Abstract
With the ability of representing structures and complex relationships between data, graph learning is widely applied in many fields. The problem of graph classification is important in graph analysis and learning. There are many popular graph classification methods based on substructures such as graph kernels or ones based on frequent subgraph mining. Graph kernels use handcraft features, hence it is so poor generalization. The process of frequent subgraph mining is NP-complete because we need to test isomorphism subgraph, so methods based on frequent subgraph mining are ineffective. To address this limitation, in this work, we proposed novel graph classification via graph structure learning, which automatically learns hidden representations of substructures. Inspired by doc2vec, a successful and efficient model in Natural Language Processing, graph embedding uses rooted subgraph and topological features to learn representations of graphs. Then, we can easily build a Machine Learning model to classify them. We demonstrate our method on several benchmark datasets in comparison with state-of-the-art baselines and show its advantages for classification tasks.
T. Huynh and T. T. T. Ho---Contributed equally to this work.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
In recent years, graph data has become increasingly popular and widely applied in many fields such as biology (Protein-Protein interaction networks) [1], chemistry (molecular structures) [2], neuroscience (brain networks) [3, 4], social networks (networks of friends) [5], and knowledge graphs [6, 7]. The power of graphs is their capacity to represent complex entities and their relationships. Graph classification is an important problem because of its wide range of applications including predicting whether a protein structure is mutated, recognizing unknown compounds, etc.
Because traditional classification algorithms cannot be applied directly to graph data, graph classification has become an independent sub-field. There are many popular graph classification methods based on substructures such as graph kernels or ones based on frequent subgraph mining. The core idea of the former is to extract information from substructures (such as subgraph, path, walk, etc.) and apply conventional classifiers. The problem with this approach is in extracting information from substructures. There are two lines of methods to extract information from substructures. First, graph kernels that are based on kernel methods, work on graph elements such as walk or path. However, these methods are difficult to find a suitable kernel function that captures the semantics of the structure while being computationally tractable. The second group of substructure methods aim at mining frequent subgraphs in graphs. The main drawback of this group is time-consuming because of the high cost of subgraph mining step.
This paper addresses these limitations by using features learned automatically from data instead of handcraft features in graph kernels. To overcome the NP-complete problem in subgraph isomorphism testing, we could mine rooted subgraphs and apply Weisfeiler-Lehman relabeling method proposed in Weisfeiler-Lehman graph kernel [8] to find subgraphs more effectively. Besides that, inspired by the recent success of doc2vec [20] in NLP, which exploits how words compose documents to learn their representation, we adopt this idea to learn graph representation as a document and rooted subgraphs as words. Specifically, we proposed a novel Graph Classification via Graph Structure Learning (GC-GSL). First, GC-GSL extracts topological attributes and builds a subgraph “vocabulary” set of graphs. Then, to train graph embedding, a neural network is designed to take a graph as input, and output is subgraphs appearing in the graph as well as topological attributes extracted from the graph. Finally, a fundamental classifier is trained for graph embedding.
We make the following main contributions:
-
We proposed a neural network graph embedding model. The neural network model will automatically learn the graph embedding corresponding to each graph. The embedding of the graph after learning not only reflects the characteristics of the graph itself but also contains the relationship between the graphs.
-
Through our experiments on several benchmark datasets, we demonstrate that GC-GSL is highly competitive compared with the graph kernels and graph classification methods based on feature vector construction.
The remainder of this article is structured as follows. Related work is listed in Sect. 2. Section 3 introduces the proposed method from extracted topological attributes vector, mining rooted subgraphs to neural networks for graph embedding. The experimental results and discussions are presented in Sect. 4. Conclusions and future works are presented in Sect. 5.
2 Related Works
Graph Kernels.
