Abstract
Genomics of cancer plays a very important role in the detection of tumour type, Treatment, involves analysis of genes that are causing cancer. DNA Microarray is used to measure the gene expression of particular tissue or cell. Data generated from DNA Microarrays is large. Machine learning can be well suited for genomics data. We used different cancer gene expression datasets such as leukaemia, Lung, Bladder, prostate, Liver and pancreas and used various Machine learning algorithms such as Decision tree, Naïve Bayes, Logistic Regression and SVM for classification of cancer types. We proposed a new classification method for gene expression dataset which can apply to any numeric dataset known as “Mean based Classification”. Mean based classification considers the mean for each class and selects the features from datasets having maximum mean difference. Classification is given by minimum distance which is similar to Rocchio classification used for text classification. Mean based classification and its variant which uses standard deviation and mean performed well and obtained high accuracy for all cancer genomics datasets. Explainable AI methods like shapely value explanations are also used for explanations of results and to understand interactions among genes and also to understand which genes are causing cancer.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Cancer is an important cause of death worldwide. Cancer is excessive, abnormal, uncontrolled growth of cells losing normal function. Cancer occurs when the mechanism that maintains normal growth rates malfunction to cause excess cell division.
Digital pathology has revolutionized the field of pathology. Digital pathology involves histopathology i.e., viewing analysing histopathological slides digitally, Cancer genomics i.e., Analysis of abnormal expression of genes [1] Various machine learning methods can be applied to the approaches mentioned above.
Deep learning for histopathological whole slides has already succussed in the detection of various cancers [2]. Deep learning approach finds intricate features of the histopathological image and uses it for classification of cancer type. Cancer classification using histopathological images has limitations as cancers having similar morphological features might be having different molecular origin therefore, cancer therapy also differs.
Cancer genomics is the study of gene expression differences between tumour cells and normal cells, it deals with understanding the genetic basis of tumour cell proliferation, for example, Tumour suppressor genes normally restrain growth so mutations that inactivate them cause inappropriate cell division. Mutations in the tumour suppressor genes BRCA1 and BRCA2 have been linked to a much higher risk of breast, ovarian and prostate cancer.
Caretaker genes normally protect the integrity of genomes when they are inactivated cell acquires additional mutations that cause cancer. HER2 positive breast cancers involve a mutated HER2 Oncogene, which produces a protein that increases the growth of cells.
The most pathologist uses histopathological slides to classify cancer, however, in certain cases the only morphological analysis is not enough as cancer showing similar morphological origins might differ in genetical origin so cancer treatment also differs. Genomic study in such cases is important to find the root cause of cancer and to provide treatment. Cancer-causing genetic mutations can be discovered and to develop potential treatment [3]. For example, in chronic lymphocytic leukaemia, the presence of a mutation in the TP53 gene means that cancer won’t respond to chemoimmunotherapy in such cases stem cell transplant might be needed. The potential personalized treatment plan for cancer by detecting mutations in the respective individual can be obtained through machine learning.
Machine learning can handle a massive amount of data. Cancer genomics contains a large amount of data on gene expression. Machine learning methods can be effectively applied to the Gene expression dataset, by applying machine learning on gene expression classification of tumour can be done [4].
Machine learning in genetic data possess a challenge as genetic data has many features also extracted genes from microarray has noise. Machine learning on DNA microarray data can be divided into four categories as Classification, Clustering, Gene identification, Gene network modelling [5]. Generally, Machine learning for DNA microarray data involves extraction of microarray data, Feature selection, Classification or clustering. Various feature selection techniques are partial least squares [6], PCA, Akaike information criterion and Bayesian information criterion [7] genetic algorithm [8] . We used the proposed method “mean based classification” for feature selection and classification.
Machine learning is a vital research tool to gain insights into cancer genomics. Interpretable AI methods can be applied to the cancer genomics problem, Genes that are most responsible for causing cancer can be studied in detail Further, how different genes interact with each other, a correlation between genes can be studied which is vital for bioinformatics research [9]. A variety of machine learning methods are well suited for genomics data.
