Abstract
Emerging viral infections have been known as a significant threat of recent years. Coronavirus-2 (SARS-CoV-2) was initially recognized in China and infected a large population through inhalation of droplets. Massive global deaths, multiple mutations, and the mysterious nature of the virus require serious actions. Transient mutations in this virus led to the production of different strains with various risks and severity of the disease, making the situation complex. Trigger of a cytokine storm after SARS-CoV-2 leads to the production of various inflammatory mediators including interleukin (IL)-6, IL-1β, tumor necrosis factor-α (TNF-α), and interferon-γ (IFN-γ),. This ultimately leads to severe lung tissue damage which is the main cause of mortality associated with this virus. Several therapeutic approaches, including pharmacological agents with anti-inflammatory and anti-viral properties, are being used to manage these patients. This chapter explains the virus, its structure, and biology. Data were collected from different clinical and animal experiments published in English (2000-April 2021), selected from Google Scholar, Scopus, PubMed, and the Cochrane library.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
Introduction
In Wuhan, China, a positive-strand RNA virus (SARS-CoV-2) was first detected in December 2019. It rapidly spread and affected populations worldwide, and case fatality rates range from 2% to 3%. SARS-CoV-2 genome comprises ssRNA (single-stranded RNA) which is approximately ∼30 kb in size [1]. Both nonstructural proteins (nsps) and structural proteins are encoded by the genome. Structural proteins include the nucleocapsid proteins (N), spike glycoproteins (S1 and S2), envelope proteins (E), and membrane proteins (M), which are all located near the 3′ end of the strand [2]. Cytokine storms are often caused by uncontrolled inflammatory responses and result in high mortality. IFN-γ, IL-1, IL-6, TNF-α, and IL-18 are crucial cytokines that are released through nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), signal transducer and activator of transcription 3 (STAT3), Toll-like receptors (TLRs), Janus kinase (JAK), protein kinase B (AKT), mammalian target of rapamycin (mTOR) signaling pathways during cytokine storms [3]. In brief, infected epithelial cells with the SARS-CoV-2 express angiotensin-converting enzyme 2 (ACE2), which induces activation of immune cells resulting in acute respiratory distress syndrome (ARDS) [4]. Beyond vaccination, there is no fully efficient agent or method recommended for pre- and post-exposure with SARS-CoV-2; however, several medications are used. For instance, Remdesivir as an anti-viral drug is prescribed for coronavirus disease 2019 (COVID-19) patients with respiratory symptoms, which causes faster recovery [5]. Hydroxychloroquine is used to prevent viral replication in severe acute respiratory syndrome (SARS), as used in the Middle East respiratory syndrome (MERS) patients years ago. Lopinavir/Ritonavir are recommended as anti-viral agents in viral infections such as HIV; corticosteroids such as dexamethasone and methylprednisolone [6] are used to reduce the mortality rates of patients and decreases the inflammatory reactions and the macrophage activation syndrome. Tocilizumab is used in patients with ARDS and reduces the elevated levels of IL-6 [7]. Aside from chemical agents, herbal medicines such as curcumin and quercetin are used to treat COVID-19 by suppressing the inflammatory signaling pathways and cytokines [8].
Method of Search
From PubMed, Google Scholar, Scopus, and Cochrane library, we collected data on published clinical and animal studies between 2000 and April 2021 in English. Also, search terms included “SARS CoV-2” or “COVID-19” and “Biology” or “Variants” or “Structure” or “Origins” or “Inflammatory response.”
The SARS-CoV-2 Biology
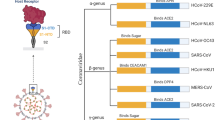
Coronaviruses are classified as a subgroup of RNA viruses with the ability to cause diseases in mammalian and avian species. They compose a positive-sense single-stranded RNA (+ssRNA) genome ranging from 26.4 to 31.7 kb in size. The genome has a 5′ methylated cap and a 3′ polyadenylated tail [9]. Coronaviruses contain the largest genome among the RNA viruses, making them capable of plastic gene adaptation and modification [10]. This family causes numerous viral infections while more lethal variants give rise to SARS, MERS, and COVID-19 [11]. Molecular biology revealed that the disease caused by the coronaviruses family is a consequence of virus genome transcription and replication and delayed or disturbed immune responses [12, 13]. Subsequently, upregulation of the inflammatory pathways and the immune cells invasion in different tissues provokes a malfunctioning cycle of the host immune response [14,15,16]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a strain of coronaviruses responsible for the ongoing COVID-19 pandemic. It is the seventh identified coronavirus capable of causing illness in humans after human coronavirus 229E (HCoV-229E), human coronavirus NL63 (HCoV-NL63), human coronavirus OC43 (HCoV-OC43), human coronavirus HKU1 (HCoV-HKU1), Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) [17]. The genome sequence of SARS-CoV-2 is 79% similar to SARS-CoV and 50% to MERS-CoV [18]. The bat coronavirus (RaTG13), spotted in