the stimulus representation to the voxel activation will not succeed because the stimulus has no influence on the variance of the voxel signal. Whole-brain evaluations of mapping models thus only have limited informative value. In previous work, different voxel selection models have been applied to analyze only a subset of interesting voxels. Wehbe et al. [32] and Brennan et al. [10] reduced the voxels by using previous knowledge about regions of interests. Restricting the brain response to voxels that fall within a pre-selected set of regions of interests can be considered as a theory-driven analysis.
Information-Driven Voxel Selection. In contrast to the theory-driven region of interest analysis, Kriegeskorte et al. [20] propose a more information-driven approach. So-called searchlight analyses move a sphere through the brain to select voxels (comparable to sliding a context window over text) and analyze the predictive power of the voxel signal within the sphere. Dehghani et al. [13] and Wehbe et al. [32] use this searchlight approach for the decoding task. In brain encoding, the predictive direction is reversed. The ability to predict voxel activation based on the stimulus is carefully interpreted as an indicator that processing the stimulus influences the activity in this particular voxel. For Words, Mitchell et al. [24] analyze all six brain responses for the same stimulus and select 500 voxels that exhibit a consistent variation in activity across all stimuli. Jain and Huth [19] calculate the model performance for a single voxel as the Pearson correlation between real and predicted responses on the test set and analyze voxels with a correlation above a threshold. Gauthier and Ivanova [17] recommend to evaluate voxels based on explained variance. We select the 500 most predictive voxels on the training set for Words by four selection methods: stability, Pearson correlation, explained variance, and random.
Results of Voxel Selection. Table 2 shows the results for different voxel selection methods. It can be seen that voxel selection by explained variance performs on par with the selection of stable voxels. We had speculated that simply reducing the number of voxels might already lead to improvements because similarity measures tend to perform better in lower-dimensional spaces [2] but a random selection of voxels has no effect. For the remainder of the paper, we report results on the 500 voxels that obtained the highest explained variance results on the training set unless indicated otherwise because the option of selecting stable voxels is not available for the other datasets.
5 Evaluation Experiments
The voxel selection results show that a small experimental parameter can have a strong effect. We thus perform three experiments using different evaluation procedures: pairwise accuracy, voxel-wise evaluation, and representational similarity analysis. We repeat each experiment with a language model that assigns a random (but fixed) vector to each word to analyze the sensitivity of the evaluation metric. Random story representations are obtained by averaging over words.
5.1 Pairwise Evaluation
As the fMRI datasets are very small for machine learning purposes, Mitchell et al. [24] introduced an evaluation procedure that maximizes the training data. Given a set of n samples, a mapping model is trained on \(n{-}2\) samples and tested on the two remaining samples. Mitchell et al. [24] call this procedure leave-two-out cross-validation but it differs from standard cross-validation setups because each sample occurs n times in the test set leading to \(\left( {\begin{array}{c}n\\ 2\end{array}}\right) \) different models. The performance is evaluated by calculating the pairwise accuracy over all models.
A pair of two test samples \((s_1,s_2)\) is considered to be classified correctly if the model prediction \(p_1\) is more similar to the true target \(s_1\) than to \(s_2\), and \(p_2\) is more similar to \(s_2\). This general idea of pairwise accuracy has been implemented in different ways. The applied similarity metrics f are cosine similarity [24], euclidean similarity [32], and Pearson correlation [11, 27]. The prediction for a pair can be considered to be correct by comparing the summed similarity of the correct alignments with the false alignments [11, 13, 24]. Wehbe et al. [32] and Wehbe et al. [33] calculate the accuracy by comparing the predictions only for the first sample. A stricter interpretation of the pairwise accuracy would only consider the prediction to be correct, if both samples are correctly matched to their prediction. We refer to the different interpretations as sum match (1), single match (2), and strict match (3):
Experimental Setup. We calculate the pairwise accuracy for all four datasets, for the two similarity metrics cosine and euclidean and for the three match definitions sum, single, and strict. The leave-two-out evaluation only works well for isolated stimuli as in Words and Stories. For the continuous stimuli, we perform standard cross-validation. The Harry data can be split into four folds according to the experimental blocks. For the Alice data, we determined six folds. The predictions for each fold are then paired with a randomly selected sample. We set a distance constraint between the two samples of at least 20 time steps to avoid overlapping response patterns. For each sample, we average the result over 1,000 random pairs as in Wehbe et al. [32].
