Abstract
Maize (Zea mays L.) is the third major and high yielding cereal crop in the world. Maize is model crop for genetic studies due to its industrial and nutritional values. So, experiments have done for production of genetically modified maize since the 1990s and adopted worldwide. Genetic techniques comprising of genome editing tools have been used including meganucleases, ZFNs, and TALENs to enhance production and to improve agronomic traits of maize. These nucleotides are protein based and thought to be perfect tools for gene editing. Various agricultural crops have been modified to produce biotic and abiotic stress-tolerant cultivars by using these tools. Crops such as Arabidopsis, cotton, and maize have been widely modified with protein-based nucleotides. Meganucleases, ZFNs, and TALENs produced efficient modifications in plants but have some limitations of time-consuming and difficult handling. To overcome the drawbacks of previously known tools, researchers developed new RNA-based CRISPR/Cas9 method for gene editing. This tool proved efficient and robust for genetic modifications in various plants. A large number of agronomic traits have been improved in maize by using CRISPR/Cas9 system.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Human population is increasing rapidly. According to the European Union, the current world population is 7.9 billion people and estimated to increase by 10 billion in the next 30 years. The FAO estimates that 2577.85 million tons of food were produced during 2016 (McIntosh et al. 2015). Current food production is not fulfilling the required demands of growing population, which leads to food shortage. It is predicted that around 9% of world population that counts for 697 million people are food insecure and around 820 million people are undernourished. For this tremendous increase in population, more food supply is required. Global food production is estimated to double by 2050 and requires a 70–85% increase in crop yield to feed undernourished.
Global food security is dependent on both adequate food production and food access, but some factors are limiting the production of food. Food production has been widely affected by abiotic and biotic factors and raised concerns of food security. Agriculture sector is facing serious problems due to negative ecological consequences. It imposes serious threats to the economy of a country (Raza et al. 2019). Agriculture-driven growth and food security are at risk due to ecological changes. Furthermore, food security is becoming a major concern around the world due to drought and heat stress that is the leading constraint of food production. Drought conditions and high temperature result in physiological, physical, and biochemical changes of plants that affect the plant growth, development, and production (Fathi and Tari 2016).
Abiotic factors are nonliving, whereas biotic factors are living factors that reduced the agricultural production to a greater extent and directly influence food supply (Razzaq et al. 2021d; Wani et al. 2022). Abiotic factors are due to environmental changes that include high or low temperature, salt stress, drought stress, relative humidity, and fluctuation of pH (Arora 2019). However, biotic factors are due to insects and pests that reduce crop yield. The role of these factors in ecosystem regulation has been extensively studied over the years, resulting in the characterization of biotic and abiotic relationships (Dresselhaus and Hückelhoven 2018). These factors are essential for energy flow and nutrient cycling of an ecosystem. Biotic and abiotic factors are mainly due to climate changes that have a significant impact on crop reduction. Temperature rising and yield losses are major challenges of climate change. Extreme weather conditions of heavy rainfall, change in CO2 concentrations, drought, alteration of host-pathogen relationship, and depletion of ozone layer cause significant reduction of crop production and adversely affect global food security (Raza et al. 2019).
Some crops are sensitive to saline environment, which develop resistance mechanisms against salt stress. Rather than developmental stages, germination stages of plants are more sensitive to salty environment. Plant uptake of nutrients, for example, nitrogen, calcium, magnesium, potassium, and iron, decreases due to high content of sodium and chloride rhizosphere (Farooq et al. 2015). Photosynthesis is enhanced by atmospheric CO2 concentrations that reduced crop water usage. Variation of CO2 levels increases global crop water productivity depending on crop type. Global yield loss could be mitigated by the elevated level of CO2 concentrations. It would reduce the consumption of water up to 4–17% (Deryng et al. 2016).
Abiotic stresses may have resulted in the aggregation of reactive oxygen species (ROS), which act as signal transduction molecules. These ROS cause cellular damage and inhibit photosynthesis. ROS are removed with antioxidants to prevent extensive cell damage (Dietz et al. 2016). Yield of staple foods is substantially reduced due to insect pests. There is a significant relationship between temperature, population growth, and metabolic rates of insects that augment grain losses. High temperature will increase insect’s population and their metabolic rates. The production losses of staple food such as rice, wheat, and maize are expected to rise by 10–15% as global warming increases (Deutsch et al. 2018). Agronomic traits of plants are different under different environmental conditions. Agronomic traits include plant height, plant biomass, harvest index (HI), spike length, and number of spikes. Plant agronomic traits appear during later stages of their growth, based on their interaction with environment (Mochida et al. 2020).
These factors alone or in combination adversely affect global food production and ultimately affect global food security. In order to overcome ecological consequences and ensure food security, scientists are forced to develop novel breeding techniques to tackle yield loss due to these stresses.
2 Significance of Maize and Global Status
Maize (Zea mays L.) is the world’s third dominant and high yielding grain crop. It is the world’s most significant crop, cultivated over more than 118 million hectares with 600 million metric tons annual production. If the world’s population grows to 9 billion people by 2050, global food demand will begin to rise rapidly. In 2020, worldwide maize demand will grow by 45%, compared to 30% for wheat and 32% for rice. This shows that the global need for maize will double, and it will become the most grown crop worldwide by 2050 (Ten Berge et al. 2019).