Graph kernels [9] are one of the prominent methods in graph classification problems. Graph kernels evaluate the similarity between a pair of graphs by recursively decomposing them into substructures (e.g. walks, paths, cycles, graphlets, etc.) and defining a similarity function over the substructures (e.g. count the number of similar substructures two graphs, etc.). Then, kernel methods (e.g. Support Vector Machines [10], etc.) could be used to perform classification tasks. Random Walk Kernels [11], the similarity of two graphs is calculated by counting the number of common walk labels of the two graphs. Shortest Path Kernels [12] first computes the shortest path for each graph in the dataset. The kernel is defined as the sum over all pairs of shortest path edges of two graphs and, using any positive definite kernel that is suitable on the edges. Nikolentzos et al. [13] proposed a method to measure the similarity between pairs of documents based on the Shortest Path Kernel method. In it, each document is represented by a graph of words. Cyclic Pattern Kernels [14] is based on the common number of cycles occurring in both graphs. Since there is no known polynomial-time algorithm to find all cycles in a graph, time-limited sampling and enumeration of cycles are used to measure the similarity of a graph. Graphlet and Subgraph Kernels [15], similar graphs should have similar subgraphs. Kernel graphlets measure the similarity of two graphs as the dot product of the count vectors of all possible connected subgraphs of degreed. Subtree Kernels [16] are based on common subtree patterns in graphs. To compare two graphs, the subtree kernel compares all pairs of vertices from two graphs by iteratively comparing their neighborhoods.
However, most of them still have some limitations. First, many of them do not provide explicit graph embedding. They allow kernelized learning algorithms such as Support Vector Machines [10] to work directly on graphs, without having to do feature extraction to transform them to fixed-length, real-valued feature vectors. Second, these graph kernels use handcrafted features (e.g., shortest paths, graphlets, etc.) i.e., features determined manually with specific well-defined functions that help to extract such substructures from graphs and thus yield poor generalization.
Graph Classification Based on Frequent Subgraph Mining.
Constructing feature vectors based on frequent subgraph mining consists of 3 steps. 1) mining the frequent subgraph of graphs (e.g. Fast Frequent Subgraph Mining [27]), 2) filtering the subgraph features (e.g. structure-based, semi-supervised learning, etc.), and 3) vectorize the graph based on subgraph features. The key issue for classification efficiency of this method is the selection of discriminative subgraph features. Fei and Huan [17] first used Fast Frequent Subgraph Mining (FFSM) [27] to mine frequent subgraphs, then proposed an embedding distance method to describe the feature consistency relationship between subgraphs, which maps all frequent subgraphs into feature consistency graphs. Then, the order of the nodes corresponding to the frequent subgraph in the consistency graph is used to extract the feature subgraphs, so that the original graph has been converted to the feature vector. Finally, Support Vector Machines [10] is used for classification. Kong and Yu [18] use the gSpan algorithm [26] to mine the subgraphs and then propose a feature evaluation criterion, called gSemi, to evaluate the subgraphs characteristic for both labeled and unlabeled graphs and get the upper bound for gSemi to reduce the subgraph search space. Then, a branch-and-bound algorithm is proposed to efficiently find the optimal set of feature subgraphs, which is useful for graph classification.
However, there are still some challenges when classifying graphs based on frequent subgraph mining. First, a large number of subgraphs are mined. Applying frequent subgraph mining algorithms on a set of graphs causes all subgraphs that occur more than a particular threshold to be detected. These samples are numerous, and the number of these samples depends on the data attribute and the defined threshold. The considerable number of samples increases the uptime, makes the selection of valuable samples more difficult and reduces scalability. Second, the process of frequent subgraph mining is NP-complete. To determine the frequency of the subgraphs, we need to test the isomorphic subgraphs. If two subgraphs are the same in terms of connectivity, then they are isomorphic. Isomorphism testing is an NP-complete problem [19], so it is expensive, especially for large graphs.
3 Proposed Method: GC-GSL
Our graph classification method, which we name GC-GSL Graph Classification via Graph Structure Learning, is based on the following key observation: two graphs are similar if they have similar graph substructures. Inspired by doc2vec [20] to learn document embedding, we extend the same to learn graph embeddings. Doc2Vec exploits the way words/word sequences compose documents to learn their embedding. Similar to doc2vec, in GC-GSL, we view a graph as a document and the rooted subgraphs in the graph as words. In addition, PV-DBOW model in doc2vec [20] ignores the local context of words, that is, the model only considers which words a document contains, not the order of words in the document. Therefore, PV-DBOW model is very suitable for graph embedding, where there is no sequential relationship between the rooted subgraphs. Besides, in GC-GSL, the topological attribute vector is added to the output layer to help the model learn the general information of the graph. And after training the graph embedding, a basic classification in Machine Learning is used for classification. In this session, we discuss the main components and complexity of GC-GSL.