Rhinolophus affinis from Yunnan province, China, shares 96.2% full-length genome sequence and 90% open reading frames in the genome with SARS-CoV-2 [19, 20]. However, recent reports are still speculating about the virus reservoir. It remains elusive how the virus was transmitted to humans and which animals acted as an intermediate reservoir [21, 22]. As Fig. 1.1 exerts [23], the virus morphology is simple. The coronavirus virion consists of the RNA genome, helical nucleocapsid, and viral membrane containing S1, S2, M, and E [24]. All coronaviruses have a similar structure. The first two-thirds of coronaviruses genomes are open reading frame (ORF) (contain ORF1a and 1b), which encode the 16 nsps [9]. The later reading frames encode S1 and S2, E, M, and N [25]. The differences between the coronaviruses are owed to the number and function of accessory proteins. The reading frames between the nonstructural and structural proteins encode the accessory proteins. The distinguishing point of these viruses is the spike that controls the virus activities and virulence and the diverse accessory proteins that combat against the host immune system [26, 27]. The differences between the functional domains of the spike protein genome of SARS-CoV and MERS-CoV have been demonstrated in Fig. 1.2 [28]. There are two subunits of spike protein, S1 and S2. The S1 subunit has a receptor-binding domain (RBD) that binds the receptor-binding motif (RBM) to the host surface. Moreover, the S2 subunit mediates the receptor attachment and the host membrane fusion [23, 29]. The primary host receptor for SARS-CoV and SARS-CoV-2 is the ACE2, while for MERS-CoV is dipeptidyl peptidase 4 (DPP4) [30,31,32,33]. Afterwards, in an outbreak of SARS-CoV in 2000, scientists started to search for other human viruses that can cause severe illnesses. In 2010, the MERS-CoV appeared, and the existing research platform from SARS-CoV empowered the scientists to develop a DNA-based vaccine against MERS-CoV infection in March 2020 [34]. When SARS-CoV-2 appeared in 2020 and caused the pandemic, the previous vaccine design methods were reproducible, and the RNA-based COVID-19 vaccine was presented in 2021 [35, 36].
The SARS-CoV-2 morphology and genome sequence. Schematic representation of the coronavirus virion entailing RNA genome, nucleocapsid (N), membrane (M), envelope (E), and the spike (S) proteins on the surface of the virus. The RNA genome has a 5′ cap and 3′ poly (A) tail. The replicas contain open reading frames (ORFs) 1a and 1b encoding 16 nonstructural proteins (nsp1-nsp16). The remaining ORFs encode the structural protein (S, E, M, and N) and accessory proteins [23]
The genome sequence of SARS-CoV and MERS-CoV, differences between the spike protein of each genome. The S protein consists of two functional subunits (S1 and S2). The S1 subunit comprises a receptor-binding domain (RBD), and the RBD comprises a receptor-binding motif (RBM). The S2 subunit includes heptad repeat regions (HR1 and HR2), fusion peptide (FP), transmembrane domain (TM), and fusion peptides (FP)
The SARS-CoV-2 Origins
Coronaviruses can typically be categorized into four different genera: alphacoronavirus, betacoronavirus, gammacoronavirus, and deltacuronavirus. Alpha coronaviruses include HCoV-229E and HCoV-NL63 [37]. Beta viruses contain HCoV-OC43, MERS-CoV, SARS-CoV-2, HCoV-HKU1, and SARS-CoV. Likewise, gamma coronaviruses are comprised of the avian infectious bronchitis virus (IBV) [38]. Delta coronaviruses categories are concluded swine delta coronavirus (PdCoV). Coronavirus is one of the newly emerging viruses that led to many deaths. Its evolution is high-speed, and the virus mutates in different ways and creates various strains [39]. Understanding the virus’s evolutionary patterns will help discover more effective treatments and vaccines. The members of the coronaviridae family have been studied in different species of fish, birds, camels, and bats [40]. The most common viruses that infect mammals are alpha and beta coronaviruses, while gamma and delta coronaviruses infect birds. Understanding the evolution or mutation pattern that the virus may have in the future may be helped by evolutionary history [41].
HCoV-229E
Initially, this coronavirus strain was called B814. Another infection with an unidentified respiratory virus led to the formation of a strain cell culture that initiated the infection. This strain eventually became a prototype for HCoV-229E [42]. Under the electron microscope, the B814 and the HCoV-229E strain are very analogous to the avian coronavirus, IBV. HCoV-229E contains ether and is composed of 89 nm particles and has single-stranded RNA, coated as genetic material, and after 6 days causes cytopathic effects. The main methods of the HCoV-229E transmission were droplet respiration and foaming [43, 44].
HCoV-OC43
HCoV-229E was discovered after virus samples from common cold patients were taken, and no antibodies were detected toward this virus, proving there was no B814 mutation equivalent to the HCoV-229E [45]. The OC43 strain was spread and eventually formed the HCoV-OC43 species. The HCoV-OC43 species is an enveloped ssRNA virus in the same way as the HCoV-229E species. HCoV-OC43 is the reason for one-third of the common colds. It is an RNA virus with a 31.5 kb size. The HCoV-229E is also involved in one-third of those cases [46].
