Abstract
In recent years, artificial intelligence (AI) has been widely used in medicine, for example, tumor screening, qualitative diagnosis, radiotherapy organ delineation, curative effect evaluation and prognosis, etc. Medical image registration is a typical problem and technical difficulty in the field of image processing. Specifically, in order to achieve the goal of information fusion, a certain spatial transformation is used to map two images in a set of image data sets to each other, so that the points corresponding to the same location in the two images are matched one by one. Most of the existing algorithms perform rigid registration based on simple features of images or non-automatic registration based on artificial markers. The registration accuracy is not ideal due to the imaging mechanism of different modal images, moreover, calculate consumption is high. In order to overcome these various problems, this essay proposes a new method of image registration based on differential homeomorphic deformation field to realize the same mode image registration between simulated CT and given CT, and that of image fusion using pixel weighted average method. Good registration and fusion imaging performance can still be achieved in the abdomen, which is extremely challenging body part in terms of medical image registration. The use of intelligent medical technology can reduce the workload of doctors and improve the accuracy of registration to make the diagnosis more Scientifically and reliably.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Intelligent medical technology
- Medical image registration
- Differential homeomorphism algorithm
- Pixel weighted average method
1 Introduction
In recent years, with the continuous growth of digital economy, artificial intelligence (AI) has been developing rapidly and now is deeply integrated with many application scenarios, Wise Information Technology of med (WITMED) has become one of research hotspots. Image registration technology has been widely used in computer vision, medical image processing and material mechanics and many other fields. Combining various images and displaying their information on the same image to provide multi-data and multi-information images for clinical medical diagnosis has become a technology with great application value, the accurate and efficient image matching criterion is both a key and a difficult point. For different medical applications, the research and development of special registration algorithms is an essential direction of development. At present, the two research directions of medical image registration are full automation and high precision. In order to avoid the over-reliance on imaging specialists in clinical practice and reduce their work intensity, image registration technology ranges from complex and laborious pre-imaging registration based on the positioning devices to a semi-automated approach to human-computer interaction, and then completely completed by the computer automatic registration, whose development is very rapid, the accuracy is gradually improved. The current registration accuracy has reached the “sub-pixel” level [1].
In order to improve the registration accuracy of Computed Tomography (CT) and Magnetic Resonance Imaging (MR), which are the two most common medical images, focusing on the abdomen, this essay proposes an indirect multimodal image registration method, which converts a multimodal image registration problem into a multiple single-modal image registration problem. The method introduces the great similarity and high precision of the single-modal image registration into the multimodal image registration. Analog CT is introduced as the intermediate medium. The MR image to be registered generates an analog CT image to be registered, and the reference image is still a CT image. The method is a quasi-single-mode image registration, where the analog CT image replaces the original MR image in the traditional registration mode as the image to be registered, and the CT image is taken as the reference image. At the same time, a semi-supervised classification model (SSC-EKE) with maximum knowledge utilization ability is introduced to generate Analog CT [2, 3]. The voxels generated by this method can correspond to the that in MRI, which is helpful to improve the registration accuracy.
In clinical diagnosis and treatment of diseases, image fusion can obtain an image that shows lesions more clearly and includes more medical information of the image [4]. Medical image fusion technology plays an important role in the localization of lesions, the diagnosis and analysis of disease, the formulation of treatment plan, and the recording and research of later pathology [5]. Therefore, since the image fusion was proposed, it has become one of hotspots of research and exploration in the field of medical imaging. In this study, we adopt the spatial domain fusion method, since it is intuitive and easy to implement.
The main content distribution of this essay is as follows:
The second part mainly introduces the related work, the third and fourth parts mainly introduce the research process and results, and then the fifth part is the summary of the proposed method.