3.1 Extracting Topological Attribute Vector
The extracted topological attribute vector includes 16 features related to many different feature groups of the graph from the statistical information, such as the number of nodes, the number of edges, to structural features such as clustering, connectivity, centrality, distance measure, and percentages of some typical node types.
-
f1 – Number of nodes.
-
f2 – Number of edges.
-
f3 – Average degree: the average value of the degree of all nodes in the graph.
-
f4 – Average neighbor degree: First, we calculate the average neighbor degree of each node. Then we take the average over all the nodes of the graph.
-
f5 – Degree assortativity coefficient: Assortativity measures how similar the connections in a graph are to the node level. It is like the Pearson correlation coefficient but measures the correlation between every pair of connected nodes.
-
f6 – Average clustering coefficient: The clustering coefficient is a measure of the degree to which nodes in a graph tend to cluster together.
-
f7 – Pagerank score: PageRank calculates the rank of nodes in a graph based on the structure of incoming links.
-
f8 – Eigenvector centrality: Eigenvector centrality is a measure of the influence of a node in a graph. It calculates the central position for a node based on the central position of the neighboring nodes.
-
f9 – Closeness centrality: Closeness centrality is a way of detecting nodes that can propagate information very efficiently through a graph. Calculating the near center of a node measures its average distance (inverse distance) to all other nodes.
-
f10 – Average betweenness centrality: Betweenness centrality measures the degree to which a vertex lies on the path between other vertices.
-
f11 – Average effective eccentricity: The eccentricity of a node is the maximum distance from that node to all other nodes in the graph, which means the longest path of all shortest paths from that node to other nodes in the graph. Average effective eccentricity is the average value of the effective eccentricity of all nodes in the graph.
-
f12 – Effective diameter: The effective diameter is the maximum effective eccentricity of the graph, defined as the maximum value of the effective eccentricity over all nodes in the graph.
-
f13 – Effective radius: The effective radius is the minimum effective eccentricity, defined as the minimum value of the effective eccentricity over all nodes in the graph.
-
f14 – Percentage of central points: A central node is a node whose eccentricity is equal to the effective radius of the graph.
-
f15 – Percentage of periphery points: Percentage of nodes in the set of nodes whose eccentricity is equal to the effective diameter.
-
f16 – Percentage of endpoints: The ratio between the number of endpoints (leaf nodes) and the total number of nodes in the graph is selected as a feature.
In extracting the topological attributes vector, if a certain graph in the dataset is disconnected and contains several components, we compute the mean for a given feature's overall components. Table 1 shows the topological attributes vector consisting of 16 features extracted from graph 0 in the MUTAG dataset.
3.2 Rooted Subgraph Mining
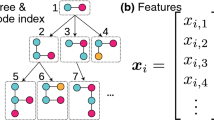
The main objective in this section is to build a “vocabulary” of rooted subgraphs (i.e., neighborhoods around every node up to a certain degree) of graphs like vocabulary in documents. Rooted subgraphs can be considered as fundamental components of any graph. Other substructures are nodes, paths, walks, etc. but this paper uses rooted subgraphs because they are non-linear and higher-order substructures, capturing more information than other substructures.
To mine rooted subgraphs in graphs, we take each node in the graph as the root node, then we find its neighborhood at a certain level d, from d = 0 (layer 1, the node itself) to d = 3 (layer 4). After that, we aggregate all rooted subgraphs of four layers and remove the repeated subgraphs to obtain a subgraph “vocabulary” set. After rooted subgraphs are mined, we have to test the isomorphism of all the subgraphs to remove repeated ones. The subgraph isomorphism test is an NP-complete problem, so it is time-consuming. To solve this problem, we follow a well-known Weisfeiler-Lehman relabeling method proposed in [8]. One iteration in Weisfeiler-Lehman relabeling consists of 4 steps:
-
Step 1: Multiset-label determination. Determine the set of multiset-label for each node in the graph. The multiset-label here is the set consisting of root node's labels and the labels of its neighbors.