Maize is valuable feed grain as it provides high energy density of 365 kcal/100 g. It contains about 72% starch, 24% carbohydrates, 10% protein, 3–5% vitamins A and B, 4% fats, and 3.7% sugars but has less amount of fiber contents (Ranum et al. 2014). Maize is a raw material and extensively used to make different products. Maize is high source of ethanol production that can be used as biofuel. As maize is a high source of energy and lowest in fiber and protein content, it can be used as livestock feedstock. Maize contributes significantly to global food security by supplying food, feedstock, and energy for the growing human population. It can grow under different environmental conditions, and this quality makes it globally desirable crop (Sanaullah et al. 2018). Maize originated from a wild grass in central Mexico around 7000 years ago. Three top maize producer countries are the United States, China, and Brazil where maize grown all around the globe. These countries produced maize worth of 563–717 million metric tons per year, grown on 70–100 million acres annually. In 2019, China harvested 41.3 million hectares of maize, 32.9 million hectares in the United States, and 17.5 million hectares in Brazil. The total maize production in the United States was 347 million metric tons, 260 million metric tons in China, and 101 million metric tons in Brazil.
In the last 10 years, 40% maize production has increased in the United States. Maize, being the world’s most produced cereal crop, is of great importance in developing countries like Pakistan, where available food supplies are not sufficient for a rapidly growing population. After wheat, cotton, and rice, maize is the fourth most grown crop in Pakistan. It is grown on an area of more than 1.4 million hectares and average grain yield of 51,203 kg/ha. In Pakistan, the harvested area is 1.4 million hectares, with an annual production of 72 million metric tons. A total of 35 and 27 million metric tons of annual maize production have been calculated in Bangladesh and in India, respectively.
The Maize extraction rate varies from 60% to 100% in different countries. Although maize is not a native crop in Africa, now 300 million Africans are growing it due to its high nutritional values. Higher extraction rates are observed in South Africa, from 62% to 99% depending on the product type. Maize varieties that grow in different seasons have lower extraction rates after the milling process (Ranum et al. 2014).
Along with its industrial and nutritional importance, maize is also a model crop for genetic research. So, experiments have been conducted, and genetically modified maize have been introduced since the 1990s and adopted worldwide. To increase maize production, genetic approaches have been applied to improve the agronomic features of the crop. Farmers have shifted from traditional approaches toward modern genetically modified techniques as they can significantly reduce the losses caused by various biotic and abiotic factors by improving agronomic traits. Traditionally, irradiation and mutagenesis were used, but nowadays, gene editing tools are used to modify the genome of maize (Razzaq et al. 2021b).
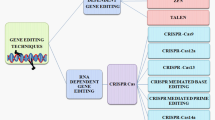
3 Genome Editing
Current crop yield has to be doubled to ensure food security in the era of growing population and developing biotic and abiotic stressors (Ray et al. 2013). To combat food insecurity concerns, it is important to develop stress-tolerant crops. The conventional approaches that are being used enhanced food production in cultivated areas but didn’t focus on stress resistance and nutritional quality. Nature has been changing the genome through natural selection from the beginning of life (Razzaq et al. 2021a). But with the advancement of knowledge, scientists developed artificial selection or selective breeding for desirable traits. Artificial selection has been done in numerous plants, such as modern corn, a descendant of teosinte (Yang et al. 2019). However, further research is in progress to combat evolving challenges and to produce stress-resistant crops.
Recent advancements in plant biotechnology have made it possible to alter the genomes of plants that influence desirable agronomic traits against biotic and abiotic stresses. The current approaches tend to increase crop yield, enhance nutritional qualities, map resistance genes, and regulate various physiological and metabolic pathways against biotic and abiotic stresses (Razzaq et al. 2021a). For nutritional improvement, it provides adequate minerals and vitamins that are beneficial for human health (Farre et al. 2012). Specific enzymes in metabolic pathways have been manipulated to increase the amount of vitamins and minerals while lowering the levels of antinutrients such as phytic acids and acrylamide-forming amino acids (Mugode et al. 2014).
Researchers target specific agronomic traits that cope with stresses to increase yield and production. For this purpose, plant structure is manipulated to introduce specific traits (Tshikunde et al. 2019). Selection of traits such as low and high temperatures, salt stress, drought stress, heavy metals, and other stresses has been targeted as abiotic factors. Resistance to bacterial, viral, and fungal pathogens against biotic stresses has become a growing concern as temperature intensifies the spread of pathogens.
A significant breakthrough in genetic engineering has focused on addition of DNA sequences into the genome (Datta 2013). Agrobacterium tumefaciens has been extensively used for gene modification in plants (Nester 2015). In 1994, the first GM crop, “Flavor Savor Tomato” was commercialized. Genetic engineering has developed crops with stress-resistant properties, enhanced nutritional content, and improved yield. However, with all the advantages and modifications, this technique has some drawbacks. The major limitation is the insertion of foreign DNA into plants for quantitative and qualitative characteristics without using the plant’s own genetic material. Public concern over GMOs is a major drawback, along with other environmental and ethical concerns. GMOs have been restricted in several countries due to nonspecificity and risk to human health.
