Abstract
Link prediction is a fundamental problem in the field of graph mining. The aim of link prediction is to infer/discover unobserved links in graphs. Link prediction in biological graphs is highly challenging. There exist many similarity-based methods in the literature for link prediction. These methods compete for victory in graphs from various domains. Unfortunately, they are efficient only in some specific graphs, and no one wins in all graphs. In this paper, we study some well-known similarity-based methods and consider them as independent features to define a feature set. The feature set is then used to train traditional supervised learning methods for link prediction in biological graphs. We evaluate the methods on ten biological graphs from different organisms. Experimental results show that the similarity-based methods collaboratively improve prediction performance, and are even comparable to high-performing embedding-based methods in some biological graphs. We compute the importance score of similarity-based features in order to explain the leading features in a graph.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Many complex biological systems can be well-represented with graphs where a node represents a biological entity (e.g. protein, gene, etc.) and a link represents the interaction between two entities. Most real-world biological graphs are incomplete in nature. For example, 99.7% of the molecular interactions in human cells are still not known [1]. The links in biological graphs must be validated by field and/or laboratory experiments, which are expensive and time consuming. Researchers have developed link prediction methods to compute the plausibility of a link between two unconnected nodes in a graph to avoid the blind checking of all possible interactions. Formally, link prediction is the task of predicting the likelihood of a link between two nodes based on available topological/attribute information of a graph [2]. Link prediction methods help us toward a deep understanding of the structure, evolution, and functions of biological graphs [3].
Similarity-based methods are the simplest and unsupervised methods of link prediction in biological graphs, which define the proximity of a link by the similarity between its end nodes. The great advantage of these methods is their interpretibility which is essential for any biological system [4]. However, each of the similarity-based methods performs well only in some particular graphs and no one wins in all graphs. These methods necessitate manually formulating various heuristics based on prior beliefs or extensive knowledge of various biological graphs. The lack of universal applicability of similarity-based methods motivates researchers to study machine learning methods to automatically learn the heuristics from a graph. To learn the appropriate heuristics automatically from a graph, researchers have developed embedding-based methods which represent nodes, edges, graphs in low dimensional vector space [5]. The embedding-based method has become a popular link prediction tool in graphs over the last decade. These methods show impressive link prediction performance in most of the graphs. The downside of embedding-based methods is that they seriously suffer from the well-known ‘black-box’ problem. As the link decisions in biological graphs are critical, a link prediction method should be sufficiently interpretable to achieve trust among stakeholders [6]. The requirement for link prediction methods to be interpretable may limit the use of embedding-based methods in real-world biological systems. Researchers are still working on opening the ‘black-box’ of embedding-based methods [9, 10].
Another group of link prediction methods is developed based on traditional supervised learning-based methods. These methods extract features from a graph and train a traditional classifier for the link prediction task [11,12,13,14,15,16,17]. These methods are nearly as performant as embedding-based methods and as interpretable as similarity-based methods in many biological graphs. These methods describe the link prediction problem as a link classification problem with two classes: existence and absence of a link. In this paper, we intend to investigate whether the existing similarity-based heuristics collaboratively improve the link prediction performance in biological graphs. We study similarity-based heuristics for feature extraction and utilize the features in supervised learning-based classifiers for link prediction in biological graphs. We find that this is not the first attempt to study supervised learning methods to link prediction problem in graphs. But there are important differences between past works [12, 18, 19] and this study. The existing methods mostly focus on node attributes for extracting features which are application dependent. However, node attributes are not available in many real-world biological graphs. In contrast, our supervised learning-based method is developed based on only the topological features (similarity-based heuristics). Kumari et al. [17] studied a few local (four) and global (three) similarity heuristics for supervised link predictions, which is the closest work in the literature to our study. However, for large graphs, global methods are not the best option as they are computationally expensive [20]. In this study, we enrich the feature set by including fourteen local similarity-based heuristics. In addition, we extract few other topological features of nodes and derive link-based features based on end node features. We study these features in supervised machine learning methods for link prediction in biological graphs. We see that supervised learning methods show comparable prediction results in many of the biological graphs. We also demonstrate the feature importance in different datasets for different supervised learning-based methods.