-
Step 2: Sorting each multiset. In this step, we sort the labels of neighboring nodes in ascending order, then combine with that root node's label and convert it into a string of characters representing that node’s label.
-
Step 3: Label compression. After we have determined and sorted the set of multiset-labels, we map each multiset-label to a numeric character that has not appeared in the previous labels to represent the label.
-
Step 4: Relabeling. In this step, we use the mapping in Step 3 to relabel all nodes in the graph.
3.3 Neural Network Graph Embedding
Figure 1 shows the architecture of the graph embedding neural network of GC-GSL, this neural network is similar to the PV-DBOW model in doc2vec [20]. The input layer gets a one-hot vector of graphs whose length is equal to the number of graphs in the dataset. Next, there is only one hidden layer in the neural network, the number of neurons of this hidden layer equals the dimensionality of feature vectors we expect after training graph embedding. The embedding matrix between the input layer and hidden is the embedding of the graphs we need to train. Finally, the output layer consists of two parts, the first part is the topological attributes vector with 16 dimensions corresponding to 16 features of the graph. And the second part of the output layer is taken from the subgraph “vocabulary” set. More formally, the second part of the output layer tries to maximize the following objective:
where \(v_g\) is the embedding of graph g, and \( v_{sg^{\prime}}\) is the embedding of a subgraph sg′ that co-occurs in the graph g. \(sg_i\) is a random sample from the subgraph “vocabulary” set and does not appear in the graph g. σ(x) is sigmoid function, i.e., σ(x) = 1/(1 + e−x).
This graph embedding training is an unsupervised learning method, it only uses the information and structures extracted from graphs including the topological attributes vectors, the rooted subgraphs in graphs, and graphs themselves to be used for training. Therefore, it does not depend on graph labels, and only learns embedding through substructures and information of graphs. Moreover, this graph embedding model automatically learns the corresponding embedding for each graph, and the embedding that we get after training not only reflects the components of the graph itself but also reflects information about relationships between graphs.
3.4 Computational Complexity
GC-GSL consists of three main parts: the topological attributes vector extraction, the subgraph “vocabulary” set construction, and graph embedding neural network.
In the algorithm to extract topological attributes vector, we use n to represent the number of nodes and m to represent the number of edges of a graph. f1 and f2 are known, so their cost is \({\text{O}}\left( {\text{1}} \right)\). Degree-based features (f3, f4, f5 and f16) can be computed in linear time \({\text{O}}\left( {{\text{m}} + {\text{n}}} \right)\). Features depend on the eigen-decomposition of the graph (f7 and f8) can be computed in \({\text{O}}\left( {n^3 } \right)\) time in the worst case. The clustering coefficient of each node calculated in the average time is is \({\text{O}}\left( {d^2 } \right) = \left( \frac{2m}{n} \right)^2\) where \(d = \frac{2m}{n}\) is the average degree. Therefore, the average clustering coefficient (f6) on all nodes can be computed in time \(O\left( {\frac{m^2 }{n}} \right)\). Features calculated based on eccentricity (f9, f10, f11, f12, f13, f14 and f15) are calculated from the SP matrix (Shortest Path Matrix) of all pairs of vertices. The SP matrix can be calculated in \({\text{O}}\left( {{ }n^2 + {\text{ mn}}} \right)\) time. From this SP matrix, the features can be computed in \({\text{O}}\left( {n^2 } \right)\) time.
Building the subgraph “vocabulary” set requires a Breadth-First Search (BFS) algorithm to mine the subgraph for each node computed in \({\text{O}}\left( {\text{n}} \right)\), then the Weisfeiler-Lahman relabeling algorithm is used to solve the subgraph isomorphism test problem computed in \({\text{O}}\left( {{\text{lt}}} \right)\) where l is the number of iterations and t is the size of multi-labels set in each iteration.