There is a need for technology that can overcome all hurdles and produce stress-tolerant crops. Mario Capecchi established gene targeting technology for genome editing in the 1980s. For example, nonhomologous end joining (NHEJ) can be used to create functional knockout of genes by inserting and deleting a few genes (Weinthal et al. 2013). Genome editing precisely alters the DNA sequence in one specific base with insertions and deletions that develop desirable agronomic traits to cope with the high and low temperatures and insect and pest attack. Functional and nutritional traits of diverse crops have been modified with genome editing (Tan et al. 2020; Sedeek et al. 2019; Razzaq et al. 2019). Genome modification by editing is among the most promising techniques of all time in applied biological and industrial research. Genetically modified plants with foreign DNA in their genomes may prove to be more sustainable for the public than genome editing. Genome editing’s accessibility has overcome all of the limitations of traditional methods (Abdallah et al. 2015). Site-specific recombinases and site-specific nucleases could be used for genome editing. The high-oleic soybean variety was the first approved gene edited product for commercialization in 2019, paving the way for commercialization of other genome edited crops.
Traditionally, irradiation and mutagenesis were used for genome modification of maize, but they have numerous limitations. This approach accuracy was limited. That’s why gene editing tools are used to modify the genome of maize to produce stress-resistant varieties with high precision. Accessibility to genomic data of maize sets new insights for genome editing.
Meganucleases, zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR/Cas systems have all been found as genome editing tools so far (Curtin et al. 2012). ZFNs and TALENs are protein-designed systems, whereas CRISPR/Cas is RNA-based system that can bind to target DNA. The main mechanism is to make double-stranded breaks (DBS) on specific sites of DNA with nucleases. Various agricultural crops have been modified to produce biotic and abiotic stress-tolerant cultivars. Many agricultural innovations have been created with this novel system.
4 Meganucleases (MegaN)
The discovery of meganucleases sparked the idea of genome editing. Meganucleases are naturally occurring endonucleases, also known as homing endonucleases due to their specific nature. Meganucleases have 12–40 bp recognition sites and make double-stranded breaks (DSBs) (Gallagher et al. 2014). This specific nature makes them perfect tools for gene editing. LAGLIDADG, GIY-YIG, HNH, His-Cys box, and PD-(D/E)XK are the five families of meganucleases based on their structural motifs (Zhao et al. 2007; Orlowski et al. 2007). LAGLIDADG is the best explored among these numerous families for genome editing. It is estimated that LAGLIDADG recognizes a DNA sequence of 14–40 bp. Gene editing with modified meganuclases has incorporated into Arabidopsis, cotton, and maize.
It is very difficult to engineer meganuclease because DNA binding domains and endonuclease catalytic domains are located in the same location and cannot be easily detached. Additionally, naturally occurring meganucleases are less in number and are not enough for potential locus. Construction of site-specific enzymes for meganucleases is time-consuming and expensive. Technically, initial protein modification is required to produce a meganuclease for target genome engineering, and this is very challenging, and patent issues have seen in cultivars of this family (Abdallah et al. 2015). As manipulation of meganucleases is difficult, additional efforts are needed to improve this approach. Therefore, researcher’s interest has shifted to more efficient and accurate methods, such as ZFNs, TALENS, and CRISPR.
5 Zinc Finger Nucleases (ZFNs)
Over the past two decades, new technologies have developed to modify plant genomes. Among these zinc finger nucleases (ZFNs) are designed enzymes that are intended to cleave DNA at certain sites or locations within the genomic structure. ZFNs are first-generation-targeted genome editing techniques that can be used for deletions, additions, and knockout of targeted genes in the genome. ZFNs are made up of a zinc finger DNA binding domain and an endonuclease (Fok1 nuclease) (Davies et al. 2017). Fok1 has distinct binding and cleavage activities. Fok1 belongs to type IIS class of endonuclease enzymes, which are commonly used cleavage domain. The cleavage domain dimerizes to cut DNA at a recognize site. A binding arrangement of four to six zinc finger protein domains has the ability to recognize about 3 bp of DNA (Carroll 2011). Binding to the appropriate targeted sequence, a pair of zinc finger arrays arrange themselves in reverse order. Two ZFAs have binding sites that are around 5–8 bp apart (each 18–24 bp in length). In ZFN design, this gap is crucial because it permits fok1 monomers to dimerize and create a DSB in the desired sequence (Ansari et al. 2020). They then employ the cellular endogenous DSB repair process to alter the target genomic region (Feng et al. 2016). To repair these double-stranded breakage (DSB), nonhomologous end joining (NHEJ) repair mechanism is used, which allows for site-specific mutagenesis (Yadava et al. 2017). It is now feasible to target a large number of DNA sequences in the genome with this technique. ZFNs can be employed in agricultural plants to induce alterations in normal genomic structure, which could be more consumer-friendly than genetically modified strains generated via gene insertion. ZFNs can be utilized to re-grow crops such as maize by changing callus or cells in culture (Townsend et al. 2009). This technology is used to modify endogenous loci in maize, but it faces many challenges. This method is time-consuming and expensive as it involves complex steps that require protein engineering. It must be genetically engineered to create double-stranded breaks (DSB) at the specific sites. It may attach to any DNA sequence in the genome rather by binding with the target location and cause a high rate of changes that make it difficult to distinguish between changed or altered alleles (Petolino 2015). Several optimizations are needed to improve editing of plant genomes via ZNFs, for example, choice of plant tissue for targeting, the lack of off-target mutagenesis, and the introduction of enzyme activity and the constant identification of mutated alleles and specialized skills required for interpretation of results, which have made it less applicable for crop genome editing (Agarwal et al. 2018).