1.1 Similarity-Based Link Prediction
Link prediction is the task of discovering or inferring a set of non-existing links in a graph based on the current snapshot of the graph. Similarity-based is the simplest category of link prediction methods, which is formulated based on the assumption that two nodes interact if they are similar in a graph [20]. Generally, these methods compute similarity scores of non-existent links, sort the links in decreasing order of their scores and top-L links are predicted as potential existent links. Defining the similarity is a crucial and non-trivial task which differs from graphs to graphs [20]. Consequently, numerous similarity-based methods exist in the literature. These methods are broadly categorized into three categories: local, global and quasi-local methods. Local methods are developed based on local topological or neighbourhood information, whereas global methods use the global topological information of graphs to define similarity functions [20]. Quasi-local methods consider the neighbourhood up-to a predefined hop for defining the similarity function. The high computational time of global methods motivates us to study only local and quasi-local methods. We study fourteen well-known local similarity-based methods for link prediction in graphs, thirteen of which are summarized in Table 1 local and one quasi-local. We summarize the similarity-based methods and the rest one (Preferential Attachment (PA)) in Table 2 with basic principles and the definition of similarity functions.
2 Methodology
In a broader sense, we consider the similarity-based heuristics as individual features to generate the feature set for a supervised learning-based classifier.
We describe each of the steps in Sects. 2.1–2.3.
2.1 Feature Extraction
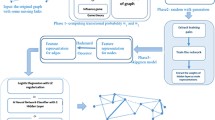
The most crucial task of a supervised learning-based classifier is to define an appropriate feature set [12]. Given a graph and a train set of links, we extract structural features for the train links. When extracting the features of a link, the link is temporarily removed from the graph and re-connected after feature extraction to ensure that the extracted features are not biased by the existence of the train link. We are motivated to use only topological features for defining our feature set as they exist in all kinds of graphs. Our feature set contains twenty topological features which are broadly categorized into two categories: similarity-based and derived link features (Fig. 1).
Similarity-Based Link Features: We define the link-based features as the features which are related to the common topological information of end-nodes of a link. We use thirteen existing similarity-based heuristics as link-based features, which are summarized in Table 1. For instance, the number of common neighbours of end nodes of a link is used as the common neighbour (CN) feature.
Derived Link Features: Few link-based features are derived from the individual features of the link’s end nodes. We summarized six derived features in Table 2. These features are related to the topological information of individual nodes only. For example, the degree of end nodes is multiplied in Preferential Attachment (PA) to define the similarity score. Note that the link features in Table 2 except PA are not directly defined in the literature. We derive the link features based on the end node feature. To compute the link feature, features of end nodes are simply added except PA. As the voterank centrality computes low ranks for high-influencing nodes in a graph, the reciprocals of the voterank scores of end nodes are summed to define the voterank centrality feature.
2.2 Feature Scaling
In general, the magnitude scale for different features in different graphs varies [7, 8]. Supervised learning-based methods are easily affected by the non-uniform scaling as there is a high chance that features with higher magnitude play a more decisive role during the training of a classifier. But, it is not desirable for the classifier to be biased towards one particular feature. Hence, we normalize each feature in the range of 0–1.
2.3 Classifier Training and Link Prediction
For the link prediction task, we train a traditional supervised machine learning classifier to classify a link into either existent or non-existent classes. There exist many classifiers in the literature which perform better than others in some particular datasets. In this paper, we study three traditional classifiers: Support Vector Machine (SVM) with RBF kernel, Decision Tree, and Logistic Regression. We extract the features of the test links and classify them into existent or non-existent classes using a trained classifier to evaluate the link prediction performance.
3 Experiments
3.1 The Baselines
To evaluate the prediction performance of supervised learning methods, we consider two categories of link prediction methods: similarity-based and embedding-based methods.