SARS-CoV
Despite discovering the role of HCoV-229E and HCoV-OC43, these species were initially thought to be the only two types of human coronaviruses. However, a new strain of coronavirus was distinguished in 2002, the SARS-CoV [21] reported in China that is transferred from palm civets to humans. SARS-CoV was found in 2003 in horseshoe bats. It was found to be an enveloping ssRNA virus. The virus has about 14 ORFs with about 30 kb RNA [47].
HCoV-NL63
An immunocompromised infant with respiratory symptoms in The Netherlands was found to have HCoV-NL63 in 2004 [48]. Studies have elucidated that HCoV-NL63 is isolated from the ancestors of HCoV-229E. Also, HCoV-NL63 possesses an ssRNA genome encased and polyadenylated with 27,553 nucleotides. The virus is more prevalent in winter and milder weather [49].
HCoV-HKU1
HCoV-HKU1 was primarily detected in Hong Kong in January 2005 that is related to the Group II prototype of HCoV-OC43. The positive samples of HCoV-HKU1 were often established in temperate countries like Italy and the USA during winter and spring [50]. The virus is a + ssRNA virus with 29,926 nucleotides. RT-PCR performs rapid diagnosis of HKU1 infections with the assistance of specific monoclonal antibodies (mAb) related to HKU1 [51].
MERS-CoV
MERS-CoV was derived from the sputum of a 60-year-old man hospitalized due to renal failure and severe acute pneumonia in 2012. Subsequent serological evidence confirmed the presence of MERS-CoV in camels in the Middle East, North Africa, and East Africa, indicating camels as a reservoir of MERS-CoV [52]. MERS-CoV has a +ssRNA genome of 30.1 kb. The MERS-CoV replicates in virus-induced bilayer vesicles lacking host pattern recognition receptors, preventing its dsRNA host from being detected [53].
SARS-CoV-2
There have been several cases of pneumonia with an unknown cause reported in Wuhan in December 2019. The virus has been renamed Wuhan coronavirus, but the ICTV (International Committee for the Classification of Viruses) named it SARS-CoV-2 and COVID-19 [35]. SARS-CoV-2 shows more minor mutations because of its corrective function. About 13 mutation sites were detected in the SARS-CoV-2 regions of ORF-1ab, −3a, −8, N, and S, including 8782 in ORF1a and 28,144 in ORF8 with mutation rates of 29.47% and 30.53%, respectively [54]. Genetic analysis of a population of 103 genomes associated with SARS-CoV-2 showed that SARS-CoV-2 advanced into two major forms, L and S, which are well characterized via two members of single nucleotide polymorphisms (SNPs) [55]. Cuttings of the spike protein are located at the S1 and S2 junction, as two significant subunits, which determine the extent of viral infection and the range of host species. The difference in mortality rates is related to viral mutations and evolutionary ability [21].
The SARS-CoV-2 Structure
Coronaviruses are large—average diameter of 80–120 nm and average molecular mass of 40,000 kDa—roughly spherical and relatively pleiomorphic with distinctive surface spikes projections [56]. Their RNA is in the center of the virus and is protected by the nucleocapsid, membrane protein, and lipid bilayer envelope [57, 58]. The viral capsid possesses a lipid bilayer and four types of structural proteins, namely, S, M, E, and N proteins—an approximate molar ratio of S:E:M is 20:1:300 (Fig. 1.1). The S protein is essential to form an interaction with the host cell. In addition to the S protein, the viral surface also encompasses hemagglutinin-esterase dimer (HE), which is not necessary for replication but is vital for the virus entry [59, 60]. The E protein is the minor structural protein and differs diversely among the coronaviruses [61]. Among the primary structural proteins, M is responsible for shaping the envelope [62]. The N protein is tied to the RNA and empowers the virus to take over the host cells [63, 64]. The genome of coronaviruses includes various ORFs. The gene order in all members is 5′-leader-UTR-replicase (ORF1ab)-S-E-M-N-3′UTR-poly (A) tail [65]. Their genome seems to have a bias against cytosine (C) and guanine (G) nucleotides with the highest composition of uracil (U) and adenosine (A) [66]. In addition to these components, 16 nsps (nsp1 to nsp16) differ between the genera of coronaviruses [9]. These nsps perform vital roles in assembling the replication/transcription complex (RTC), RNA polymerization, RNA proofreading, mRNA capping, allosteric activation, and repression of the host immune system [67, 68].
The coronaviruses spike (S) protein anchors to the ACE2 receptors for viral entrance, expressed on numerous cell surfaces. The transmembrane protease serine 2 (TMPRSS2) and lysosomal proteases also play significant roles in the SARS-CoV-2 entry [69]. Following the cytoplasm entry, the virus induces spatial alteration in the endosome, uncoating the viral nucleocapsid (N). Finally, the viral genome is ultimately released within the cytoplasm, and the RTC initiates [70]. Moreover, the SARS-CoV-2 sustains the largest genome with 30,000 bases in the RNA sequence length. A unique feature of SARSCoV2 is its capacity to cleave the spike protein at its polybasic site through furin-mediated cleavage, which increases its virulence. Moreover, it was proposed that the furin-cleavage region at the SARS-CoV-2 spike protein was needed to enable the virus to infect humans as well as animals [21].