DNA microarray is used to deduce gene expression [10]. DNA microarrays are used to analyse the genes which are expressed in tissue or cell. Mutations in genes lead to cancer, DNA microarray is a tool to detect which genes are mutating and causing cancer. DNA microarray works as follows. Figure 1 outlines the working of DNA microarray. The patients’ tissue sample which is having a tumour is taken and DNA is cut into small pieces known as fragments which represent genes. The control sample which is having normal tissue is taken and it is cut into fragments. Both tissue samples are labelled with a fluorescent dye, Patients sample having red dye and the normal sample having green dye. Both sets of genes are inserted in the microarray chip and allowed to hybridize. Genes having mutation doesn’t hybridize well to the normal sample which can be detected in a microarray chip.
Machine learning methods such as SVM, Decision tree, naïve Bayes, Logistic regression are used for classification in the dataset used for this article. We proposed and used a novel classification approach known as “mean based classification” which considers the mean value of a specific feature for two classes present in the dataset. Here, all cancer gene expression datasets contain two class labels. Mean based classification algorithm takes consideration of the mean difference in classes. The main reason behind considering mean difference is when mean difference is maximum, feature shows distinct signature or range for different classes hence could be useful for classification. Classification in mean-based classification is done by finding difference from mean for respective classes and considering minimum among them, the class label will be assigned to the class which is having a minimum distance between the class mean and feature value which is similar to Rocchio classification [11]. We also proposed variant for mean based classification which uses standard deviation. Standard deviation for each class for a feature is calculated. Two values for each standard deviation is added known as sum of standard deviation. Algorithm aims to find feature which has maximum mean difference and minimum sum of standard deviation. Finding minimum of sum of standard deviation basically means that distributions for two classes are closer to mean and have less chance of overlapping values.
Explainable AI also has great potential in the field of genomics. The understanding black box of a particular model is essential as it could shed light on many important research questions such as which genes are contributing most to particular cancer, how different genes are interacting to affect the model’s behaviour. Shapely value-based explainable ai methods are used.
2 Proposed Approach
2.1 Overview
The overview of the proposed method used in this article is shown in Fig. 2. Gene expression microarray data is used for classification. Classification methods such as SVM, Naïve Bayes, Decision tree, logistic regression and novel centroid classification is used for classification. Interpretation methods such as shapely value explanations are used to understand genes that are most important for the classification of cancer type.
2.2 Machine Learning Algorithms
Naïve bayes: Naïve bayes is supervised learning method. It is based on Bayes (Eq. 1). P(C|x) is the probability of instance x belongs to class c which is known as posterior probability. P(x|C) is the probability of having instance x given class c known as likelihood. P (C) is a number of instances of class C known as prior.
P(C│x)=(P(x│C)P(C))/(P(x))
Equation 1 – Bayes Theorem
Naïve Bayes algorithm assumes that the presence of a particular feature in a class is unrelated to the presence of any other feature [12].
Decision Tree:
Decision Tree uses a tree-like structure. Each node in the Decision tree indicates a test at that node, each branch represents an outcome of the test. A terminal node in the decision tree indicates the class label.
A tree can be “learned” by splitting the source set into subsets based on an attribute value test. This process is repeated on each derived subset in a recursive manner called recursive partitioning. The recursion is completed when the subset at a node all has the same value of the target variable, or when splitting no longer adds value to the predictions. The construction of a decision tree classifier does not require any domain knowledge or parameter setting, and therefore is appropriate for exploratory knowledge discovery. Entropy is used to measure homogeneity. If data is completely homogeneous then the entropy is 0, else if data is divided 50-50 entropy is 1.
Information gain is used for the splitting of the decision tree.
SVM
Support vector machine algorithm is linear classification. If some data points are given and the task is to classify them. Linear classifier approaches this problem by finding a hyperplane that divides the classes. However, there exists a set of hyperplanes that can classify the data. SVM not only finds a hyperplane that divides the classes but find the best hyperplane which is having a maximum margin from the classes, It takes the help of a support vector to achieve this [13].
Logistic regression
Logistic regression is an asymmetric statistical model used for mapping numerical value to probability. Logistic regression uses a logistic function to model a binary dependent model. The binary logistic model has two possible values. In binary logistic regression model, the dependent variable has two levels. Log odds are an alternate way of expressing probabilities. Odds are the ratio of something happening to something not happening [14].