For the similarity-based category, we consider all the heuristics in Table 1 in Table 2. For the embedding-based methods, we choose two popular methods: Node2Vec [40] and SEAL [41]. We shortly describe Node2Vec and SEAL methods. For more details, we refer to the original papers. Node2Vec [40] is a classical skip-gram model-based graph embedding method which learns node embeddings by optimizing a neighbourhood preserving objective function. It makes an interpolation between BFS (Breadth First Search) and DFS (Depth First Search) to define a 2\(^{nd}\) order random walk. A fixed size neighbourhood is sampled using the 2\(^{nd}\) order random walk and fed into the well-known skip-gram model [42] to learn the node embedding. The link embedding is then computed as the Hadamard product of the end node embeddings. A logistic regression-based classifier is then trained for the link prediction task. SEAL, the second embedding-based approach, is based on neural networks (NN). Learning from Sub-graphs, Embeddings and Attributes (SEAL) utilizes the latent and explicit features of end nodes and structural information of the graph to learn the link embedding. SEAL starts with extracting a h-hop neighbouring sub-graph and node labeling by a double radius node labeling (DRNL) algorithm. In the second step, the labelled sub-graph is then used to generate the structural encoding. The link embedding is the concatenation of structural encoding, pre-computed latent encoding and explicit feature encoding. In the final step, a neural network (NN) is trained for link prediction task.
3.2 Experimental Datasets
In this study, we focus on only biological graphs. For evaluating performance, we collect six biological graphs from the Network RepositoryFootnote 1. Table 3 summarizes the topological statistics and descriptions of the graph datasets.
The link prediction performance is evaluated using a random sampling validation protocol [7, 8, 41]. For a graph dataset, train and test sets are prepared by splitting the existent links. The train set consists of 90% existent and an equal number of non-existent links. The test set contains the remaining 10% existent and equal number of non-existent links. To prepare five train and five test sets for each graph, we repeat the link splitting operation five times independently. The datasets are available in a GitLab repositoryFootnote 2.
3.3 Evaluation Metrics
The link prediction problem is considered as a binary classification problem [46]. A traditional classifier, in general, learns a threshold to classify links as existent or non-existent. However, for similarity-based link classification methods, we find no standard approach for computing the threshold. The threshold is calculated in an optimistic manner. We first normalize the link scores to a range of 0–1 and then use the normalized scores to compute a ROC curve. The curve gives the true positiverate (TPR) and false positive rate (FPR) for different score threshold settings. The threshold point with the highest [TPR + (1 − FPR)] is computed as the threshold as we want to maximize TPR as well as minimize FPR. We classify links based on this threshold. A link with a \(score>=threshold\) is classified as existent and non-existent otherwise. Based on the true and predicted classes of links, we define four metrics: true positive (TP), true negative (TN), false positive (FP), and false negative (FN). TP is the number of existent links predicted to be existent, TN is the number of non-existent links predicted to be non-existent, FP is the number of non-existent links predicted to be existent, and FN is the number of existent links predicted to be non-existent links. We compute the following three well-known metrics using these four metrics.
3.4 Results and Discussion
In this section, we describe the prediction performance of supervised learning-based methods on six biological graphs. We also illustrate the importance of the features in graphs.
Prediction Performance: The prediction performance is computed for all methods over all the five sets for each graph, and the average scores are recorded. We do not include the standard deviation results as the values are very low in all the experiments. The precision, recall and F1 scores are tabulated Table 4, where the best two similarity-based methods are denoted with \(Sim^1\) and \(Sim^2\). We compute the precision scores of similarity-based methods in a optimistic way. The precision scores of similarity-based methods (best and second best) are very high and highest among all the methods in all the graphs, as shown in the table. This demonstrates the ability of similarity-based methods to predict high-quality links. However, the recall scores are low, implying that these methods identify the majority of existing test links as non-existent. As a result, the F1 score for similarity-based methods is very low. We also see that, as expected, the two best-performing similarity-based methods differ for different datasets. Among the supervised learning methods (SVM, DT, LR), DT shows the worst prediction results, but it is still much better than similarity-based methods. The other two classifiers have similar performance scores. The performance of the other two classifiers in terms of prediction scores is impressive. Yet in many graphs, supervised learning-based classifiers show superior prediction performance than embedding-based methods. Relating the performance to graph properties, we see that traditional classifiers outperform embedding-based methods in dense graphs. This is intuitive as the majority of the studied similarity-based heuristics are based on common neighbours (see Table 1). The performance scores of traditional classifiers are worse in the sparse graphs (CE-HT, CE-LC, Yeast), where embedding-based methods show better performance scores.