6 Transcription Activator-Like Effector Nucleases (TALENs)
TALENs uses Fok1 nucleases in the same way as ZFNs do, but its mechanism for recognizing the target site is totally different. TALEN has emerged as a potential genetic technique for mutagenesis of specific genes. Each of two monomers of TALEN has the TALE DNA binding domain (highly conserved repeats) linked to the Fok1 domain. The DNA binding domain is derived from TALE produced by Xanthomonas spp. that is used to change the gene transcription in host plants (Khan et al. 2017).
TALENs belong to first generation genome editing tool and recognize the 14–20 bp recognition site of target DNA (Kumar et al. 2019). TALE domain recognizes the defined specific DNA site, and Fok1 nuclease dimerizes to create DSBs. These DSBs can be repaired by NHEJ and HR (Char et al. 2015). These mechanisms have potential for deletions, insertions, rearrangements, or replacement of chromosomes. NHEJ mostly leads to insertion and deletions into the genome and leads to loss of gene function, while DSBs are accurately repaired in HR repair mechanism by using a DNA template from homologous donor (Sprink et al. 2015).
In contrast to ZFN, TALEN has many advantages. It is more efficient, flexible, and less toxic. It is easier to engineer. The main advantage is that they can bind with specific DNA sequence, while in the case of the zinc finger, it chooses a library of fingers that have the necessary binding characteristics. As an efficient tool, TALENs have been applied to create useful traits and qualities in crop plants and make the crops resilient to withstand the enormous biotic and abiotic stresses (Malzahn et al. 2017). But still, this technique has many limitations. The creation of TALE repetitions is still a challenge, and the effectiveness of TALEN gene targeting varies.
It is time-consuming, expensive, and difficult to handle and can never be used on large scale. There are still some questions that needed to be answered to get desired outcomes, for example, which vector transformation method has to be used and the choice of explant (Khan et al. 2017). Scientists are optimistic that targeted genome engineering will have a bright future and will bring solutions to our issues.
7 CRISPR/Cas9: A Robust Genome Editing Tool
Zinc finger nucleases and TALENS were extensively utilized genome editing techniques until 2013. However, with the discovery of CRISPR/Cas from the last few decades, scientific advances have ushered in a revolution in genome editing. CRISPR is more simple, reliable, and robust method of specific mutagenesis than previously used tools. This second-generation tool is simple to use, versatile, accurate, and economical than ZFN and TALEN tools of first generation. CRISPR opens up new opportunities against biotic and abiotic factors and produced high-quality resistant crops and improved yield traits. The implementation of this robust tool has provided great insights in agricultural biotechnology and improved crop production (Razzaq et al. 2019).
CRISPR/Cas9 was extracted from immune system of bacteria (Jinek et al. 2012) and initially was classified into three types and ten subtypes (Makarova and Koonin 2015).
After that, it grew to include two classes with further five types and sixteen subtypes (Makarova et al. 2015). With two previously known classes, the classification of CRISPR was extended to six types and 33 subtypes (Makarova et al. 2018, 2020). CRISPR systems that can target multiple genes and improve the specificity of the CRISPR toolbox include type II, V, and VI of class 2 (Cong et al. 2013; Tang and Fu 2018). CRISPR repeats range in size from 23 to 55 in different organisms and have average length of 32 bp. In certain animals, each repeat contains a unique nucleotide sequence that is highly conserved and partly palindromic (Karimi et al. 2018). Streptococcus pyogenes-derived Cas9 (SpCas9) endonuclease has been widely used in crop improvement (Le Rhun et al. 2019) via different mechanisms including site-specific CRISPR system and precise base editing (Zong et al. 2017).
CRISPR can manipulate genome without the insertion of foreign DNA or exogenous via ribonucleoprotein complex (RNP) and thus results in transgene free plants (He and Zhao 2020; Tsanova et al. 2021). The RNA-driven DNA endonucleases that produce double-stranded breaks are used in this gene editing technique (Jinek et al. 2012).
A single guide (sgRNA) and Cas9 nuclease combine to make up the CRISPR/Cas9 system. The linker loop of the guide sequence contains crRNA and tracrRNA (Cong et al. 2013). The guide sequence is recognized with duplex of CRISPR RNA (crRNA) and trans-encoded CRISPR RNA (tracrRNA) by binding while engineering. crRNA hybridize to the complementary strand of targeted genome adjacent to PAM sites. DNA cleavage is carried out with crRNA and tracrRNA on guided RNA (gRNA) and enhanced the cleavage frequency (Jinek et al. 2012).
The Cas9 endonuclease contains HNH and RuvC domains. sgRNA combine or pair up with cas9 endonuclease and results in the formation of Cas9 complex. Cas9 paired with gRNA on targeted complementary sequence and generate double-stranded breaks at three to four nucleotides that are upstream of the PAM site of targeted genomic locus. Thus, with accurate insertion of sgRNAs, Cas9-mediated cleavage has the ability to initiate site-specific genome editing (O’Connell et al. 2014; Hanna and Doench 2020).