SARS-CoV-2 Variants
Coronaviruses belong to the sub-family of Orthocoronavirinae in the family Coronaviridae, order Nidovirales, and realm Riboviria [71, 72]. As mentioned, the coronaviruses are sorted into four genera: deltacoronavirus, gammacoronavirus, betacoronavirus, and alphacoronavirus. However, the number of species increases and many coronaviruses are unspecified [71, 73]. The betacoronavirus and alphacoronavirus uniquely infect mammalian species, while deltacoronavirus and gammacoronavirus infect both mammalian and avian species. The coronavirus infection mostly leads to respiratory, gastrointestinal, and neurologic disorders [74, 75]. Several variants of SARS-CoV-2 are of interest and concern. Generally, a variant is called a variant of interest when it displays evidence of mutation, which is expected to circulate broadly. The Mu and Lambda variants are currently the World Health Organization (WHO) variants of interest. When a variant of interest is more transmissible and detrimental, it becomes a variant of concern. The recently acknowledged variants of concern are presented in Table 1.1 [76, 77].
Conclusion
In December 2019, the outbreak of SARS-CoV-2 spread in Wuhan, China. This virus causes various diseases, from the common cold to ARDS [78, 79]. The prevalence of ARDS also increases with the rise of inflammatory cytokines. The activation of the ACE2 and TMPRSS2 receptors are the main mechanisms of the cytokine storm [80, 81]. High levels of inflammatory cytokines and chemokines in COVID-19 patients are accounted for more elevated levels of IL-6, IL-1β, IL-10, TNF-α, and IFN-γ through the activation of the various signaling pathways such as NF-κB, STAT3, JAK, AKT, and mTOR pathways [82, 83]. Different variants of coronavirus are determined and classified into Alpha coronaviruses (HCoV-229E and HCoV-NL63), Beta viruses (HCoV-OC43), SARS-CoV, HCoV-HKU1, MERS-CoV, and SARS-CoV-2, Gamma coronaviruses (avian IBV), and Delta coronaviruses (PdCoV) [84, 85]. Nevertheless, several types of coronavirus would be distinguished after mutation in humans because of adapting coronaviruses to their human hosts. Genetic evolution in coronaviruses results in mutant versions of coronaviruses that may differ from their ancestral strains in various ways. During this pandemic, several variants of SARS-CoV-2 have been described. Recently, different therapeutic approaches have been examined to elucidate precise treatment protocols. Therefore, various medications such as Lopinavir/Ritonavir, Hydroxychloroquine, Tocilizumab, Remdesivir, corticosteroids, as well as methylprednisolone, and dexamethasone resulted in a reduction of symptoms and improved outcomes. Furthermore, some herbal medicines such as quercetin, resveratrol, curcumin, have been tried in the treatment of COVID-19 because of their anti-inflammatory characteristics [86]. Currently, multiple vaccines are developed and distributed worldwide, such as Oxford-AstraZeneca, Pfizer- BioNTech, CoronaVac, and COVID Shield, which support people worldwide and decrease the rate and prevention of getting infected with COVID-19; however, even after injection coronavirus vaccines, with different mechanisms of action, it is possible to be infected with new variant of coronavirus due to mutation in different regions of the virus, particularly structural protein areas.
Abbreviations
- +ssRNA:
-
Positive-sense single-stranded RNA
- ACE2:
-
Angiotensin-converting enzyme 2
- ADE:
-
Antibody-dependent enhancement
- AKT:
-
Protein kinase B
- APC:
-
Antigen-presenting cells
- APN:
-
Aminopeptidase N
- ARDS:
-
Acute respiratory distress syndrome
- CBP:
-
Convalescent blood products
- CEACAM1:
-
Carcino embryonic antigen-related cell adhesion molecule 1
- CFDA:
-
China food and drug administration
- CFR:
-
Case fatality ratio
- COVID-19:
-
The Coronavirus disease-2019
- CRISPR:
-
Clustered regularly interspaced short palindromic repeats
- CT:
-
Computerized tomography
- DIC:
-
Disseminated intravascular coagulation
- DMV:
-
Double-membrane vesicles
- DPP4:
-
Dipeptidyl peptidase 4
- DRF:
-
Damage response framework
- dsRNA:
-
Double-stranded RNA
- ERGIC:
-
Endoplasmic reticulum golgi intermediate compartment
- FDA:
-
Food and drug administration
- HCoV-229E:
-
Human coronavirus 229E
- HCoV-HKU1:
-
Human coronavirus HKU1
- HCoV-NL63:
-
Human coronavirus NL63
- HCoV-OC43:
-
Human coronavirus OC43
- HLA:
-
Human leukocyte antigen
- IBV:
-
Infectious bronchitis virus
- ICTV:
-
International committee on taxonomy of viruses
- IFN-γ:
-
Interferon-gamma
- IL:
-
Interleukin
- JAK:
-
Janus kinase
- kb:
-
Kilobases
- mAb:
-
monoclonal antibodies
- MERS:
-
Middle East respiratory syndrome
- MERS-CoV:
-
Middle East respiratory syndrome coronavirus
- MHC:
-
Major histocompatibility complex
- MODS:
-
Multiple organ dysfunction syndromes
- MOF:
-
Multiple organ failure
- mTOR:
-
Mammalian target of rapamycin
- NF-κB:
-
Nuclear Factor κ-light-chain-enhancer of activated B cells
- nsps:
-
Nonstructural proteins
- ORF:
-
Open Reading Frame
- PdCoV:
-
Porcine delta coronavirus
- RAS:
-
Renin-angiotensin system
- RBD:
-
Receptor-binding domain
- RBM:
-
Receptor-binding motif
- RER:
-
Rough endoplasmic reticulum\
- RTC:
-
Replicase-transcriptase complex
- RT-qPCR:
-
Real-time quantitative polymerase chain reaction
- SARS:
-
Severe acute respiratory syndrome
- SARS-CoV:
-
Severe acute respiratory syndrome coronavirus
- SARS-CoV-2:
-
Severe acute respiratory syndrome coronavirus 2
- SARSCoV-2:
-
Severe acute respiratory syndrome coronavirus-2
- ssRNA:
-
Single-stranded RNA
- STAT3:
-
Signal transducer and activator of transcription 3
- TLRs:
-
Toll-like receptors
- TMPRSS2:
-
Transmembrane protease serine 2
- TNF-α:
-
Tumor Necrosis Factor-alpha
- WHO:
-
World Health Organization
References
Wang M-Y, Zhao R, Gao L-J, Gao X-F, Wang D-P, Cao J-M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:587269.