Results. The results in Table 3 are averaged over all subjects. It can be seen that the differences between the three similarity metrics and the sum and the single match are very small. The strict match is consistently more rigorous than the other match types. This indicates that both predictions would often be matched to the same stimulus when ignoring the pairwise exclusivity constraint. We conclude that the other two match types tend to slightly overestimate the discriminability of the stimulus. We also note that the difference to the random language model is more pronounced for the strict match for Words and Stories. For these two datasets, the results vary strongly across subjects. Subjects 1,3 and 4 in Words yield high accuracy results (0.87, 0.87, 0.76 for the cosine sum match) whereas the prediction for subject 6 is below chance level. We provide violin plots in Fig. 1 for a better impression of the variance across subjects in Stories. Although the results are worse than for Words, the accuracy is quite high for some subjects (0.80, 0.78, 0.7). The results obtained for the isolated stimuli are comparable to those reported previously by Mitchell et al. [24] and Dehghani et al. [13]. For the continuous stimuli, the encoding model is not able to learn a robust signal. Wehbe et al. [32] reported better results for the Harry data but they performed the decoding task. Brennan et al. [10] did not report encoding or decoding results but focused on correlating the fMRI signal with computational models for surprisal.
5.2 Voxel-Wise Evaluation
The pair-wise distance measures are an abstraction over all voxels. A model that mostly predicts constant values and only varies a few indicative voxels could perform well. As the mapping model independently predicts each voxel, we can take a closer look at the predictability of each voxel. This procedure accounts for the assumption that not every voxel in our brain will be influenced by the stimulus. In previous work, prediction results have often been reported only over significant voxels.
Experimental Setup. The explained variance (EV) and the coefficient of determination (\(R^2\)) are the most common metrics for evaluating linear regression. They measure the proportion of the variance in the dependent variable that is predictable by the model. The two metrics are closely related but explained variance also accounts for the mean error. We use the implementation of these scores in the python library scikit-learn [26]. Jain and Huth [19] calculate a different \(r^2\) value: they multiply the Pearson correlation between the predictions and the observed activations for voxel \(v_i\) with the absolute correlation (\(r^2(v_i) = r_{v_i}{\times }\vert r_{v_i} \vert \)). We refer to this measure as \(r^2 simple\). We calculate all three metrics and compare the results for the whole brain with a selection of the 500 best-performing voxels on the training and on the testing set respectively. Selection on the test set is not recommended but added to compare previous work.
Results. Tables 4 shows the results for the voxel-wise evaluation averaged over all subjects and over all voxels. It can be seen that the models are highly overfitted as we get much better results when voxels are directly selected on the test results than when they are pre-selected on the training data. In the conditions which control for overfitting, the explained variance and the \(R^2\) are always negative. A value of zero for explained variance is obtained for a model that constantly predicts the mean. It is almost impossible to identify which one of two very negative models performs less bad based on this value alone. The prediction quality should generally be interpreted with caution as the number is averaged over all voxels, all folds and all subjects. Both, the inter-subject variance and the variance in voxel predictability are very high, so that positive and negative results cancel each other out. The \(r^2 simple\) metric almost always returns a positive score. This might be a more satisfying result when evaluating the encoding quality; however, the metric also returns high positive scores for the random language model in some cases.
Accumulation Method. Instead of averaging the encoding quality over voxels, Jain and Huth [19] report the sum. For comparison, the summed results are provided in the appendix in Table 6. Sum metrics depend on the number of voxels over which they are calculated. For the whole brain analysis, averaged sum metrics are thus not interpretable in absolute terms because the number of voxels in the brain varies between subjects (see Fig. 2 for an illustration). When accumulating the sum over a fixed set of selected voxel, we see that the results for the \(r^2 simple\) metric are consistently better (3) but the extreme change on the x-axis in the two figures indicates that sum scores should be interpreted with caution.
Violin plots of the voxel-wise results (summed over all voxels) for all subjects in Stories for all voxels. Note the extreme change in the scale of the y-axis compared to Fig. 2 due to the number of voxels. If the number of voxels over which the sum is calculated is unknown, the result cannot be interpreted.
Model-Driven Voxel Selection. We additionally determine the voxels with the highest explained variance on the test set when training on 80% of the data. We set a threshold (0.3 for Stories and Words, 0 for Alice) and plot predictive voxels for the subjects for which we obtained highest accuracy in the pairwise comparison in Fig. 4. The results are rather inconclusive. There is almost no overlap in the voxels and they are spread over several brain regions. This indicates that model-driven voxel information should only be interpreted on larger datasets.
