Abstract
Robotic cochlear implantation is an effective way to treat deafness and severe losses in hearing, which can reduce errors caused by human factors. It requires the pre-operative CT and intra-operative CBCT image registration to map the preoperatively computed drilling trajectory into the intra-operative space, and has extremely high requirements for registration speed and accuracy. At present, the research on the registration method is mature, while the evaluation method is not effective. The current evaluation metrics are mostly limited to the similarity, lacking of geometric information. Whereas in clinical surgery, we are more concerned with the target registration error (TRE). In this work, we complete the CT-CBCT registration by the commonly used intensity-based method and do the process with the open source tool Elastix. We do experiment on 2 cadaver head datasets with 8 screws implanted and 14 human head datasets. We calculate the centroid distance of the screws in CBCT image and registered CT image. Meanwhile, we use SIFT to extract key points in images and calculate the average Euclidean distance between corresponding points. Results show that the registration time is less than one minute. The average centroid distances of the screws in two cadaver heads are 0.19 mm and 0.12 mm, and the average Euclidean distances of the key points in two cadaver heads are 0.196 mm and 0.239 mm. TRE of all 16 datasets are within one voxel. The TRE calculated by SIFT key points is very close to the result obtained from implanted screws. We can use SIFT feature extraction method to evaluate the registration accuracy instead of implanting screws into the patient’s head during pre-operation period, which will greatly simplify surgical procedure and avoid unnecessary injury.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Thousands of people around the world suffer from cochlear damage which affects their hearing ability. Cochlear implant (CI) surgery is currently an effective method for the treatment for deafness and severe losses in hearing [1]. Doctors use a manual procedure, which costs time and effort, to get relevant information from medical images of the cochlea before CI surgery, and then work at the limits of their visual-tactile feedback and dexterity. Robotic cochlear implantation can reduce the instability of man-made operations in traditional surgery [2]. While automating this manual procedure is a challenging problem due to the small size and complicated structure of the cochlea. Combining cochlea images from different modalities using image registration and fusion techniques may help in the automation of cochlea structure identification, more accurate measurements of the cochlea, drilling path planning and implementation relevant to CI surgery.
Preoperative imaging is performed before CI surgery. A CT scan of the patient is done to identify ear anatomy and segment facial nerve [3]. An optimally safe drilling trajectory is computed based on the identified structures. During surgery, a Cone-beam computed tomography (CBCT) scan of the patient is obtained to help ensure the patient positioning and present the anatomy during treatment, which is a common on-treatment imaging method owning to its fast acquisition, cost-effectiveness, and low dose to patients. Registration between the planning CT and the intra-operative CBCT is crucial to match the drilling path and cochlear structures between two time points.
In general, the existing medical image registration methods are mainly classified into feature-based, intensity-based, segmentation-based and fluoroscopy-based [4]. Segmentation-based registration need to define a region of interest, and fluoroscopy-based registration is used for 2D-3D registration. So we mainly talk about the first two kinds. Feature-based registration has been used for CT-CBCT or other modalities image registrations in [5,6,7], it is computationally efficient, but the quality of registration largely depends on the accuracy of extracting and matching features. Usually, manual participation is required, even not, how to precisely match corresponding points is a problem to be solved. Intensity-based registration directly operate on voxel values, thus without manual intervention. This method achieves the purpose of aligning the two images by maximizing the similarity measures of the two images. These similarity measures include the sum of squared differences (SSD) for monomodal registration, mutual information (MI) or correlation coefficient (CC) for multimodal registration. CBCT intensity is inconsistent with CT due to artifacts from various sources such scatter, truncation. So even though CT and CBCT use the same imaging modality, X-ray, the CT-CBCT registration can be regarded as a special case of multimodal registration [8], where MI or CC is widely used. Even though intensity-based registration is widely used for its simplicity and easy-operability, it is limited by time and computing resources. Fortunately, with a hierarchical system (adoption of pyramid structure on images, eg. the method in [9]), computing time and memory can be greatly reduced. Furthermore, a large number of software have been presented for medical image registration based on intensity, such as ITK, 3D Slicer, Elastix or other commercial software. Most of them are based on C++ or shell language to improve coding performance and speed. In our study, we used Elastix for intensity-based CT-CBCT registration.
How to measure the registration effect is a more critical issue, because if there is no accurate value, no matter how advanced the algorithm is, it has no practical value. The most common method of performance evaluation is similarity measures between the whole images or outlined structures, it is the criteria to evaluate how much two or more images are similar. Among various similarity measures, root mean square error (RMSE) is the simplest, others include structural similarity index (SSIM), dice similarity coefficient (DSC), etc. While study shows that these measures, even when used in combination, cannot distinguish accurate from inaccurate registrations [10]. In addition, these measures often have no geometric significance. More reliable measure is target registration error (TRE). It evaluates the registration accuracy based on points correspondence, by computing Euclidean distance between corresponding points. It has more physical meaning, but how to choose and correspond to these points is a problem. In our study, we will comprehensively use the similarity measures and TRE to evaluate the registration error. Among them, the similarity measure is used to evaluate the overall registration effect, and the TRE is used to make up for the limitations of the similarity measures and provide a more intuitive judgment. We will discuss the methods in detail in the following sections.