DSBs are produced on HNH domain by cleaving the complementary strands, while RuvC domains produce DSBs by cleaving the noncomplementary strands. As a result, these DSBs trigger DNA repair mechanism either through nonhomologous end joining (NHEJ) that stands a chance of error or through homology-directed repair (HDR) for accurate repair. Many outstanding repairs of single base substitution, gene replacement, and targeted knock-in were achieved via HDR (Schiml et al. 2014). Transformation methods are used to deliver the expression cassette of sgRNA and Cas9 into desired cells via PEG-mediated, agrobacterium-mediated, and biolistic-mediated transformation (Sandhya et al. 2020). This cassette delivery into cell can be stable or involve transient transformation. Ability of CRISPR to cleave DNA at specific site with remarkably efficient targeting makes it a more flexible and robust approach for plant genome engineering than ZFNs and TALENs (Bortesi and Fischer 2015).
8 Applications of CRISPR Cas9 in Maize Improvement
CRISPR/Cas has changed genome editing approaches since its discovery. To address the obstacles, this new technique is utilized to produce specific targeted mutations in living organisms. It is a robust method with a lot of potential for crop improvement. CRISPR/Cas has several applications in maize improvement, including biotic/abiotic stress tolerance, and the production of novel agronomic traits to boost yield and nutrition (Zhang et al. 2021).
Teng et al. (2020) employed CRISPR/Cas9 to disrupt the Dicer-like 5 (Dcl5) which is supposed to be involved in the generation of secondary small interfering RNAs (phasiRNAs) and has major function in anther and flower development in maize. The mutant lines showed temperature-sensitive male fertility, defective tapetal cells, and short anthers. This suggested that (Dcl5) in crucial in heat tolerance during anthers development and can be targeted to develop heat tolerant maize lines in future.
ARGOS8 gene is the negative regulator of ethylene response in maize. The overexpression of the ARGOS8 gene in transgenic plants reduces ethylene sensitivity, resulting in increased grain yield under drought stress. To check the response of maize to overexpression ARGOS8 under drought stress and measure the abiotic stress tolerance, CRISPR/Cas9-mediated targeted mutagenesis was performed on about 400 inbred maize lines to produce novel variants. The native maize GOS2 promoter was inserted into the 5′ untranslated region of the native ARGOS8 gene, resulting in a moderate level of constitutive expression. PCR and RNA sequencing revealed genomic DNA alterations at the maize gene locus (ARGOS8). The results revealed that transgenic plants with the GOS2 promoter overexpressed the ARGOS8 gene, indicating an increase in grain production under drought stress conditions as compared to natural cultivars. As a consequence, the findings highlight the necessity of using CRISPR/Cas9 to create novel variations for breeding drought-tolerant maize cultivars (Shi et al. 2017).
One of the primary factors of limiting maize production across the world is salinity. Maize is a glycophyte plant that is extremely vulnerable to salt stress (Razzaq et al. 2021c). Maize has two phases of physiological challenges. The first is osmotic, whereas the second is ion toxic. Plant growth is slowed in the osmotic phase due to reduction in environmental water potential. In ion toxic, the sodium ions accumulate in the plant at toxic level and compete with potassium ions. Maintaining plant salt tolerance requires a balance of sodium potassium ions in the cytoplasm and a low concentration of Na ions. Salt tolerance-QTL, ZmHKT1 is linked to sodium ions and is an essential salt tolerance-QTL. Retrotransposon incorporation in maize and function of gene loss provide enhanced Na+ ion concentrations in leaves, which leads to high salt sensitivity. CRISPR-/Cas-mediated genome editing of ZmHKT1 was achieved. The results showed increased concentration of Na+ in xylem that transfer from root to shoot. Hence, it was concluded that ZmHKT1 plays an essential role in maize salinity stress tolerance and produce new varieties of maize with improved salinity tolerance (Zhang et al. 2018).
Executed CRISRP-/Cas9-mediated genome editing to target ZmHKT2 and study the salinity tolerance mechanism in maize. The edited lines indicated reduced level of K+ transports and conder salinity tolerance. For this, recombinant inbred lines (RILs) were developed and mapped the novel QTL ZmHKT2, which controls the K+ concentration through HKT transporters under salinity stress. Furthermore, K+ concentrations in xylem sap has been increased in the absence of ZmHKT2, which offer salt tolerance.
Schwartz et al. (2020) demonstrated the targeted inversion of specific region of 75.5 Mb on chromosome 2 in superior maize inbred line. This resulted in the opening of a huge chromosomal segment which harbors significant genetic diversity for recombination and can provide excellent platform to develop elite maize lines with desired traits. In another study, Gao et al. (2020) used CRISPR/Cas9 to disrupt the two waxy allele in maize inbred lines to develop waxy corn hybrids that showed superior field performance. This procedure was very quick in contrast to marker-assisted selection and backcrossing for conventional trait introgression. The field trails indicated that the engineered waxy hybrid lines produce higher yield and exhibited elite agronomic traits.
Developed a novel prime editing system for maize by improving the pegRNA expression. Two genes such as ZmALS1 and ZmALS2 related to herbicide tolerance were mutated by designing the highly efficient pegRNA prime editor vector. The results revealed that the pZ1WS prime editor was very beneficial to develop transgene-free multiple herbicide-resistant maize progenies by targeting the two ALS genes. Development of male sterile lines is always of great interest for the plant breeders to produce hybrid seed development. Chen et al. (2018) used a CRISPR/Cas9 vector to create male sterile lines in maize by mutating the MS8 gene via agrobacterium transformation. Male sterile transgene-free lines were captured by screening of sterile population, and male sterility can be achieved in other superior inbred maize varieties for hybrid development.