Feature Importance: In this section, we investigate the influence of each feature in a classifier for the link prediction task. To compute the feature importance coefficient, we use the Permutation importance module from the sklearn python-based machine learning toolFootnote 3. When a feature is unavailable, the coefficient is calculated by looking at how much the score (accuracy) drops [47]. The higher the coefficient, the higher the importance of the feature. In Fig. 2, we demonstrate the feature importance in the HS-HT biological graph in the logistic regression (LR) classifier to investigate how the importance of features differs in different sets of the same biological graph. In the LR classifier for the HS-HT biological graph, four features dominate. The dominance of multiple heuristics or features in a graph shows that heuristics that work collaboratively perform better than heuristics that work alone. We can also find that the feature importance coefficients in all five sets in the HS-HT graph are substantially identical.
We further investigate the importance score of features in three classifiers (SVM, DT, LR) for three different datasets. We evaluate the importance score of features for only one set for each graph. We see that different classifiers give different importance coefficients to different features in different datasets. In DM-HT dataset, all the classifiers compute high coefficient for LPI feature and they have close prediction performance (in Table 4). In the Celegans dataset, the HPI feature dominates in SVM and LR classifiers whereas LPI dominates in the DT classifier. In the Celegans dataset, SVM and LR outperform DT in terms of prediction (in Table 4), demonstrating that LR and SVM compute feature importance scores more correctly. Surprisingly, we see that DT has a tendency to give more importance to the LPI feature in these three datasets (Fig. 3).
4 Conclusion
Do similarity-based heuristics compete or collaborate for link prediction task in graphs? In this article, we study this question. We study fourteen similarity-based heuristics in six biological graph from three different organisms. As expected, we observe they perform well only in some particular biological graphs and no one wins in all graphs. Rather than using them as standalone link prediction methods, we consider them as features for supervised learning methods. In addition, we derive six link features based on the node’s topological information. Based on the twenty features, we train three traditional supervised learning methods: SVM, DT and LR-based classifiers. We see that the similarity-based heuristics collaboratively improve link prediction performance remarkably, even outperforming embedding-based methods in some graphs.
We propose three future dimensions of this study. Firstly, studying collaboration of similarity-based heuristics in large scale biological as well as social graphs could be a potential future work as the graphs in the current study are small/medium in size. Secondly, exploring some other heuristics might improve prediction performance in sparse graphs. The final future research could be studying other classifiers like Random Forest, AdaBoost, K-Neighbors for the link prediction task in graphs.
References
Stumpf, M.P., et al.: Estimating the size of the human interactome. Proc. Natl. Acad. Sci. 105(19), 6959–6964 (2008)
Xu, Z., Pu, C., Yang, J.: Link prediction based on path entropy. Phys. A 456, 294–301 (2016)
Shen, Z., Wang, W.X., Fan, Y., Di, Z., Lai, Y.C.: Reconstructing propagation networks with natural diversity and identifying hidden sources. Nat. Commun. 5(1), 1–10 (2014)
Zhou, T., Lee, Y.L., Wang, G.: Experimental analyses on 2-hop-based and 3-hop-based link prediction algorithms. Phys. A Stat. Mech. Appl. 564, 125532 (2021)