Li C-X, Chen J, Lv SK, Li JH, Li LL, Hu X. Whole-transcriptome RNA sequencing reveals significant differentially expressed mRNAs, miRNAs, and lncRNAs and related regulating biological pathways in the peripheral blood of COVID-19 patients. Mediat Inflamm. 2021;2021:6635925.
Moradian N, Gouravani M, Salehi MA, Heidari A, Shafeghat M, Hamblin MR, Rezaei N. Cytokine release syndrome: inhibition of pro-inflammatory cytokines as a solution for reducing COVID-19 mortality. Eur Cytokine Netw. 2020;31(3):81–93.
Fara A, Mitrev Z, Rosalia RA, Assas BM. Cytokine storm and COVID-19: a chronicle of pro-inflammatory cytokines. Open Biol. 2020;10(9):200160.
Varghese PM, Tsolaki AG, Yasmin H, Shastri A, Ferluga J, Vatish M, Madan T, Kishore U. Host-pathogen interaction in COVID-19: pathogenesis, potential therapeutics and vaccination strategies. Immunobiology. 2020;225(6):152008.
Nile SH, Nile A, Qiu J, Li L, Jia X, Kai G. COVID-19: pathogenesis, cytokine storm and therapeutic potential of interferons. Cytokine Growth Factor Rev. 2020;53:66–70.
Chi Z, Wu Z, Li J, Zhao H, Wang G. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist tocilizumab may be the key to reduce the mortality. Int J Antimicrob Agents. 2020;55(5):105954.
Noor H, Ikram A, Rathinavel T, Kumarasamy S, Nasir Iqbal M, Bashir Z. Immunomodulatory and anti-cytokine therapeutic potential of curcumin and its derivatives for treating COVID-19–a computational modeling. J Biomol Struct Dyn. 2021:1–16.
Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, editors. Coronaviruses: methods and protocols. New York, NY: Springer New York; 2015. p. 1–23. https://doi.org/10.1007/978-1-4939-2438-7_1.
Woo PCY, Huang Y, Lau SKP, Yuen K-Y. Coronavirus genomics and bioinformatics analysis. Viruses. 2010;2(8):1804–20. https://doi.org/10.3390/v2081803.
Chathappady House NN, Palissery S, Sebastian H. Corona viruses: a Review on SARS, MERS and COVID-19. Microbiology Insights. 2021;14:11786361211002481. https://doi.org/10.1177/11786361211002481.
Wong L-YR, Perlman S. Immune dysregulation and immunopathology induced by SARS-CoV-2 and related coronaviruses — are we our own worst enemy? Nat Rev Immunol. 2022;22(1):47–56. https://doi.org/10.1038/s41577-021-00656-2.
Boechat JL, Chora I, Morais A, Delgado L. The immune response to SARS-CoV-2 and COVID-19 immunopathology—Current perspectives. Pulmonology. 2021;27(5):423–37. https://doi.org/10.1016/j.pulmoe.2021.03.008.
Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, Levantovsky R, Malle L, Moreira A, Park MD, Pia L, Risson E, Saffern M, Salomé B, Esai Selvan M, Spindler MP, Tan J, van der Heide V, Gregory JK, Alexandropoulos K, Bhardwaj N, Brown BD, Greenbaum B, Gümüş ZH, Homann D, Horowitz A, Kamphorst AO, Curotto de Lafaille MA, Mehandru S, Merad M, Samstein RM, Sinai Immunology Review P. Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910–41. https://doi.org/10.1016/j.immuni.2020.05.002.
Weatherhead JE, Clark E, Vogel TP, Atmar RL, Kulkarni PA. Inflammatory syndromes associated with SARS-CoV-2 infection: dysregulation of the immune response across the age spectrum. J Clin Invest. 2020;130(12):6194–7. https://doi.org/10.1172/JCI145301.