9 Conclusion and Future Perspectives
Maize is the most important staple crop and contributes significantly to global food security by supplying food. It provides higher amount of energy to human, and its global demand is increasing as the human population is increasing. Due to continuous climate change, many biotic and abiotic factors impose serious threats to maize production and globally affect the food security. Crop output must be increased in order to ensure food security in the era of growing population and biotic and abiotic challenges. To control these factors and ensure food security, breeding methods develop to make the crops resistant by targeting specific traits to cope with stresses to increase yield and production. Genetic engineering is used to make genetically modified corps that are resistant to stresses and enhance the agronomic traits, but this approach has drawbacks along with other environmental and ethical concerns. To overcome all these hurdles, other genome editing technologies are used like meganucleases, zinc finger nucleases, and TALENs that are protein based. These techniques provide efficient genome changes by targeted modifications and produce better agronomic traits. But these techniques have also some limitations. These are time-consuming, expensive, and difficult to handle and can never be used on large scale. The most powerful and excellent tool paves its ways to cope with all these stresses is CRISPR/Cas9, which is RNA-based technology. CRISPR technology is used to produce better varieties with remarkably efficient targeting that makes it more versatile and robust technique for plant genome engineering than others. This technique has many applications in maize improvement and used to create stress-tolerant cultivars and the production of new agronomic traits to increase the yield and nutrition.
References
Abdallah NA, Prakash CS, McHughen AG (2015) Genome editing for crop improvement: challenges and opportunities. GM Crops Food 6(4):183–205. https://doi.org/10.1080/21645698.2015.1129937
Agarwal A, Yadava P, Kumar K, Singh I, Kaul T et al (2018) Insights into maize genome editing via CRISPR/Cas9. Physiol Mol Biol Plants 24(2):175–183
Ansari WA, Chandanshive SU, Bhatt V, Nadaf AB, Vats S et al (2020) Genome editing in cereals: approaches, applications and challenges. Int J Mol Sci 21(11):4040
Arora NK (2019) Impact of climate change on agriculture production and its sustainable solutions. Environ Sustain 2(2):95–96. https://doi.org/10.1007/s42398-019-00078-w
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33(1):41–52. https://doi.org/10.1016/j.biotechadv.2014.12.006
Carroll D (2011) Genome engineering with zinc-finger nucleases. Genetics 188(4):773–782
Char SN, Unger-Wallace E, Frame B, Briggs SA, Main M et al (2015) Heritable site-specific mutagenesis using TALEN s in maize. Plant Biotechnol J 13(7):1002–1010
Chen R, Xu Q, Liu Y, Zhang J, Ren D, Wang G, et al (2018) Generation of transgene-free maize male sterile lines using the CRISPR/Cas9 system. Front Plant Sci 9:1180. https://doi.org/10.3389/fpls.2018.01180
Cong L, Ran FA, Cox D, Lin S, Barretto R, Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.). Science 339(6121):819–823. https://doi.org/10.1126/science.1231143.Multiplex
Curtin SJ, Voytas DF, Stupar RM (2012) Genome engineering of crops with designer nucleases. Plant Genome 5(2):42–50. https://doi.org/10.3835/plantgenome2012.06.0008
Datta A (2013) Genetic engineering for improving quality and productivity of crops. Agric Food Sec 2(1):2–4. https://doi.org/10.1186/2048-7010-2-15
Davies JP, Kumar S, Sastry-Dent L (2017) Use of zinc-finger nucleases for crop improvement. Prog Mol Biol Transl Sci 149:47–63
Deryng D, Elliott J, Folberth C, Müller C, Pugh TAM et al (2016) Regional disparities in the beneficial effects of rising CO2 concentrations on crop water productivity. Nat Clim Chang 6(8):786–790. https://doi.org/10.1038/nclimate2995
Deutsch CA, Tewksbury JJ, Tigchelaar M, Battisti DS, Merrill SC et al (2018) Increase in crop losses to insect pests in a warming climate. Science (80-.) 361(6405):916–919. https://doi.org/10.1126/science.aat3466
Dietz KJ, Mittler R, Noctor G (2016) Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol 171(3):1535–1539. https://doi.org/10.1104/pp.16.00938
Dresselhaus T, Hückelhoven R (2018) Biotic and abiotic stress responses in crop plants. Agronomy 8(11):8–13. https://doi.org/10.3390/agronomy8110267
Farooq M, Hussain M, Wakeel A, Siddique KHM (2015) Salt stress in maize: effects, resistance mechanisms, and management. A review. Agron Sustain Dev 35(2):461–481. https://doi.org/10.1007/s13593-015-0287-0
Farre G, Gomez-Galera S, Naqvi S, Bai C, Sanahuja G et al (2012) Biotechnologycrop/croppingbiotechnologyand nutritional Improvementcrop/croppingnutritional improvementof Cropscrop/cropping. In: Meyers RA (ed) Encyclopedia of sustainability science and technology. Springer, New York, pp 1676–1723