2 Related Work
2.1 Laplace Support Vector Machine (LAPSVM)
The learning performance of traditional SVM depends on the quality and quantity of training samples to a great extent. When the training label data is insufficient but the quantity of non-label data is sufficient, the classification accuracy is low. Transforming the geometric structure information of edge distribution of labeled data and unlabeled data into manifold regular terms and adding them to the traditional classification supervision algorithm SVM, LAPSVM algorithm extends SVM to a semi-supervised learning algorithm.
Let be a training dataset, which contains n labeled data and n unlabeled data. The label corresponding to the tag data is denoted as yi. The popular regularization framework can be expressed as:
where \({\text{V}}\left( \cdot \right)\) is the loss function, \({\upgamma }_{\text{A}}\) and \({\upgamma }_{\text{I}}\) are the coefficients of the two regularization terms.
The first term in Eq. (1) controls the empirical risk expressed as a hinge loss function, and the second item avoids over-fitting by applying a smoothing condition to the possible solutions in the regenerated kernel Hilbert space R KHS. Fitting problem, the third term is based on manifold learning to exploit the inherent geometric distribution of all data strengths and bases on manifold learning. Use the adjacency data graph to \({\text{G}} = \left( {{\text{w}},{\text{f}}} \right)\) characterize the intrinsic manifold of the data distribution:
At this point \({\text{f}} = \left[ {{\text{f}}\left( {{\text{x}}_1 } \right), \ldots ,{\text{f}}\left( {{\text{x}}_{{\text{l}} + {\text{u}}} } \right)} \right]^{\text{T}}\), \({\text{W}}_{{\text{ij}}} \in {\text{W}},{\text{i}},{\text{j}} = 1, \ldots ,{\text{u}} + {\text{l}}\), it represents the edge weights in the data adjacency graph. \({\text{L}} = {\text{S}} - {\text{W}}\) Refers to the Laplacian plot, where \(\mathrm{D}\) represents the degree matrix, D the mid-diagonal elements \({\text{D}}_{{\text{ii}}} = \sum_{{\text{j}} = 1}^{{\text{l}} + {\text{u}}} {{\rm{W}}_{{\text{ij}}} }\), and the rest are 0 in matrix D.
From formula (1), the framework of Laplacian support vector machine (LapSVM) can be obtained:
The solution of the Eq. (3) is \({\text{f}}^{*} \left( {\text{x}} \right) = \sum\nolimits_{{\text{i}} = 1}^{{\text{l}} + {\text{u}}} {{\upalpha }_{\text{i}} {\text{K}}\left( {{\text{x}},{\text{x}}_{\text{i}} } \right)}\), introducing slack variables \({\upxi }_{\text{i}} ,{\text{i}} = 1, \ldots ,{\text{l}}\), Eq. (3) can be rewritten as:
Based on the Karush-Kuhn-Tucker (KKT) condition, the dual condition of Eq. (4) is as follow:
where \(Q = YJK(2\gamma_A I + (2\gamma_I /(u + l)^2 LK)^{ - 1} J^T Y\), J = [I 0] is a \(l \times (l + u)\) matrix of size \(l \times (l + u)\), I is the identity matrix, \(Y = diag(y_1 ,y_2 , \ldots ,y_l )\) and K is \((l + u) \times (l + u)\) the kernel matrix with the size of \((l + u) \times (l + u)\).
According to formula (5), it can be obtained that the solution of formula (4) is as follow:
2.2 Semi-supervised Classification Maximum Knowledge Utilization (SSC-EKE)
Semi-Supervised Classification Algorithm with Maximum Knowledge Utilization (SSC-EKE) is a semi-supervised learning algorithm [3], which effectively combines the manifold regularization mechanism of Laplacian support vector and fully mining useful knowledge in samples and a large number of unlabeled data samples to improve the classification performance, and achieved remarkable results.