Graph embedding neural network complexity of \({\text{O}}\left( {{\text{ES}}\left( {{\text{NH }} + {\text{ NHlog}}\left( {{\text{V }} + {\text{ F}}} \right)} \right)} \right)\), where E is the number of epochs to train the model, S is the number of graphs in dataset, N is the number of negative samples, H is the number of neurons in the hidden layer, V is the size of the subgraph “vocabulary” set, F is the dimensionality of the topological attributes vector.
4 Experiments
Datasets.
We use seven popular benchmark datasets, including 5 bioinformatics datasets of MUTAG, PROTEINS, NCI1, NCI109 and PTC_MR, and 2 social network datasets of IMDB-BINARY and IMDB-MULTI [21], whose characteristics are summarized in Table 2. MUTAG is a dataset of 188 nitro compounds labeled with respect to whether they have mutagenic effects on bacteria. NCI1 and NCI109 datasets are two subsets of the balanced dataset of screened chemical compounds for activity against non-small cell lung cancer and ovarian cancer cell lines. PROTEINS is a dataset where nodes are Secondary Structural Elements (SSEs), and edges represent neighborhood relationships in amino acid sequences or in 3-dimension space. PTC_MR dataset records the carcinogenicity of 344 chemical compounds in male rats. IMDB-BINARY and IMDB-MULTI are ego-network collection of actors, where two actors in the same movies make an edge, and the task is to infer the genre of an ego-network.
Experiments and Configurations.
In our experiment, the dimension of the hidden layer in the neural network is chosen to be 128, the best results are obtained when the topological attributes vector normalization method is z-score, the learning rate value is 0.003 and the number of epochs on seven data sets is 3000. The learning algorithm used is Adam. The size of a mini-batch for all datasets is 512. The base classifier is Support Vector Machines (SVM) [10]. Evaluation results are based on the results of 10 runs of each graph classification algorithm, with each run using 10-fold cross-validation. The final evaluation results are the mean and standard deviation of accuracy across all runs of each algorithm on each dataset.
Baselines.
Our method is compared with state-of-the-art baselines including Weisfeiler-Lehman kernel (WL), [8] Deep WL [21], Deep Divergence Graph Kernels (DDGK) [22], Anonymous walk embeddings (AWE) [23], Methods based on frequent subfragment mining (FSG) [24, 25].
4.1 Results
Accuracy.
Table 3 shows the average classification accuracy and standard deviation of the three graph classification methods on five bioinformatics datasets and two social network datasets. Overall, GC-GSL gives the best results on all the datasets compared with the traditional graph kernels and graph classification based on frequent subgraph mining. The accuracy of GC-GSL is better than FSG-Bin on all 7 datasets. Compared with the other combination graph kernels and neural networks approaches such as Deep WL, DDGK and AWE, results of GC-GSL are also good, especially on PROTEINS, NCI1 and NCI109 datasets. In addition, 3 datasets MUTAG, NCI1 and NCI109 have accuracy greater than 80%.
In summary, GC-GSL is highly effective on large datasets such as PROTEINS and NCIs with the graph classification accuracy better than the other methods. Moreover, it is robust and stable, which is implied by its small standard deviations. GC-GSL is the most effective because it automatically learns information and structures of the graph at both local and global levels. Moreover, in training graph embedding, GC-GSL also learns the relationship between graphs.
Embedding.
Through the graph embedding neural network, the graph dataset is transformed into a set of embedding vectors corresponding to each graph, where each vector has a length of 128. Therefore, to observe this result, the t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm is used for nonlinear dimensionality reduction on the 128-dimensional embedding vectors into 2-dimensional vectors. Figure 2 shows the visualization results after the graph embedding of 7 datasets in 2-dimensions with different colored circles representing different categories of graphs in the dataset. We can see that the visualization results of MUTAG and PROTEINS datasets have a clear distinction between the two classes. The visualization of NCI1 and NCI109 datasets, although as explicit as MUTAG, is that a class is subdivided into many other sub-clusters and there is a distinction between sub-clusters of two different classes. Particularly, PTC_MR dataset, which distributes data of two different classes, is complex and intertwined. IMDB-BINARY dataset has visual results quite like the NCI sets but is sparser. With IMDB-MULTI dataset, it is easy to see the different partitions of the blue circles, the green and orange circles distributed around the partitions of the blue circles. In conclusion, graph embedding is trained based on substructures and feature information in the graph, so graphs with similar substructures and information will be closer together.