Lai C, Liu X, Yan Q, Lv H, Zhou L, Hu L, Cai Y, Wang G, Chen Y, Chai R, Liu Z, Xu Y, Huang W, Xiao F, Hu L, Li Y, Huang J, Zhou Q, Li L, Peng T, Zhang H, Zhang Z, Chen L, Chen C, Ji T. Low innate immunity and lagged adaptive immune response in the re-tested viral RNA positivity of a COVID-19 patient. Front Immunol. 2021;12:664619. https://doi.org/10.3389/fimmu.2021.664619.
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W. China novel coronavirus I, research T (2020) a novel coronavirus from patients with pneumonia in China. N Engl J Med. 2019;382(8):727–33. https://doi.org/10.1056/NEJMoa2001017.
Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–74. https://doi.org/10.1016/s0140-6736(20)30251-8.
Liu K, Pan X, Li L, Yu F, Zheng A, Du P, Han P, Meng Y, Zhang Y, Wu L, Chen Q, Song C, Jia Y, Niu S, Lu D, Qiao C, Chen Z, Ma D, Ma X, Tan S, Zhao X, Qi J, Gao GF, Wang Q. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184(13):3438–3451.e3410. https://doi.org/10.1016/j.cell.2021.05.031.
Wrobel AG, Benton DJ, Xu P, Roustan C, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol. 2020;27(8):763–7. https://doi.org/10.1038/s41594-020-0468-7.
Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19(3):141–54. https://doi.org/10.1038/s41579-020-00459-7.
Mohammed MEA. The percentages of SARS-CoV-2 protein similarity and identity with SARS-CoV and BatCoV RaTG13 proteins can be used as indicators of virus origin. J Proteins Proteom. 2021;12(2):81–91. https://doi.org/10.1007/s42485-021-00060-3.
Rastogi M, Pandey N, Shukla A, Singh SK. SARS coronavirus 2: from genome to infectome. Respir Res. 2020;21(1):318. https://doi.org/10.1186/s12931-020-01581-z.
Cherry J, Demmler-Harrison GJ, Kaplan SL, Steinbach WJ, Hotez P. Feigin and Cherry's textbook of pediatric infectious diseases E-book. Elsevier Health Sciences. Philadelphia: Elsevier; 2017.
Snijder EJ, Bredenbeek PJ, Dobbe JC, Thiel V, Ziebuhr J, Poon LLM, Guan Y, Rozanov M, Spaan WJM, Gorbalenya AE. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003;331(5):991–1004. https://doi.org/10.1016/s0022-2836(03)00865-9.
Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22. https://doi.org/10.1016/j.jsb.2010.11.021.
Cruz CAK, Medina PMB. Diversity in the accessory proteins of SARS-CoV-2, SARS-CoV, and MERS-CoV Betacoronaviruses. Curr Protein Pept Sci. 2021;22(10):695–715. https://doi.org/10.2174/1389203722666210910111055.
Li F, Du L. MERS-CoV. Basel: MDPI - Multidisciplinary Digital Publishing Institute; 2019.
Tortorici MA, Veesler D. Structural insights into coronavirus entry. Adv Virus Res. 2019;105:93–116. https://doi.org/10.1016/bs.aivir.2019.08.002.
Zhu Z, Lian X, Su X, Wu W, Marraro GA, Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21(1):224. https://doi.org/10.1186/s12931-020-01479-w.
Samukawa K. Use of stable isotopes in life science (III). Measurement of 15N abundance in amino acids with gas chromatography-mass spectrometry (author's transl). Radioisotopes. 1982;31(3):166–74.
Dooley DC. Glycerol permeation of the human granulocyte. Exp Hematol. 1982;10(5):413–22.
Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500(7461):227–31. https://doi.org/10.1038/nature12328.
Butler D. SARS veterans tackle coronavirus. Nature. 2012;490(7418):20. https://doi.org/10.1038/490020a.
Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–27.
Fiorino G, Allocca M, Furfaro F, Gilardi D, Zilli A, Radice S, Spinelli A, Danese S. Inflammatory bowel disease care in the COVID-19 pandemic era: the Humanitas, Milan, experience. J Crohn's Colitis. 2020;14(9):1330–3.
Bloom JD, Chan YA, Baric RS, Bjorkman PJ, Cobey S, Deverman BE, Fisman DN, Gupta R, Iwasaki A, Lipsitch M. Investigate the origins of COVID-19. Science. 2021;372(6543):694.
Heiat M, Heiat F, Halaji M, Ranjbar R, Yaali-Jahromi E, Azizi M, Badri T. Phobia and fear of COVID-19: origins, complications and management, a narrative review. Ann Ig. 2021;33(4):360–70.
Otto SP, Day T, Arino J, Colijn C, Dushoff J, Li M, Mechai S, Van Domselaar G, Wu J, Earn DJ. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr Biol. 2021;31(14):R918–29.
Forestieri S, Pintus R, Marcialis MA, Pintus MC, Fanos V. COVID-19 and developmental origins of health and disease. Early Hum Dev. 2021;155:105322.
Mallapaty S. Meet the scientists investigating the origins of the COVID pandemic. Nature. 2020;588(7837):208–9.