Cui, P., Wang, X., Pei, J., Zhu, W.: A survey on network embedding. IEEE Trans. Knowl. Data Eng. 31(5), 833–852 (2018)
Gerke, S., Minssen, T., Cohen, G.: Ethical and legal challenges of artificial intelligence-driven healthcare. In: Artificial Intelligence in Healthcare, pp. 295–336. Academic Press (2020)
Islam, M.K., Aridhi, S., Smail-Tabbone, M.: Appraisal study of similarity-based and embedding-based link prediction methods on graphs. In: Proceedings of the 10th International Conference on Data Mining & Knowledge Management Process, pp. 81–92 (2021)
Islam, M.K., Aridhi, S., Smaïl-Tabbone, M.: An experimental evaluation of similarity-based and embedding-based link prediction methods on graphs. Int. J. Data Min. Knowl. Manag. Process 11, 1–18 (2021)
Faber, L., Moghaddam, A.K., Wattenhofer, R.: Contrastive graph neural network explanation. In: Proceedings of the 37th Graph Representation Learning and Beyond Workshop at ICML 2020, p. 28. International Conference on Machine Learning (2020)
Yuan, H., Yu, H., Wang, J., Li, K., Ji, S.: On explainability of graph neural networks via subgraph explorations. In: Proceedings of the 38th International Conference on Machine Learning (2021)
Cukierski, W., Hamner, B., Yang, B.: Graph-based features for supervised link prediction. In: The 2011 International Joint Conference on Neural Networks, pp. 1237–1244. IEEE, July 2011
Al Hasan, M., Chaoji, V., Salem, S., Zaki, M.: Link prediction using supervised learning. In: SDM 2006: Workshop on Link Analysis, Counter-Terrorism and Security, vol. 30, pp. 798–805, April 2006
Berton, L., Valverde-Rebaza, J., de Andrade Lopes, A.: Link prediction in graph construction for supervised and semi-supervised learning. In: 2015 International Joint Conference on Neural Networks (IJCNN), pp. 1–8. IEEE, July 2015
Benchettara, N., Kanawati, R., Rouveirol, C.: A supervised machine learning link prediction approach for academic collaboration recommendation. In: Proceedings of the Fourth ACM Conference on Recommender Systems, pp. 253–256, September 2010
Ahmed, C., ElKorany, A., Bahgat, R.: A supervised learning approach to link prediction in Twitter. Soc. Netw. Anal. Min. 6(1), 1–11 (2016). https://doi.org/10.1007/s13278-016-0333-1
Shibata, N., Kajikawa, Y., Sakata, I.: Link prediction in citation networks. J. Am. Soc. Inform. Sci. Technol. 63(1), 78–85 (2012)
Kumari, A., Behera, R.K., Sahoo, K.S., Nayyar, A., Kumar Luhach, A., Prakash Sahoo, S.: Supervised link prediction using structured-based feature extraction in social network. Concurr. Comput. Pract. Exp. 34(13), e5839 (2020)
Liben-Nowell, D., Kleinberg, J.: The link-prediction problem for social networks. J. Am. Soc. Inform. Sci. Technol. 58(7), 1019–1031 (2007)
De Sá, H.R., Prudêncio, R.B.: Supervised link prediction in weighted networks. In: The 2011 International Joint Conference on Neural Networks, pp. 2281–2288. IEEE, July 2011
Martínez, V., Berzal, F., Cubero, J.C.: A survey of link prediction in complex networks. ACM Comput. Surv. (CSUR) 49(4), 1–33 (2016)
Lorrain, F., White, H.C.: Structural equivalence of individuals in social networks. J. Math. Sociol. 1(1), 49–80 (1971)
Adamic, L.A., Adar, E.: Friends and neighbors on the web. Soc. Netw. 25(3), 211–230 (2003)
Zhou, T., Lü, L., Zhang, Y.C.: Predicting missing links via local information. Eur. Phys. J. B 71(4), 623–630 (2009). https://doi.org/10.1140/epjb/e2009-00335-8
Jaccard, P.: Étude comparative de la distribution florale dans une portion des Alpes et des Jura. Bull Soc. Vaudoise. Sci. Nat. 37, 547–579 (1901)
Salton, G., McGill, M.J.: Introduction to Modern Information Retrieval. McGraw-Hill (1983)