4.2 Discussions
We discuss in terms of parts of GC-GSL. First, with the proposed topological attributes vector, it helps the neural network that trains the graph embedding to learn more general information about the graph. Although the topological attributes vector carries a variety of information from many distinct aspects of the graph, these features only revolve around the topology in the graph. In the graph, there is still other useful information that has not been considered such as properties of nodes, edges, etc. For example, with the analysis of social network problems, in addition to the connections between people, personal information of each person such as gender, age, etc. is also extremely necessary.
Second, the rooted subgraph mining is more effective when applying the Weisfeiler-Lehman relabeling method. However, these subgraphs are only local. This is the main reason the topological attributes vector is proposed. It helps the neural network to train graph embedding to learn more global information, but it only solves a part of the problem that cannot be solved yet thoroughly.
Finally, the neural network used to train graph embedding, the training results are effective. As we can see in Fig. 2, graphs with similar substructures will be closer to each other, and graphs with different substructures will be far away from each other. Therefore, with these graph embedding results, the classification results on the embedding vectors of the graphs will give satisfactory results (see Table 3). The results of the evaluation of the measures on experimental datasets have confirmed this with some datasets having an accuracy of over 80% and better than the other graph classification methods. Moreover, with the results of graph embedding, in addition to being used for graph classification, we can also use it for many other tasks at the graph level such as clustering, community detection.
5 Conclusion
Based on the original idea of doc2vesc in NLP to automatically learn document embedding, we applied it to graph data and proposed a novel graph classification method based on graph structure learning named GC-GSL. GC-GSL helps us to solve the problem of subgraph isomorphism testing as well as easily applied to real-world problems with large datasets and highly scalable without the need for complex implementation as in graph kernels or graph classification based on frequent subgraph mining methods. Experiments on bioinformatics data sets and social networks have both good and effective results.
Although the results of GC-GSL are effective, there will still be limitations as mentioned in the discussion such as local subgraph problems, some other information in the graph such as node, edge, etc. is still unexplored. Further improvement in classification results by overcoming the above limitations is one of the probable future development directions. On the other hand, this novel graph classification method also needs further improvements in practical terms such as speed and scalability for larger datasets.
References
Szklarczyk, D., et al.: STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47(D1), D607–D613 (2019)
Trinajstic, N.: Chemical Graph Theory. CRC Press (2018)
Siew, C.S., Wulff, D.U., Beckage, N.M., Kenett, Y.N.: Cognitive network science: a review of research on cognition through the lens of network representations, processes, and dynamics. Complexity 2019, 2108423 (2019)
Lanciano, T., Bonchi, F., Gionis, A.: Explainable classification of brain networks via contrast subgraphs. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, pp. 3308–3318 (2020)
Tabassum, S., Pereira, F.S., Fernandes, S., Gama, J.: Social network analysis: an overview. Wiley Interdisc. Rev.: Data Min. Knowl. Discovery 8(5), e1256 (2018)
Chen, X., Jia, S., Xiang, Y.: A review: knowledge reasoning over knowledge graph. Expert Syst. Appl. 141, 112948 (2020)
Domingo-Fernández, D., et al.: COVID-19 knowledge graph: a computable, multi-modal, cause-and-effect knowledge model of COVID-19 pathophysiology. Bioinformatics 37(9), 1332–1334 (2021)