Li Z, Tomlinson AC, Wong AH, Zhou D, Desforges M, Talbot PJ, Benlekbir S, Rubinstein JL, Rini JM. The human coronavirus HCoV-229E S-protein structure and receptor binding. elife. 2019;8:e51230.
Müller C, Ulyanova V, Ilinskaya O, Pleschka S, Shah Mahmud R. A novel antiviral strategy against MERS-CoV and HCoV-229E using binase to target viral genome replication. Bionanoscience. 2017;7(2):294–9.
Zhang W, Zheng Q, Yan M, Chen X, Yang H, Zhou W, Rao Z. Structural characterization of the HCoV-229E fusion core. Biochem Biophys Res Commun. 2018;497(2):705–12.
St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol. 2004;78(16):8824–34.
Schirtzinger EE, Kim Y, Davis AS. Improving human coronavirus OC43 (HCoV-OC43) research comparability in studies using HCoV-OC43 as a surrogate for SARS-CoV-2. J Virol Methods. 2022;299:114317.
Ludwig S, Zarbock A. Coronaviruses and SARS-CoV-2: a brief overview. Anesth Analg. 2020;131(1):93–6.
Arden KE, Nissen MD, Sloots TP, Mackay IM. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005;75(3):455–62.
Aldridge RW, Lewer D, Beale S, Johnson AM, Zambon M, Hayward AC, Fragaszy EB, Group FW. Seasonality and immunity to laboratory-confirmed seasonal coronaviruses (HCoV-NL63, HCoV-OC43, and HCoV-229E): results from the flu watch cohort study. Wellcome Open Res. 2020;5:52.
Zhao Q, Li S, Xue F, Zou Y, Chen C, Bartlam M, Rao Z. Structure of the main protease from a global infectious human coronavirus, HCoV-HKU1. J Virol. 2008;82(17):8647–55.
Liu DX, Liang JQ, Fung TS. Human coronavirus-229e,-oc43,-nl63, and-hku1 (coronaviridae). Encyclopedia Virol. 2021;2021:428.
Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21(2):131–43.
Chafekar A, Fielding BC. MERS-CoV: understanding the latest human coronavirus threat. Viruses. 2018;10(2):93.
Hasöksüz M, Kiliç S, Saraç F. Coronaviruses and sars-cov-2. Turk J Med Sci. 2020;50(SI-1):549–56.
Harrison AG, Lin T, Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41(12):1100–15.
Sender R, Bar-On YM, Gleizer S, Bernshtein B, Flamholz A, Phillips R, Milo R. The total number and mass of SARS-CoV-2 virions. Proc Natl Acad Sci U S A. 2021;118(25):e2024815118. https://doi.org/10.1073/pnas.2024815118.
Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, Rota PA, Bankamp B, Bellini WJ, Zaki SR. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10(2):320–6. https://doi.org/10.3201/eid1002.030913.
Kumar S, Nyodu R, Maurya VK, Saxena SK. Morphology, genome organization, replication, and pathogenesis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Coronavirus Disease 2019 (COVID-19). 2020;2020:23–31. https://doi.org/10.1007/978-981-15-4814-7_3.
Ke Z, Oton J, Qu K, Cortese M, Zila V, McKeane L, Nakane T, Zivanov J, Neufeldt CJ, Cerikan B, Lu JM, Peukes J, Xiong X, Kräusslich H-G, Scheres SHW, Bartenschlager R, Briggs JAG. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588(7838):498–502. https://doi.org/10.1038/s41586-020-2665-2.
Hurdiss DL, Drulyte I, Lang Y, Shamorkina TM, Pronker MF, van Kuppeveld FJM, Snijder J, de Groot RJ. Cryo-EM structure of coronavirus-HKU1 haemagglutinin esterase reveals architectural changes arising from prolonged circulation in humans. Nat Commun. 2020;11(1):4646. https://doi.org/10.1038/s41467-020-18440-6.
Mandala VS, McKay MJ, Shcherbakov AA, Dregni AJ, Kolocouris A, Hong M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol. 2020;27(12):1202–8. https://doi.org/10.1038/s41594-020-00536-8.
Hu Y, Wen J, Tang L, Zhang H, Zhang X, Li Y, Wang J, Han Y, Li G, Shi J, Tian X, Jiang F, Zhao X, Wang J, Liu S, Zeng C, Wang J, Yang H. The M protein of SARS-CoV: basic structural and immunological properties. Genomics Proteomics Bioinformatics. 2003;1(2):118–30. https://doi.org/10.1016/s1672-0229(03)01016-7.
Lissenberg A, Vrolijk MM, van Vliet AL, Langereis MA, de Groot-Mijnes JD, Rottier PJ, de Groot RJ. Luxury at a cost? Recombinant mouse hepatitis viruses expressing the accessory hemagglutinin esterase protein display reduced fitness in vitro. J Virol. 2005;79(24):15054–63. https://doi.org/10.1128/jvi.79.24.15054-15063.2005.
Boopathi S, Poma AB, Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2021;39(9):3409–18. https://doi.org/10.1080/07391102.2020.1758788.
de Haan CAM, Volders H, Koetzner CA, Masters PS, Rottier PJM. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J Virol. 2002;76(24):12491–502. https://doi.org/10.1128/jvi.76.24.12491-12502.2002.