On the basis of LAPSVM, SSC-EKE uses MS and CS to represent two collections----must-link and cannot-link, and then defines a matrix that is as follow:
After combining this matrix with the LAPSVM part, the definition of the joint regularization formula for the manifold and pairwise constraints can be obtained:
where \({\text{Z}} = {\text{H}} - {\text{Q}}\) and \(H = diag(Q \cdot 1_{(l + u) \times 1} )\), similar to the definition of L in LapSVM, so in this formula, the range of values for \({\text{f}}^{\text{T}} {\rm{L}}^{\prime}{\text{f}}\) and \({\text{f}}^{\text{T}} {\rm{Z}}^{\prime}{\text{f}}\) are same, \({\uptau } \in \left. {0,1} \right)\) is the dimension trade-off coefficient, which can control their individual significance in any data scenario.
Following the SVM minimum risk structure, the expression definition of SSC-EKE can be obtained from formula (6):
On the basis of Eq. (7), a bias term b is Introduced, so that Eq. (4) can be rewritten as:
Lagrange multiplier is expressed by, \({\upbeta } = \left( {{\upbeta }_1 , \ldots ,{\upbeta }_{\text{l}} } \right)\), and then according to the K KT condition, the dual form of formula (8) is as follow:
According to the solution of Eq. (9), the initial solution of Eq. (8) can be expressed as:
Ultimately, the final form of the classification decision function of the S SC-EKE algorithm is as follow:
2.3 Medical Image Registration Based on Phase Difference
In order to achieve the effect of registration without changing the overall topological information of the image, we use the phase difference between multiple CT image data to minimize the objective function in the differential homeomorphic space, so as to improve its accuracy and improve the effect of image registration for large deformation. The phase difference is the wavefront aberration extracted from several different images by using the wavefront detection technology.
This algorithm is mainly divided into three steps:
Deformation field calculation: transform the phase difference between the floating image and the reference image into deformation field by using orthogonal filter.
-
(1)
Deformation field calculation: Transform the phase difference between the floating image and the reference image into deformation field by using orthogonal filter. During each iteration, updating the deformation field (Du) is through the function (θ) calculated from the floating image (m) driven by the reference image (f) and the deformation field (Da), Δa and Δu represents deformation field Da, Du. The calculation form is:
$$ \rm{D}_{\rm{u}} \leftarrow {{\theta}}(\rm{f,m} \circ {\Delta a})\rm{D}_{\rm{a}} $$ -
(2)
Deformation field superposition: The differential homeomorphism algorithm is used to map the deformation field exponentially and add the new deformation field to the original deformation field. After calculating the deformation field, increase the total displacement Da by updating the field Du:
$$ \rm{D}_{\rm{a}} \leftarrow \Phi (\rm{D}_{\rm{a}} ,\rm{D}_{\rm{u}} ) $$ -
(3)
Deformation field regularization: The main purpose is to obtain smooth transformation and reduce the influence of image noise on the registration output. Its mathematical expression is: \( \rm{D}_{\rm{a}} \leftarrow \Psi (\rm{D}_{\rm{a}} ) \). Operation \( \Psi \) is achieved by applying a low-pass filter to each component of the deformation field. The specific implementation is carried out by using the convolution of the deformation field through the Gaussian kernel. At the same time, considering the deterministic mapping mentioned above, in order to ensure the high certainty of the position, the corresponding relationship between the displacement vector and its corresponding determined position is maintained, and the deterministic mapping is regularized as the deformation field mapping.
2.4 Pixel Weighted Average Image Fusion Algorithm
Multimodal medical image fusion is based on the registration of two or more different modes of medical images, which can achieve the purpose of synthesizing different modes of medical images [6].