Fathi A, Tari DB (2016) Effect of drought stress and its mechanism in plants. Int J Life Sci 10(1):1–6. https://doi.org/10.3126/ijls.v10i1.14509
Feng C, Yuan J, Wang R, Liu Y, Birchler JA et al (2016) Efficient targeted genome modification in maize using CRISPR/Cas9 system. J Genet Genomics 43(1):37–43
Gallagher RR, Li Z, Lewis AO, Isaacs FJ (2014) Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat Protoc 9(10):2301–2316. https://doi.org/10.1038/nprot.2014.082
Gao H, Gadlage MJ, Lafitte HR et al (2020) Superior field performance of waxy corn engineered using CRISPR–Cas9. Nat Biotechnol 38:579–581. https://doi.org/10.1038/s41587-020-0444-0
Hanna RE, Doench JG (2020) Design and analysis of CRISPR–Cas experiments. Nat Biotechnol 38(7):813–823. https://doi.org/10.1038/s41587-020-0490-7
He Y, Zhao Y (2020) Technological breakthroughs in generating transgene-free and genetically stable CRISPR-edited plants. aBIOTECH 1(1):88–96. https://doi.org/10.1007/s42994-019-00013-x
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA et al (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (80-.) 337(6096):816–821. https://doi.org/10.1126/science.1225829
Karimi Z, Ahmadi A, Najafi A, Ranjbar R (2018) Bacterial CRISPR regions: general features and their potential for epidemiological molecular typing studies. Open Microbiol J 12(1):59–70. https://doi.org/10.2174/1874285801812010059
Khan Z, Khan SH, Mubarik MS, Sadia B, Ahmad A (2017) Use of TALEs and TALEN technology for genetic improvement of plants. Plant Mol Biol Report 35(1):1–19
Kumar R, Kaur A, Pandey A, Mamrutha HM, Singh GP (2019) CRISPR-based genome editing in wheat: a comprehensive review and future prospects. Mol Biol Rep:1–13
Le Rhun A, Escalera-Maurer A, Bratovič M, Charpentier E (2019) CRISPR-Cas in Streptococcus pyogenes. RNA Biol 16(4):380–389. https://doi.org/10.1080/15476286.2019.1582974
Makarova KS, Koonin EV (2015) Annotation and classification of CRISPR-Cas systems. Methods Mol Biol 1311:47–75. https://doi.org/10.1007/978-1-4939-2687-9_4
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA et al (2015) An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13(11):722–736. https://doi.org/10.1038/nrmicro3569
Makarova KS, Wolf YI, Koonin EV (2018) Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 1(5):325–336. https://doi.org/10.1089/crispr.2018.0033
Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS et al (2020) Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18(2):67–83. https://doi.org/10.1038/s41579-019-0299-x
Malzahn A, Lowder L, Qi Y (2017) Plant genome editing with TALEN and CRISPR. Cell Biosci 7(1):1–18
McIntosh RA, Williamson PM, Wrigley CW (2015) The nature, causes, and control of grain diseases in the major cereal species. Encycl Food Grains Second Ed 4–4:74–82. https://doi.org/10.1016/B978-0-12-394437-5.00183-2
Mochida K, Nishii R, Hirayama T (2020) Decoding plant–environment interactions that influence crop agronomic traits. Plant Cell Physiol 61(8):1408–1418. https://doi.org/10.1093/pcp/pcaa064
Mugode L, Ha B, Kaunda A, Sikombe T, Phiri S et al (2014) Carotenoid retention of biofortified provitamin a maize (Zea mays L.) after Zambian traditional methods of milling, cooking and storage. J Agric Food Chem 62(27):6317–6325. https://doi.org/10.1021/jf501233f
Nester EW (2015) Agrobacterium: Nature’s genetic engineer. Front Plant Sci 5(JAN):1–16. https://doi.org/10.3389/fpls.2014.00730
O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M et al (2014) Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516(7530):263–266. https://doi.org/10.1038/nature13769
Orlowski J, Boniecki M, Bujnicki JM (2007) I-Ssp6803I: the first homing endonuclease from the PD-(D/E)XK superfamily exhibits an unusual mode of DNA recognition. Bioinformatics 23(5):527–530. https://doi.org/10.1093/bioinformatics/btm007
Petolino JF (2015) Genome editing in plants via designed zinc finger nucleases. In Vitr Cell Dev Biol Plant 51(1):1–8
Ranum P, Peña-Rosas JP, Garcia-Casal MN (2014) Global maize production, utilization, and consumption. Ann N Y Acad Sci 1312(1):105–112
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop Production by 2050. PLoS One 8(6):e66428. https://doi.org/10.1371/journal.pone.0066428
Raza A, Razzaq A, Mehmood SS, Zou X, Zhang X et al (2019) Impact of climate change on crops adaptation and strategies to tackle its outcome: a review. Plan Theory 8(2). https://doi.org/10.3390/plants8020034
Razzaq A, Saleem F, Kanwal M, Mustafa G, Yousaf S, Imran Arshad HM et al (2019) Modern trends in plant genome editing: an inclusive review of the CRISPR/Cas9 toolbox. Int J Mol Sci 20(16):4045
Razzaq A, Kaur P, Akhter N, Wani SH, Saleem F (2021a) Next-generation breeding strategies for climate-ready crops. Front Plant Sci 12