3 Mean Based Classification
Shows the distribution for each class i.e., class 1 and class 2. For each class mean is calculated i.e., mean 1 and mean 2 respectively. Mean difference is the absolute difference between two means. The Mean based classification algorithm aims to find out a feature which is having a maximum mean difference.
Mean based classification is based on average, here mean is an average value for a particular feature. Mean based classification is an easily interpretable algorithm.
The best features are those which vary by different classes. Mean based classification selects the features which are varying across classes. The detailed procedure is shown in Fig. 4. In the first step i.e. Feature selection phase For the respective feature calculate average for respective classes i.e. if dataset is having two classes then for each class average for respective feature will be calculated so two averages per feature will be generated.
In the next step, the Difference between Averages is calculated for each feature. The feature having maximum difference is varying across classes thus it is most suitable for classification will be selected for classification.
In the classification step, the Feature which is selected from the feature selection phase is considered, for this feature difference from average to feature is calculated and the class is assigned to test instance according to minimum difference. The classification step of mean-based classification is similar to the Rocchio classifier which is used for text classification. Figure 5 shows, the concept of the Rocchio classification algorithm.
Figure 3 depicts the concept of the mean difference. Mean 1 and mean 2 are calculated for class 1 and class 2 respectively and the mean difference is an absolute difference between mean 1 and means 2. The feature which has the largest mean difference is selected for classification Classification is similar to Rocchio classification which is depicted in Fig. 5.
Algorithm
Step 1 and step 2 of the mean-based classifier algorithm are for feature selection. Step 3 and step 4 are for classification.
Step 1) For all features in Feature set F = {f1, f2, f3, f4—fn} is set of input features in dataset. Class set = {y1, y2 } is set of classes in dataset. µ(i, j) will be the mean for feature fi of all training instances having class yi. For each feature in the feature set, there will be two averages as mean based classification works for two classes i.e. µ(i, 1) and µ(i, 2). Mean difference of I’th feature is MD(i) = |µ(i, 1)−µ(i, 2)| is calculated for each feature so Mean difference set MD={MD(1), MD(2), MD(n)} is obtained.
Step 2) For set MD max {MD(1), MD(2),…MD(n)} = MD(i) is obtained, where MD(i) is maximum element in set MD and i corresponds to fi feature will be selected for classification in set F.
Step 3) If X be instance to be classified and have feature X ={xf1, xf2,..xfn}. I’th feature selected from step 2 will be selected from set X i.e. xfi.
Step 4) class1 difference is cd1 = |µ(i, 1)−xfi| and class 2 difference is cd2 = |µ(i, 2)−xfi| , X will be assigned to class by selecting minimum from cd1 and cd2 i.e., min {cd1, cd2} and respective class corresponding to that difference will be assigned.
Mean based classification algorithm. a) Feature selection phase. For every feature class-wise average (see Box 1) is calculated and mean difference is calculated. The feature which is having a maximum mean difference is selected. b) Classification phase. For novel instance select value of feature from feature selected in the feature selection phase. Calculate the difference of feature value of the novel subject average for each class. Assign the class having minimum difference from the respective average.
Rocchio Classification. Blue triangles represent class 1 while red rectangles represent class 2. Query instance is shown in yellow. Distance 1 is the distance from centroid 1 to query instance while distance 2 is the distance between centroid 2 and query instance. Query instance will be assigned to class 2 since distance 2 is smaller than distance 1. (Color figure online)
4 Mean Based Classification (Standard Deviation Variant)
General Mean-based classification suffers from feature having maximum mean difference may not be best as there may be values in both classes that are overlapping, to minimize overlapping standard deviation variant is considered. Mean based classification using standard deviation considers standard deviation for each class. The Sum of standard deviation for classes is calculated. Standard variation-based variant of mean-based classification selects feature for classification which have maximum mean separation across classes and have minimum standard deviation across classes, these two characteristics refers to features will have minimum overlapping across classes will be given high priority.
A schematic diagram for mean based classification using standard deviation variant is shown in Fig. 6.