5.3 Representational Similarity Analysis
The previous methods indicate that the continuous stimuli cannot be well encoded. In order to be able to attribute this flaw more directly to the language model, we perform representational similarity analysis [21] to compare the relations between brain activation vectors to the relations between stimulus representations without the intermediate mapping model. The approach assumes that similar brain activation patterns are caused by strongly related stimuli. The quality of the computational representation of the stimuli can then be assessed by its ability to model these relations [3, 11, 34]. As commonly performed in previous work, we measure the relations between vectors by the cosine distance and compare brain scans and representations by Spearman correlation and Pearson correlation.
Results. At first glance, the results in Table 5 seem to confirm the impression that the encoding model performs better for the isolated stimuli. However, the same results can be obtained with the random language model. The random model could capture word identity (recall that the same random vector is assigned to different occurrences of the same word) which might serve as a relevant signal for the story stimuli but this would not explain the results for the Words dataset with 60 different words. It can be seen that generally the more conservative rank-based Spearman correlation is much lower than the Pearson correlation. For the current setup, the representational similarity analysis results are unsatisfactory. However, the methodology largely reduces the number of parameters and facilitates the comparison of different computational models. We thus think that it could be a promising analysis method for future experiments.
6 Discussion
The setup of encoding experiments requires many modelling decisions for the stimulus representation, the stimulus–response alignment, the mapping model and its learning parameters, the noise reduction techniques for the brain responses, the voxel selection, and the evaluation metric. Experimenting with a single dataset bears the danger of overfitting the experimental setup. We have seen that different evaluation metrics can interpret the predictive power of an encoding model very differently. Encoding results should thus always be compared to a reasonable baseline and hypotheses should be tested over several datasets. In this comparison, we intentionally restricted the experimental setup by choosing the same language model for all datasets. At this point, it remains unclear, whether the close to random results in many settings result from an unfortunate choice of the language model parameters or from a noisy signal. Our experimental pipeline is modular and provides a useful testbed for future experiments with alternative stimuli representations.
More sophisticated context models might increase the number of dimensions. From a machine learning perspective, most encoding experiments are problematic because the number of features is often higher than the number of samples. In addition, similarity metrics are known to sometimes behave unexpectedly when applied on high-dimensional data [2]. One could apply dimensionality reduction on the language representations but these methods change the structure of the representation and make it difficult to derive cognitive insights for the original model. For future data collections, it would be important to obtain more data points from fewer subjects to facilitate more powerful pattern analyses.
FMRI encoding is an intriguing but also very challenging task because of the noisy signal. Within the current state of the art, even a tiny signal that is significantly different from chance, can be seen as a success. The pairwise estimation measures can present the results in a more pronounced way. However, as our analysis with the strict match have shown, the other match definitions tend to give an overly optimistic impression of the discriminability of the stimuli. A similar problem occurs, when summing the \(r^2 simple\) value only over predictive voxels. We are convinced that in the long run, the field benefits from a more conservative estimate of the predictive power of the developed models.
7 Conclusions
We have performed a robust comparison for language–brain encoding experiments and receive very diverse results for different evaluation metrics. It is our hope that our experimental framework can pave the way for future experiments to gradually determine the optimal encoding parameters. We plan to extend our experiments to the datasets by Pereira et al. [27] and to other languages. We can already provide a set of practical recommendations for evaluation: 1. For the pairwise evaluation, it is helpful to additionally report the strict match to put the results in perspective. 2. Averaging over subjects is not very informative, violin plots can give a better impression of the variance. 3. For sum metrics, it is important to clearly specify the number of voxels that are taken into consideration. 4. Voxel selection methods should only be performed on the training set and should be transparently documented because they have a strong effect on the results.
Notes
- 1.
The code is available at https://github.com/beinborn/brain-lang.
- 2.
Whether a linear model is a plausible choice is debatable. We use it here for comparison with previous work.
- 3.