Kandeel M, Ibrahim A, Fayez M, Al-Nazawi M. From SARS and MERS CoVs to SARS-CoV-2: moving toward more biased codon usage in viral structural and nonstructural genes. J Med Virol. 2020;92(6):660–6. https://doi.org/10.1002/jmv.25754.
Gorkhali R, Koirala P, Rijal S, Mainali A, Baral A, Bhattarai HK. Structure and function of major SARS-CoV-2 and SARS-CoV proteins. Bioinform Biol Insights. 2021;15:11779322211025876. https://doi.org/10.1177/11779322211025876.
Romano M, Ruggiero A, Squeglia F, Maga G, Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9(5):1267. https://doi.org/10.3390/cells9051267.
Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727. https://doi.org/10.1073/pnas.2003138117.
van Hemert MJ, van den Worm SH, Knoops K, Mommaas AM, Gorbalenya AE, Snijder EJ. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4(5):e1000054. https://doi.org/10.1371/journal.ppat.1000054.
Fan Y, Zhao K, Shi Z-L, Zhou P. Bat Coronaviruses in China. Viruses. 2019;11(3):210. https://doi.org/10.3390/v11030210.
Payne S. Chapter 17 - family Coronaviridae. In: Payne S, editor. Viruses. Cambridge: Academic Press; 2017. p. 149–58. https://doi.org/10.1016/B978-0-12-803109-4.00017-9.
"International Committee on Taxonomy of Viruses (ICTV)" (Retrieved 2020-09-14). talk.ictvonline.org.
Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100:163–88. https://doi.org/10.1016/bs.aivir.2018.01.001.
Pal M, Berhanu G, Desalegn C, Kandi V. Severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3):e7423. https://doi.org/10.7759/cureus.7423.
World Health Organization ((27 November 2021)) "Tracking SARS-CoV-2 variants". World Health Organization. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants.
Konings F, Perkins MD, Kuhn JH, Pallen MJ, Alm EJ, Archer BN, Barakat A, Bedford T, Bhiman JN, Caly L, Carter LL, Cullinane A, de Oliveira T, Druce J, El Masry I, Evans R, Gao GF, Gorbalenya AE, Hamblion E, Herring BL, Hodcroft E, Holmes EC, Kakkar M, Khare S, Koopmans MPG, Korber B, Leite J, MacCannell D, Marklewitz M, Maurer-Stroh S, Rico JAM, Munster VJ, Neher R, Munnink BO, Pavlin BI, Peiris M, Poon L, Pybus O, Rambaut A, Resende P, Subissi L, Thiel V, Tong S, van der Werf S, von Gottberg A, Ziebuhr J, Van Kerkhove MD. SARS-CoV-2 variants of interest and concern naming scheme conducive for global discourse. Nat Microbiol. 2021;6(7):821–3. https://doi.org/10.1038/s41564-021-00932-w.
V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19(3):155–70.
Dong N, Yang X, Ye L, Chen K, Chan EW-C, Yang M, Chen S (2020) Genomic and protein structure modelling analysis depicts the origin and infectivity of 2019-nCoV, a new coronavirus which caused a pneumonia outbreak in Wuhan, China. BioRxiv.
Haas P, Muralidharan M, Krogan NJ, Kaake RM, Hüttenhain R. Proteomic approaches to study SARS-CoV-2 biology and COVID-19 pathology. J Proteome Res. 2021;20(2):1133–52.
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–20.
Khan S, Siddique R, Shereen MA, Ali A, Liu J, Bai Q, Bashir N, Xue M. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J Clin Microbiol. 2020;58(5):e00187–20.
Tomohiro S, Takaaki A. What triggers inflammation in COVID-19? Elife. 2022;11:e76231.
Joob B, Wiwanitkit V. New COVID-19 variant, VUI-202012/01: molecular change, epitope alteration, and implication for vaccine efficacy. Int J Prev Med. 2022;12(12):12–172. https://doi.org/10.4103/IJPVM.IJPVM_2708_2020.
Vasireddy D, Vanaparthy R, Mohan G, Malayala SV, Atluri P. Review of COVID-19 variants and COVID-19 vaccine efficacy: what the clinician should know? J Clin Med Res. 2021;13(6):317–25.
Wu CR, Yin WC, Jiang Y, Xu HE. Structure genomics of SARS-CoV-2 and its omicron variant: drug design templates for COVID-19. Acta Pharmacol Sin. 2022:1–13.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Lashgari, NA. et al. (2022). Biology of SARS-CoV-2 Coronavirus; Origin, Structure, and Variants. In: Banach, M. (eds) Cardiovascular Complications of COVID-19. Contemporary Cardiology. Humana, Cham. https://doi.org/10.1007/978-3-031-15478-2_1
Download citation
DOI: https://doi.org/10.1007/978-3-031-15478-2_1
Published:
Publisher Name: Humana, Cham
Print ISBN: 978-3-031-15477-5
Online ISBN: 978-3-031-15478-2
eBook Packages: MedicineMedicine (R0)