Algorithm
Step 1) For all features in Feature set F = {f1, f2, f3, f4—fn} is set of input features in dataset where n is number of input features. Class set = {y1, y2} is set of classes in dataset. µ(i,j) will be the mean for feature fi of all training instances having class yi. For each feature in the feature set, there will be two averages as mean based classification works for two classes i.e. µ(i, 1) and µ(i, 2). Mean difference of I’th feature is MD(i) = | µ(i, 1)−µ(i, 2)| is calculated for each feature so Mean difference set MD = {MD(1), MD(2), MD(n)} is obtained.
Step 2) For all features in Feature set F = {f1, f2, f3, f4—fn} is set of input features in dataset. Class set = {y1, y2 } is set of classes in dataset. σ (i, j) will be standard deviation for feature fi of all training instances having class yj. as mean based classification works for two classes i.e. σ (i, 1) and σ (i, 2) . Standard deviation sum will be given as SD(i) = σ (i, 1) + σ (i, 2) calculated for each feature so Set SD = {SD(1), SD(2),…SD(n)}
Step 3)
Find out feature from F which maximizes MD and minimizes SD will have corresponding mean difference as MD(i) and SD(i) so feature fi will be selected for classification.
Step 4)
If X be instance to be classified and have feature X = {xf1, xf2,..xfn}. I’th feature selected from step3 will be selected from set X i.e. xfi.
Step 5)
class1 difference is cd1 = |µ(i, 1)−xfi| and class 2 difference is cd2 = |µ(i, 2)−xfi| , X will be assigned to class by selecting minimum from cd1 and cd2 i.e., min {cd1, cd2} and respective class corresponding to that difference will be assigned.
Mean based classification using standard deviation. 1) For given features mean difference and standard deviation sum (see Box 1) are calculated. 2) Features are stored in the array and sorted for mean difference and sorted in decreasing order so that the first feature in the array will have maximum mean difference. Till this point steps are similar to the mean-based algorithm discussed in Fig. 1. 3) Feature which is having maximum mean difference and the minimum standard deviation is selected iteratively and used for classification discussed in Fig. 4b.
5 Shapely Value-Based Interpretation
For any model, certain numbers of features are involved in model prediction. The basic Intuition behind shape values is to consider each feature and to explain the model further by feature interaction and impact and explaining global model behaviour.
SHAP (Shapely Additive Explanation) by Lundberg and Lee (2016) [15] is a method to explain individual prediction. It is based on Shap values from game theory. This method can be used on Tabular data and also on unstructured data.
Shapely values are based on game theory. The goal of shapely value-based prediction is to computer contribution of each feature to the final prediction. It is based on coalition game theory. The feature values of a data instance act as players in a coalition. Shapely value tells us about how to fairly distribute the payout among features. The player can be individual feature value e.g., Tabular data. The player can also be a group of features. A group of features also can be regarded as a player.
6 Results and Discussions
6.1 Dataset Used
We have considered 5 different cancer genomic datasets. All dataset was obtained from SBCB (https://sbcb.inf.ufrgs.br/cumida). The genomic dataset consists of gene expression values for genes and corresponding cancer class labels.
Prostate Cancer gene dataset: is having 51 subjects having 54677 features i.e., genes, it has two classes tumoral and normal.
Pancreas Cancer gene dataset: is having 48 subjects having 54677 features i.e., genes, it has two classes tumoral and normal.
Liver Cancer gene dataset: is having 357 subjects having 22279 features i.e., genes, it has two classes HCC and normal.
Blood Cancer gene dataset: is having 52 subjects having 22647 features i.e., genes, it has two classes CLL and normal_B_Cell
6.2 Machine Learning Algorithms Results
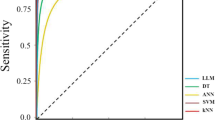
We used various machine learning algorithms and also novel Mean based classification. Accuracies for all machine learning models are shown in Table 1. Mean based classification algorithm and centroid classification (Standard deviation variant) and logistic regression obtained high accuracy for all cancer datasets. Mean based classification with standard deviation obtained high accuracy almost for all datasets.
Figure 7 shows, comparison of accuracies between SVM, Logistic regression, mean based classification and mean based classification standard deviation variant. Mean based classification-std and Logistic regression obtained the highest accuracy among other classification algorithms.