2 Methods
In this section, we present a detailed description of the process we use to perform automatic registration of pre-operative CT images and intra-operative CBCT images. Most importantly, we propose specific evaluation metrics for the registration accuracy requirements, which needs to pay attention to in the actual surgery.
2.1 CT-CBCT Registration
Image Preprocessing.
The raw data has different voxel size and dimensions, and contains patient bed which may be a disruptive factor in subsequent registration processes. So firstly, we need to resample the data to the same resolution. And then, remove patient bed from CT and CBCT images, consist of intensity normalization, binarization, and morphological processing operations, as shown in Fig. 1.
Intensity-Based Image Registration.
The algorithm and the components of intensity-based image registration used in our study are described in the flowchart in Fig. 2.
The preprocessed images act as input images. When starting the iteration process, it is necessary to sample, that is to say, adopt a hierarchical strategy. If not, it is time-consuming for large images. Then, in each level of the pyramid, the images go into the registration process, computing the cost function, e.g. the advanced mattes’s mutual information (AMMI). The regular step gradient descent (RSGD) optimizer modifies parameters of the affine transform to minimize the cost function. When AMMI is maximum or it has reached the maximum iteration, optimization process ends, and outputs the transformation matrix. The transformation matrix is applied to the moving image to obtain the registered image.
We do this process using Elastix [11], which is an open-source intensity-based medical image registration software, based on the well-known Insight Segmentation and Registration Toolkit (ITK). The software allows the user to set various parameters to quickly configure, test, and compare different registration methods for a specific application. In previous studies, Elastix has been widely used for mono-modal or multi-modal, rigid or non-rigid registration [12,13,14,15], but rarely used for CT and CBCT registration.
2.2 Evaluation
For robotic cochlear implant surgery, the registration speed and accuracy are the issues we focus on, otherwise, a little carelessness can cause damage to the nerve around the cochlea, since for a facial nerve a margin of up to 1.0 mm is available and an accuracy of at least 0.3 mm is required, depending on the navigation system [16]. In terms of time, from the start of timing after reading the dicom image to the stop of generating the registration image or registration matrix, this time should be less than 2 min as a project metric. There is currently no convincing gold standard for measuring registration accuracy. As is mentioned above, similarity measures cannot always distinguish accurate from inaccurate registrations, only used as a reference indicator. At present, the more reliable, intuitive, and widely used evaluation metric is target registration error (TRE). While in our study, in order to comprehensively evaluate the registration results, we use both similarity measures and TRE to evaluate registration result.
For TRE, we evaluate registration accuracy from the following two aspects: on the one hand, segment a specific structure in the two images and calculate the distance of centroid distance [17]. To ensure the accuracy of the segmentation, we used implanted titanium screws as the target structure, because in the image, the brightness and contrast of the screws is much higher than the surrounding tissue. On the other hand, determine the corresponding points in the two images and calculate the average Euclidean distance. Points can usually be manually selected by experienced doctors, however, this is more affected by human factors and cannot be accurate to a single voxel. So we adopted the method of automatically selecting points, based on the SIFT feature operator. Then we filter out some of these landmarks based on manual experience. We will discuss these two aspects in detail in the sections below.
Similarity Metrics.
Similarity metrics include root mean square error (RMSE), correlation coefficient (CC), normalized mutual information (NMI), structural similarity index (SSIM). The calculation formula of each metric is as follows. X, Y represent the two images, \({x}_{i}\) and \({y}_{i}\) are the gray value of the \(\mathrm{i}\)th voxel, \({\mu }_{x}\) and \({\mu }_{y}\) are the mean gray value of the two images, \(\mathrm{p}\) stands for gray value distribution probability, \({\sigma }_{x}^{2}\) and \({\sigma }_{y}^{2}\) are the variances, and \({\sigma }_{xy}\) is the cross-covariance. \({C}_{1}\) and \({C}_{2}\) are regularization constants for the luminance and contrast respectively.
Screws Centroid Position.
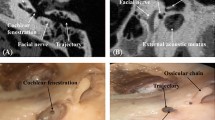
The TRE is defined as the mean Euclidean distance between the centroid of the eight corresponding screws implanted in the specimens. The local positions of eight screws are shown in Fig. 3. They can be easily identified using threshold segmentation [18]. Then centroid position in a volume image is calculated as the equation below, where g(i, j, k) is the gray value at the voxel (i, j, k).
Feature Points Extraction.