Razzaq A, Mustafa G, Ali MA, Khan MS, Joyia FA (2021b) 23 CRISPR-mediated genome editing in maize for improved abiotic stress tolerance. In: Molecular breeding in wheat, maize and sorghum: strategies for improving abiotic stress tolerance and yield. CAB International, p 405
Razzaq A, Saleem F, Wani SH, Abdelmohsen SA, Alyousef HA, Abdelbacki AM et al (2021c) De-novo domestication for improving salt tolerance in crops. Front Plant Sci 1623
Razzaq A, Wani SH, Saleem F, Yu M, Zhou M, Shabala S (2021d) Rewilding crops for climate resilience: economic analysis and de novo domestication strategies. J Exp Bot 72(18):6123–6139
Sanaullah AB, Khan A, Rahman WU, Nasrullah A, Khan MAR et al (2018) Estimating cost and net return: a profitability comparison of maize and potato in district upper Dir of Khyber Pakhtunkhwa, Pakistan. Econ Surv:19
Sandhya D, Jogam P, Allini VR, Abbagani S, Alok A (2020) The present and potential future methods for delivering CRISPR/Cas9 components in plants. J Genet Eng Biotechnol 18(1):25. https://doi.org/10.1186/s43141-020-00036-8
Schiml S, Fauser F, Puchta H (2014) The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J 80(6):1139–1150. https://doi.org/10.1111/tpj.12704
Schwartz C, Lenderts B, Feigenbutz L, Barone P, Llaca V, Fengler K, Svitashev S (2020) CRISPR-Cas9-mediated 75.5-Mb inversion in maize. Nat Plants 6(12):1427–1431. https://doi.org/10.1038/s41477-020-00817-6
Sedeek KEM, Mahas A, Mahfouz M (2019) Plant genome engineering for targeted improvement of crop traits. Front Plant Sci 10(February):1–16. https://doi.org/10.3389/fpls.2019.00114
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL et al (2017) ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J 15(2):207–216
Sprink T, Metje J, Hartung F (2015) Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr Opin Biotechnol 32:47–53
Tan YY, Du H, Wu X, Liu YH, Jiang M et al (2020) Gene editing: an instrument for practical application of gene biology to plant breeding. J Zhejiang Univ Sci B 21(6):460–473. https://doi.org/10.1631/jzus.B1900633
Tang Y, Fu Y (2018) Class 2 CRISPR/Cas: an expanding biotechnology toolbox for and beyond genome editing 06 biological sciences 0604 genetics. Cell Biosci 8(1):1–13. https://doi.org/10.1186/s13578-018-0255-x
Ten Berge HFM, Hijbeek R, van Loon MP, Rurinda J, Tesfaye K (2019) Maize crop nutrient input requirements for food security in sub-Saharan Africa. Glob Food Sec 23:9–21
Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML et al (2009) High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459(7245):442–445
Tsanova T, Stefanova L, Topalova L, Atanasov A, Pantchev I (2021) DNA-free gene editing in plants: a brief overview. Biotechnol Biotechnol Equip 35(1):131–138. https://doi.org/10.1080/13102818.2020.1858159
Tshikunde NM, Mashilo J, Shimelis H, Odindo A (2019) Agronomic and physiological traits, and associated quantitative trait loci (QTL) affecting yield response in wheat (Triticum aestivum L.): a review. Front Plant Sci 10(November):1–18. https://doi.org/10.3389/fpls.2019.01428
Wani SH, Samantara K, Razzaq A, Kakani G, Kumar P (2022) Back to the wild: mining maize (Zea mays L.) disease resistance using advanced breeding tools. Mol Biol Rep:1–17
Weinthal DM, Taylor RA, Tzfira T (2013) Nonhomologous end joining-mediated gene replacement in plant cells. Plant Physiol 162(1):390–400. https://doi.org/10.1104/pp.112.212910
Yadava P, Abhishek A, Singh R, Singh I, Kaul T et al (2017) Advances in maize transformation technologies and development of transgenic maize. Front Plant Sci 7:1949
Yang CJ, Samayoa LF, Bradbury PJ, Olukolu BA, Xue W et al (2019) The genetic architecture of teosinte catalyzed and constrained maize domestication. Proc Natl Acad Sci U S A 116(12):5643–5652. https://doi.org/10.1073/pnas.1820997116
Zhang M, Cao Y, Wang Z, Wang Z, Shi J et al (2018) A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol 217(3):1161–1176
Zhang D, Zhang Z, Unver T, Zhang B (2021) CRISPR/Cas: a powerful tool for gene function study and crop improvement. J Adv Res 29:207–221
Zhao L, Bonocora RP, Shub DA, Stoddard BL (2007) The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J 26(9):2432–2442. https://doi.org/10.1038/sj.emboj.7601672
Zong Y, Wang Y, Li C, Zhang R, Chen K et al (2017) Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35(5):438–440. https://doi.org/10.1038/nbt.3811
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Farooq, S., Shahzadi, A., Razzaq, A., Saleem, F., Wani, S.H., Sandhu, K. (2023). Advances in Genome Editing for Maize Improvement. In: Wani, S.H., Dar, Z.A., Singh, G.P. (eds) Maize Improvement. Springer, Cham. https://doi.org/10.1007/978-3-031-21640-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-21640-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-21639-8
Online ISBN: 978-3-031-21640-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)