The spatial domain fusion method is to directly process the pixel values of the image and fuse them in the gray space of the image pixels [7,8,9]. The spatial domain fusion methods suitable for medical image processing mainly include: the pixel gray value selection method, the pixel weighted average method [10], the pixel insertion method and the method based on Neural Network [4]. After comprehensive consideration of the effect and time of several algorithms, we choose to use the pixel weighted average method [5], which can process the fusion between different modal images in real time, and simply and intuitively express the characteristics of different modal images. The mathematical expression is as follows:
3 Experimental Details
3.1 Generate Analog CT
This article mainly discusses the accuracy of multimodal medical image registration with simulated CT as the intermediate medium, so we choose to use the current method with better effect to generate simulated CT of lower abdomen to generate simulated CT [8]. The following is a brief description of this method:
The first stage is to extract features from MR images. Because there is some noise in MR images, extracting feature data directly will have an unpredictable impact on subsequent experiments. Therefore, a filter matrix is set to smooth MR images to reduce the impact of noise. We obtained abdominal texture features from Dixon-water sequence, Dixon-fat sequence, Dixon-IP sequence and Dixon-OP sequence [11]. Considering the low bone and air signals in the mDixon sequence, the mesh generation strategy is used to obtain the location information. Finally, texture features and position information are combined to obtain seven dimensional feature vectors.
The second stage is to generate simulated CT. Read the circled data, adjust the super parameters by grid search and cross validation, and train eight SSC-EKE classifiers. After inputting the required image, about 400 points are extracted from it to obtain the average probability. Combined with the seven dimensional characteristics of the image, all voxel tissue types are estimated by KNN nearest neighbor method, and the corresponding CT values are given. The values of bone, fat, air and soft tissue were set as 380, −98, 700 and 32 [12] respectively.
3.2 Image Registration
Based on the simulated CT image generated in Sect. 3.1, this method changes the floating image into simulated CT, and the reference image is still the real CT image. The phase difference between simulated CT images and CT images will produce corresponding deformation on MR images.
The first step is to transform the phase difference between analog CT and real CT into deformation field by using orthogonal filter.
The second step is to use the differential homeomorphism algorithm to map the deformation field exponentially and superimpose the new deformation field with the original deformation field.
The third step is to use the Gaussian kernel to regularize the deformation field by using the convolution of the deformation field, so as to obtain a smooth transformation and reduce the impact of noise.
4 Experimental Results
4.1 Environment Configuration
The experimental environment and configuration of the system are as follows: the operating system is Windows 11, the CPU model is AMD Ryzen 54600H, the CPU memory size is 16 GB, the GPU model is NVIDIA Geforce GTX 1650, and the GPU memory size is 11 GB. The project operation effect may be different under different configurations.
4.2 A Subsection Sample
This chapter focuses on measuring the performance of the proposed method through performance indicators of some internationally recognized technical indicators. The specific performance indicators are shown in the following figure (Table 1):
The technical indicators used to evaluate the quality of the images after registration have the following specific meanings:
LP index: expressed as local phase difference, this index is used to describe the registration effect between two images. The lower the value is, the better the registration effect is.
CC index: indicates the degree of correlation between the image and the reference image and the degree of matching in the relative position between the images. It is also used to describe the registration effect between the image and the CT image, the closer a value is to 0, the better the registration.
MI index: indicates the mutual information between the image to be registered and the reference image, and is also used to describe the registration effect between the MR image and the CT image. The closer the value is to 1, the better the registration effect.
4.3 Evaluation of Results
We recruited 8 subjects using a protocol approved by the Cleveland Medical Center Institutional Review Board to obtain their lower abdomen as well as CT data. Using the known method to generate the corresponding simulated CT, and then based on the simulated CT multi-modality medical image registration, thus indirectly achieve the purpose of registration with CT.
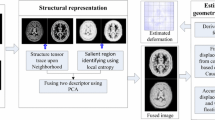
The following table shows the three index values of this method and the traditional multimodal registration method, through observation, we can find that the registration method based on simulated CT can achieve smaller phase difference and maximum mutual information than that based on local phase difference and maximum mutual information. It can be seen that the registration method based on simulated CT is an effective registration method (Fig. 1).