Sorensen, T.A.: A method of establishing groups of equal amplitude in plant sociology based on similarity of species content and its application to analyses of the vegetation on Danish commons. Biol. Skar. 5, 1–34 (1948)
Ravasz, E., Somera, A.L., Mongru, D.A., Oltvai, Z.N., Barabási, A.L.: Hierarchical organization of modularity in metabolic networks. Science 297(5586), 1551–1555 (2002)
Leicht, E.A., Holme, P., Newman, M.E.: Vertex similarity in networks. Phys. Rev. E 73(2), 026120 (2006)
Cannistraci, C.V., Alanis-Lobato, G., Ravasi, T.: From link-prediction in brain connectomes and protein interactomes to the local-community-paradigm in complex networks. Sci. Rep. 3(1), 1–14 (2013)
Wu, Z., Lin, Y., Wang, J., Gregory, S.: Link prediction with node clustering coefficient. Phys. A 452, 1–8 (2016)
Wu, Z., Lin, Y., Wan, H., Jamil, W.: Predicting top-L missing links with node and link clustering information in large-scale networks. J. Stat. Mech: Theory Exp. 2016(8), 083202 (2016)
Lü, L., Jin, C.H., Zhou, T.: Similarity index based on local paths for link prediction of complex networks. Phys. Rev. E 80(4), 046122 (2009)
Barabási, A.L., Albert, R.: Emergence of scaling in random networks. Science 286(5439), 509–512 (1999)
Langville, A.N., Meyer, C.D.: A survey of eigenvector methods for web information retrieval. SIAM Rev. 47(1), 135–161 (2005)
Onnela, J.P., Saramäki, J., Kertész, J., Kaski, K.: Intensity and coherence of motifs in weighted complex networks. Phys. Rev. E 71(6), 065103 (2005)
Dubitzky, W., Wolkenhauer, O., Cho, K.H., Yokota, H. (eds.): Encyclopedia of Systems Biology, vol. 402. Springer, New York (2013). https://doi.org/10.1007/978-1-4419-9863-7
Bonacich, P.: Power and centrality: a family of measures. Am. J. Sociol. 92(5), 1170–1182 (1987)
Freeman, L.: Centrality in networks: I. Conceptual clarifications. Soc. Netw. 1(3), 215–239 (1979)
Zhang, J.X., Chen, D.B., Dong, Q., Zhao, Z.D.: Identifying a set of influential spreaders in complex networks. Sci. Rep. 6, 27823 (2016)
Grover, A., Leskovec, J.: node2vec: scalable feature learning for networks. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 855–864, August 2016
Zhang, M., Chen, Y.: Link prediction based on graph neural networks. Adv. Neural. Inf. Process. Syst. 31, 5165–5175 (2018)
Mikolov, T., Chen, K., Corrado, G., Dean, J.: Efficient estimation of word representations in vector space. In: International Conference on Learning Representations (2013)
Duch, J., Arenas, A.C.: Community identification using extremal optimization. Phys. Rev. E 72, 027104 (2005)
Cho, A., et al.: WormNet v3: a network-assisted hypothesis-generating server for Caenorhabditis elegans. Nucleic Acids Res. 42(W1), W76–W82 (2014)
Goh, K.I., Cusick, M.E., Valle, D., Childs, B., Vidal, M., Barabási, A.L.: The human disease network. Proc. Natl. Acad. Sci. 104(21), 8685–8690 (2007)
Kumar, A., Singh, S.S., Singh, K., Biswas, B.: Link prediction techniques, applications, and performance: a survey. Phys. A 553, 124289 (2020)
Breiman, L.: Random forests. Mach. Learn. 45(1), 5–32 (2001). https://doi.org/10.1023/A:1010933404324
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Islam, M.K., Aridhi, S., Smail-Tabbone, M. (2022). From Competition to Collaboration: Ensembling Similarity-Based Heuristics for Supervised Link Prediction in Biological Graphs. In: Islam, A.K.M.M., Uddin, J., Mansoor, N., Rahman, S., Al Masud, S.M.R. (eds) Bangabandhu and Digital Bangladesh. ICBBDB 2021. Communications in Computer and Information Science, vol 1550. Springer, Cham. https://doi.org/10.1007/978-3-031-17181-9_10
Download citation
DOI: https://doi.org/10.1007/978-3-031-17181-9_10
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-17180-2
Online ISBN: 978-3-031-17181-9
eBook Packages: Computer ScienceComputer Science (R0)