References
Abnar, S., Ahmed, R., Mijnheer, M., Zuidema, W.: Experiential, distributional and dependency-based word embeddings have complementary roles in decoding brain activity. In: Proceedings of the 8th Workshop on Cognitive Modeling and Computational Linguistics (CMCL’18), pp. 57–66. Association for Computational Linguistics (2018). http://aclweb.org/anthology/W18-0107
Aggarwal, C.C., Hinneburg, A., Keim, D.A.: On the surprising behavior of distance metrics in high dimensional space. In: Van den Bussche, J., Vianu, V. (eds.) Database Theory – ICDT 2001, pp. 420–434. Springer, Berlin Heidelberg, Berlin, Heidelberg (2001). http://kops.uni-konstanz.de/bitstream/handle/123456789/5715/On_the_Surprising_Behavior_of_Distance_Metric_in_High_Dimensional_Space.pdf?sequence=1
Anderson, A.J., Bruni, E., Bordignon, U., Poesio, M., Baroni, M.: Of words, eyes and brains: Correlating image-based distributional semantic models with neural representations of concepts. In: Proceedings of the 2013 Conference on Empirical Methods in Natural Language Processing, pp. 1960–1970. Association for Computational Linguistics (2013). http://aclweb.org/anthology/D13-1202
Anderson, A.J., Kiela, D., Clark, S., Poesio, M.: Visually grounded and textual semantic models differentially decode brain activity associated with concrete and abstract nouns. Trans. Assoc. Comput. Linguist. 5, 17–30 (2017). http://aclweb.org/anthology/Q17-1002
Artstein, R., Poesio, M.: Inter-coder agreement for computational linguistics. Comput. Linguist. 34(4), 555–596 (2008). https://www.mitpressjournals.org/doi/pdfplus/10.1162/coli.07-034-R2
Athanasiou, N., Iosif, E., Potamianos, A.: Neural activation semantic models: computational lexical semantic models of localized neural activations. In: Proceedings of the 27th International Conference on Computational Linguistics, pp. 2867–2878 (2018). http://www.aclweb.org/anthology/C18-1243
Barrett, M., Bingel, J., Hollenstein, N., Rei, M., Søgaard, A.: Sequence classification with human attention. In: Proceedings of the 22nd Conference on Computational Natural Language Learning, pp. 302–312 (2018). http://www.aclweb.org/anthology/K18-1030
Beinborn, L., Zesch, T., Gurevych, I.: Predicting the difficulty of language proficiency tests. Trans. Assoc. Comput. Linguist. 2(1), 517–529 (2014). http://www.aclweb.org/anthology/Q14-1040
Bingel, J., Barrett, M., Søgaard, A.: Extracting token-level signals of syntactic processing from fMRI - with an application to pos induction. In: Proceedings of the 54th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), 1, pp. 747–755 (2016). http://www.aclweb.org/anthology/P16-1071
Brennan, J.R., Stabler, E.P., Van Wagenen, S.E., Luh, W.M., Hale, J.T.: Abstract linguistic structure correlates with temporal activity during naturalistic comprehension. Brain Lang. 157, 81–94 (2016). https://www.sciencedirect.com/science/article/pii/S0093934X1530068
Bulat, L., Clark, S., Shutova, E.: Speaking, seeing, understanding: correlating semantic models with conceptual representation in the brain. In: Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, pp. 1081–1091. Association for Computational Linguistics (2017). http://aclweb.org/anthology/D17-1113
Carroll, L.: Alice’s Adventures in Wonderland. Macmillan, London (1865)
Dehghani, M., et al.: Decoding the neural representation of story meanings across languages. Human Brain Mapp. 38(12), 6096–6106 (2017). https://www.ncbi.nlm.nih.gov/pubmed/28940969
Frank, S.L., Otten, L.J., Galli, G., Vigliocco, G.: Word surprisal predicts n400 amplitude during reading. In: Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics (Volume 2: Short Papers), 2, pp. 878–883 (2013). https://www.semanticscholar.org/paper/Word-surprisal-predicts-N400-amplitude-during-Frank-Otten/0998e0763328764935e74db7c124ee4ee277c360
Fyshe, A., Sudre, G., Wehbe, L., Rafidi, N., Mitchell, T.M.: The semantics of adjective noun phrases in the human brain. bioRxiv (2016). https://www.biorxiv.org/content/biorxiv/early/2016/11/25/089615.full.pdf
Fyshe, A., Talukdar, P.P., Murphy, B., Mitchell, T.M.: Interpretable semantic vectors from a joint model of brain-and text-based meaning. In: Proceedings of the Conference. Association for Computational Linguistics. Meeting, vol. 2014, p. 489. NIH Public Access (2014). http://aclweb.org/anthology/P14-1046
Gauthier, J., Ivanova, A.: Does the brain represent words? An evaluation of brain decoding studies of language understanding. arXiv:1806.00591 (2018). https://arxiv.org/pdf/1806.00591.pdf