From Fig. 2, we can find that the multimodal registration method based on simulated CT can more accurately identify the position, shape and tissue and organs of the bones compared with the two traditional registration methods mentioned above. Competently meet the registration requirements of the lower abdomen.
5 Conclusion
In this paper, we introduce simulated CT as an intermediate medium for multimodal registration of MR and CT images, and select two traditional registration methods for comparison. The traditional multi-modal registration problem is transformed into single-modal registration, and the high-precision effect of single-modal registration is used to indirectly achieve high-precision registration between multi-modalities and obtain high-quality registration images. After comparing various fusion algorithms, we choose the pixel weighted average algorithm, which is simple, intuitive and easy to implement. It can realize the fusion of two images with different degrees of fusion, and the fusion effect is better.
References
Alafeef, M., Fraiwan, M.: On the diagnosis of idiopathic Parkinson’s disease using continuous wavelet transform complex plot. J. Ambient Intell. Hum. Comput. 10, 2805–2815 (2019). https://doi.org/10.1007/s12652-018-1014-x
Iizuka, S., Simo-Serra, E., Ishikawa, H.: Globally and locally consistent image completion. ACM Trans. Graph. (TOG) 36, 1–14 (2017)
Guo, K., Cao, R., Kui, X., Ma, J., Kang, J., Chi, T.: LCC: towards efficient label completion and correction for supervised medical image learning in smart diagnosis. J. Netw. Comput. Appl. 133, 51–59 (2019)
Raja, N.S.M., Fernandes, S.L., Dey, N., et al.: Contrast enhanced medical MRI evaluation using Tsallis entropy and region growing segmentation. J. Ambient Intell. Hum. Comput. (2018). https://doi.org/10.1007/s12652-018-0854-8
Alvén, J., Norlén, A., Enqvist, O., Kahl, F.: Überatlas: fast and robust registration for multi-atlas segmentation. Pattern Recogn. Lett. 80, 249–255 (2016)
Fernandez-de-Manuel, L., et al.: Organ-focused mutual information for nonrigid multimodal registration of liver CT and Gd–EOB–DTPA-enhanced MRI. Med. Image Anal. 18(1), 22–35 (2014)
Ahmad, S., Khan, M.F.: Multimodal non-rigid image registration based on elastodynamics. Vis. Comput. 34(1), 21–27 (2018)
Qian, P., Xi, C., et al.: SSC-EKE: semi-supervised classification with extensive knowledge exploitation. Inf. Sci.: Int. J. 422, 51–76 (2018)
Engelen, J.E.V., Hoos, H.H.: A survey on semi-supervised learning. Mach. Learn. 109, 373–440 (2019)
Wald, L.: Some terms of reference in data fusion. IEEE Trans. Geosci. Remote Sens. 37(3), 1190–1193 (1999)
Eggers, H., Brendel, B., Duijndam, A.: Dual-echo Dixon imaging with flexible choice of echo times. Magn. Reson. Med. 65(1), 96–107 (2011)
Schneider, W., et al.: Correlation between CT numbers and tissue parameters needed for Monte Carlo simulations of clinical dose distributions. Phys. Med. Biol. 45(2), 459–478 (2000). https://doi.org/10.1088/0031-9155/45/2/314
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Wang, X., Su, Y., Liu, R., Qu, Q., Liu, H., Gu, Y. (2022). Medical Image Registration Method Based on Simulated CT. In: Huang, DS., Jo, KH., Jing, J., Premaratne, P., Bevilacqua, V., Hussain, A. (eds) Intelligent Computing Methodologies. ICIC 2022. Lecture Notes in Computer Science(), vol 13395. Springer, Cham. https://doi.org/10.1007/978-3-031-13832-4_59
Download citation
DOI: https://doi.org/10.1007/978-3-031-13832-4_59
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-13831-7
Online ISBN: 978-3-031-13832-4
eBook Packages: Computer ScienceComputer Science (R0)