6.3 Shapely Value-Based Explanations on Pancreas Dataset
Shapely value-based explanations can be given on the pancreas dataset. Figure 8 shows a shap summary plot for the pancreas dataset. Gene 1552295_a_at is having the highest impact classification. Shap summary plot summarizes the impact of each feature on the overall classification.
Prostate cancer
An explainable AI approach is applied to the prostate cancer gene expression dataset. Gene 121_at is having the highest impact on the model. Higher the value of gene 121_at lowers its impact on the model while lower value has a higher impact on the model (Fig. 9).
7 Conclusion and Future Scope
Machine learning algorithms applied successfully on various cancer genomics datasets. All machine learning algorithms performed better in terms of accuracy. The proposed method for classification “Mean based classification and its variant” obtained high accuracy for all dataset. Performance for Mean based classification for standard deviation is better than mean based classification. The shapely value-based interpretation method successfully applied to the dataset. The overall impact of an induvial gene can be predicted from shap interaction plots. Mean based classification can be extended to a multilabel dataset. The algorithm for finding the best feature in feature space in the case of mean-based classification-standard deviation is a linear optimised algorithm needed to be developed to further optimise this method. To conclude, Machine learning can be used in cancer genomics and interactable methods can be applied successfully to further understand results.
References
Jahn, S.W., Plass, M., Moinfar, F.: Digital pathology: advantages, limitations and emerging perspectives. J. Clin. Med. 9, 3697 (2020)
LeCun, Y., Bengio, Y., Hinton, G.: Deep learning. Nature 521, 436–444 (2015)
Nogrady, B.: How cancer genomics is transforming diagnosis and treatment. Nature 579, S10 (2020)
Pirooznia, M., Yang, J.Y., Yang, M.Q., et al.: A comparative study of different machine learning methods on microarray gene expression data. BMC Genomics (2008). https://doi.org/10.1186/1471-2164-9-S1-S13
Cho, S.B., Won, H.H.: Machine learning in DNA microarray analysis for cancer classification. In: Proceedings of the First Asia-Pacific Bioinformatics Conference on Bioinformatics, vol. 19 (2003)
Nguyen, D.V., Rocke, D.M.: Tumor classification by partial least squares using microarray gene expression data. Bioinformatics 18(1), 39–50 (2002)
Li, W., Yang, Y.: How many genes are needed for a discriminant microarray data analysis. In: Lin, S.M., Johnson, K.F. (eds.) Methods of Microarray Data Analysis, pp. 137–149. Springer, Boston (2002). https://doi.org/10.1007/978-1-4615-0873-1_11
Li, L., et al.: Gene selection for sample classification based on gene expression data: study of sensitivity to choice of parameters of the GA/KNN method. Bioinformatics 17, 1131–1142 (2001)
Lundberg, S.M.: From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2, 56–67 (2020)
Heller, M.J.: DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 4, 129–153 (2002)
Rocchio, J.J.: Relevance feedback in information retrieval (1965)
Bayes, P.: An essay towards solving a problem in the doctrine of chance. Philos. Trans. R. Soc. London 53, 370–418 (1763)
Cortes, C., Vapnik, V.: Support-vector networks. Mach. Learn. 20, 273–297 (1995). https://doi.org/10.1007/BF00994018
Walker, S.H., Duncan, D.B.: Estimation of the probability of an event as a function of several independent variables. Biometrika 54(1–2), 167–179 (1967)
Lundberg, S.M., Lee, S.I.: A unified approach to interpreting model predictions. In: Advances in Neural Information Processing Systems (NIPS), vol. 17 (2017)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Joshi, H., Rajeswari, K., Joshi, S. (2022). Machine Learning in Cancer Genomics. In: Raman, I., Ganesan, P., Sureshkumar, V., Ranganathan, L. (eds) Computational Intelligence, Cyber Security and Computational Models. Recent Trends in Computational Models, Intelligent and Secure Systems. ICC3 2021. Communications in Computer and Information Science, vol 1631. Springer, Cham. https://doi.org/10.1007/978-3-031-15556-7_7
Download citation
DOI: https://doi.org/10.1007/978-3-031-15556-7_7
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-15555-0
Online ISBN: 978-3-031-15556-7
eBook Packages: Computer ScienceComputer Science (R0)