Hale, J., Dyer, C., Kuncoro, A., Brennan, J.R.: Finding syntax in human encephalography with beam search. In: Proceedings of the 56th Annual Meeting of the Association for Computational Linguistics, Volume 1 (Long Papers), pp. 2727–2736. Association for Computational Linguistics (2018). http://aclweb.org/anthology/P18-1254
Jain, S., Huth, A.: Incorporating context into language encoding models for fMRI. bioRxiv (2018). https://www.biorxiv.org/content/early/2018/05/21/327601
Kriegeskorte, N., Goebel, R., Bandettini, P.: Information-based functional brain mapping. Proc. National Acad. Sci. 103(10), 3863–3868 (2006). http://www.pnas.org/content/103/10/3863.full
Kriegeskorte, N., Mur, M., Bandettini, P.A.: Representational similarity analysis-connecting the branches of systems neuroscience. Front. Syst. Neurosci. vol. 2, p. 4 (2008). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2605405/
Li, J., Fabre, M., Luh, W.M., Hale, J.: The role of syntax during pronoun resolution: evidence from fMRI. In: Proceedings of the 8th Workshop on Cognitive Aspects of Computational Language Learning and Processing, pp. 56–64. Association for Computational Linguistics (2018). http://aclweb.org/anthology/W18-2808
Miezin, F.M., Maccotta, L., Ollinger, J., Petersen, S., Buckner, R.: Characterizing the hemodynamic response: effects of presentation rate, sampling procedure, and the possibility of ordering brain activity based on relative timing. Neuroimage 11(6), 735–759 (2000). https://doi.org/10.1006/nimg.2000.0568
Mitchell, T.M., et al.: Predicting human brain activity associated with the meanings of nouns. science 320(5880), 1191–1195 (2008). https://www.cs.cmu.edu/tom/pubs/science2008.pdf
Monsalve, I.F., Frank, S.L., Vigliocco, G.: Lexical surprisal as a general predictor of reading time. In: Proceedings of the 13th Conference of the European Chapter of the Association for Computational Linguistics, pp. 398–408. Association for Computational Linguistics (2012). https://aclanthology.info/pdf/E/E12/E12-1041.pdf
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). http://www.jmlr.org/papers/volume12/pedregosa11a/pedregosa11a.pdf
Pereira, F., et al.: Toward a universal decoder of linguistic meaning from brain activation. Nat. Commun. 9(1), 1–13 (2018). https://doi.org/10.1038/s41467-018-03068-4
Peters, M., et al.: Deep contextualized word representations. In: Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers), 1, pp. 2227–2237 (2018). http://www.aclweb.org/anthology/N18-1202
Resnik, P., Lin, J.: Evaluation of NLP systems. The Handbook of Computational Linguistics and Natural Language Processing, vol. 57, pp. 271–295 (2010). https://pdfs.semanticscholar.org/41ef/e3fb47032d609bbb13b7c850bb8b1dbd544d.pdf
Rowling, J.K.: Harry Potter and the Sorcerer’s Stone. Levine Books, Arthur A (1998)
Sudre, G., et al.: Tracking neural coding of perceptual and semantic features of concrete nouns. NeuroImage 62(1), 451–463 (2012). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4465409/
Wehbe, L., Murphy, B., Talukdar, P., Fyshe, A., Ramdas, A., Mitchell, T.: Simultaneously uncovering the patterns of brain regions involved in different story reading subprocesses. PLoS One 9(11), e112575 (2014). https://doi.org/10.1371/journal.pone.0112575
Wehbe, L., Vaswani, A., Knight, K., Mitchell, T.: Aligning context-based statistical models of language with brain activity during reading. In: Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP). Association for Computational Linguistics (2014). https://doi.org/10.3115/v1/d14-1030
Xu, H., Murphy, B., Fyshe, A.: Brainbench: a brain-image test suite for distributional semantic models. In: Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, pp. 2017–2021 (2016). http://www.aclweb.org/anthology/D16-1213
Acknowledgements
The work presented here was funded by the Netherlands Organisation for Scientific Research (NWO), through a Gravitation Grant 024.001.006 to the Language in Interaction Consortium.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Appendix
Appendix
Rights and permissions
Copyright information
© 2023 Springer Nature Switzerland AG
About this paper
Cite this paper
Beinborn, L., Abnar, S., Choenni, R. (2023). Robust Evaluation of Language–Brain Encoding Experiments. In: Gelbukh, A. (eds) Computational Linguistics and Intelligent Text Processing. CICLing 2019. Lecture Notes in Computer Science, vol 13451. Springer, Cham. https://doi.org/10.1007/978-3-031-24337-0_4
Download citation
DOI: https://doi.org/10.1007/978-3-031-24337-0_4
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-24336-3
Online ISBN: 978-3-031-24337-0
eBook Packages: Computer ScienceComputer Science (R0)