Shervashidze, N., Schweitzer, P., Van Leeuwen, E.J., Mehlhorn, K., Borgwardt, K.M.: Weisfeiler-lehman graph kernels. J. Mach. Learn. Res. 12(9), 2539–2561 (2011)
Kriege, N.M., Johansson, F.D., Morris, C.: A survey on graph kernels. Appl. Netw. Sci. 5(1), 1–42 (2019). https://doi.org/10.1007/s41109-019-0195-3
Chang, C.C., Lin, C.J.: LIBSVM: a library for support vector machines. ACM Trans. Intell. Syst, Technol. (TIST) 2(3), 1–27 (2011)
Vishwanathan, S.V.N., Schraudolph, N.N., Kondor, R., Borgwardt, K.M.: Graph kernels. J. Mach. Learn. Res. 11, 1201–1242 (2010)
Borgwardt, K.M., Kriegel, H.P.: Shortest-path kernels on graphs. In: Fifth IEEE International Conference on Data Mining (ICDM’05), pp. 8-pp. IEEE (2005)
Nikolentzos, G., Meladianos, P., Rousseau, F., Stavrakas, Y., Vazirgiannis, M.: Shortest-path graph kernels for document similarity. In: Proceedings of the 2017 Conference on Empirical Methods in Natural Language Processing, pp. 1890–1900 (2017)
Horváth, T., Gärtner, T., Wrobel, S.: Cyclic pattern kernels for predictive graph mining. In: Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 158–167 (2004)
Shervashidze, N., Vishwanathan, S. V. N., Petri, T., Mehlhorn, K., Borgwardt, K.: Efficient graphlet kernels for large graph comparison. In: Artificial Intelligence and Statistics, pp. 488–495. PMLR (2009)
Ramon, J., Gärtner, T.: Expressivity versus efficiency of graph kernels. In: Proceedings of the First International Workshop on Mining Graphs, Trees and Sequences, pp. 65–74 (2003)
Fei, H., Huan, J.: Structure feature selection for graph classification. In: Proceedings of the 17th ACM Conference on Information and Knowledge Management, pp. 991–1000 (2008)
Kong, X., Yu, P.S.: Semi-supervised feature selection for graph classification. In: Proceedings of the 16th ACM SIGKDD international conference on Knowledge discovery and data mining, pp. 793–802 (2010)
Schöning, U.: Graph isomorphism is in the low hierarchy. J. Comput. Syst. Sci. 37(3), 312–323 (1988)
Le, Q., Mikolov, T.: Distributed representations of sentences and documents. In: International Conference on Machine Learning, pp. 1188–1196. PMLR (2014)
Yanardag, P., Vishwanathan, S.V.N.: Deep graph kernels. In: Proceedings of the 21th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1365–1374 (2015)
Al-Rfou, R., Perozzi, B., Zelle, D.: Ddgk: Learning graph representations for deep divergence graph kernels. In: The World Wide Web Conference, pp. 37–48 (2019)
Ivanov, S., Burnaev, E.: Anonymous walk embeddings. In: International conference on machine learning, pp. 2186–2195. PMLR (2018)
Rousseau, F., Kiagias, E., Vazirgiannis, M.: Text categorization as a graph classification problem. In: Proceedings of the 53rd Annual Meeting of the Association for Computational Linguistics and the 7th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), pp. 1702–1712 (2015)
Wang, H., et al.: Incremental subgraph feature selection for graph classification. IEEE Trans. Knowl. Data Eng. 29(1), 128–142 (2016)
Yan, X., Han, J.: gSpan: graph-based substructure pattern mining. In: 2002 IEEE International Conference on Data Mining, 2002 Proceedings, pp. 721–724. IEEE (2002)
Huan, J., Wang, W., Prins, J.: Efficient mining of frequent subgraphs in the presence of isomorphism. In: Third IEEE International Conference on Data Mining, pp. 549–552. IEEE (2003)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Huynh, T., Ho, T.T.T., Le, B. (2022). Graph Classification via Graph Structure Learning. In: Nguyen, N.T., Tran, T.K., Tukayev, U., Hong, TP., Trawiński, B., Szczerbicki, E. (eds) Intelligent Information and Database Systems. ACIIDS 2022. Lecture Notes in Computer Science(), vol 13758. Springer, Cham. https://doi.org/10.1007/978-3-031-21967-2_22
Download citation
DOI: https://doi.org/10.1007/978-3-031-21967-2_22
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21966-5
Online ISBN: 978-3-031-21967-2
eBook Packages: Computer ScienceComputer Science (R0)