To compute the average distance between the points of the reference and the registered images, we need to extract feature points and match corresponding points. The scale invariant feature transform (SIFT) is invariant to rotation, scaling, and brightness changes [19]. So it is capable of extracting and matching stable and characteristic points between two images. The SIFT-feature-based registration has been used in [7, 20, 21]. In our study, we use SIFT feature not for registration, but for evaluation.
SIFT feature extraction includes extreme detection in scale space, keypoints localization, orientation and generating a features vector called “descriptors”. The whole process is shown in Fig. 4 and Fig. 5. Scale-space refers to the space formed by the convolution of a Gaussian function with the original image at different resolutions called ‘octave’. The general principle of extreme value detection is to find local extremes based on the difference of Gaussians (DoG) in each octave, as shown in Fig. 4(b). The points corresponding to these found extremes, comparing each voxel to its neighbors, are called keypoints, shown in Fig. 4(c). In order to match feature points in two images, the directions of keypoints should firstly be determined, that is, the direction in which the gray value decreases the fastest, and the gradient direction and amplitude of all voxels within a certain range with the feature point as the center of the circle are counted. The angle with the highest amplitude is the main direction (Fig. 4(d)), (in order to increase robustness, an auxiliary direction is usually determined). Rotate the image to the main orientation, calculate the gradient direction histogram of eight directions in sub-region, and draw the accumulated value of each gradient direction to for a seed point (Fig. 4(e)).
For registration accuracy, the most similar SIFT descriptors in reference images and registered images need to be identified (Fig. 5). We computed the nearest and the second nearest distance neighbor in two feature descriptors. If the ratio is below a threshold [7], the feature having the lowest distance value is chosen to corresponding points, and the value is TRE. Otherwise, no association is identified.
(a) Original image; (b) DoG in each octave: each column stands for a resolution (from left to right, the resolution decreases) and each row stands for a Gaussian coefficient (from top to bottom, the coefficients increase, and the image becomes blurry); (c) Keypoints detection. (d) Orientation; (e) Generate descriptors (one of descriptors is shown)
3 Experiments
3.1 Datasets
In this study, we conducted experiments on 16 pairs of pre-operative CT scans and intra-operative CBCT scans, of which 14 pairs were human data and 2 pairs were cadaveric data. Detailed data information is shown in Table 1. The institutional review board has approved this study.
3.2 Experimental Setup
Registration was done by using Melastix Toolbox, which is a collection of MATLAB wrappers for Elastix. Program runs on MATLAB version 2021a based on Intel(R) Core(TM) i7-9750H CPU (2.60 GHz, 2592 MHz, 6 cores, 12 logical processors).
3.3 Registration Parameters
The main registration parameters we use based on Elastix are shown in Table 2.
4 Results
Some visual results of the registration are shown in Fig. 6, showing three slices of corpse_head1 and corpse_head2, human4 and human10. From left to right, they are the reference images, registered images and fusion display of them.
In the fusion image map, gray regions have the same intensities, while magenta and green regions show where the intensities are different. For the reason that CT and CBCT have different gray value ranges, we can see most of the area is colorful. The images are aligned, which is reflected in the overlapping of magenta and green. Among them, the corpse_head2 was deformed due to the long soaking time and incorrect placement. We used affine transformation because in the actual operation, the head will not deform greatly.
The centroid distances of the eight screws implanted in the two cadaver heads are shown in Table 3, and Table 4 shows registration time, average distance of the corresponding feature points (AveDis) and similarity of the two cadaver heads. Human’s results are shown In Table 5.
To sum up, first of all, the registration speed depends largely on the hardware equipment. In our experiments, the entire registration process can be completed within one minute for all 16 sets of data. Secondly, in terms of accuracy, we calculated the similarity metrics, the centroid distance of the implanted screws and average distance of feature points. About the similarity metrics, the larger the value, the higher the grayscale similarity of the two. We found that the 14 human datasets are significantly higher than that of 2 corpse head datasets. One of the corpse heads was deformed due to the long soaking time, and the other had limited field of view during scanning, and part of the voxel information was missing. These may be the reasons for the low similarity metrics.
For corpse data, from Table 3 and Table 4, a good alignment is achieved during the registration process. The average centroid distance of implanted screws is 0.19 mm and 0.12 mm respectively, which can basically meet the requirements of surgical precision. In the extraction of corresponding feature points, key points are automatically extracted and matched based on SIFT. After obtaining the corresponding points, combined with manual experience, another selection is carried out to remove the obviously non-corresponding points on the image to ensure that the final results are not affected by individual abnormal points. Results show that TRE obtained by implanted titanium screws is very close to that obtained by SIFT feature points. So SIFT feature extraction can be used to replace titanium screws implanting during the pre-operation period for registration results evaluation, which will greatly simplify surgical procedure and avoid unnecessary injury.
For human data, the average feature points distances of 14 datasets are less than one voxel. Furthermore, as can be seen from Table 5, although the similarity metrics seem great, the TRE results do not coincide with them. Therefore, we cannot estimate registration quality only from the similarity, for it reflects the difference of the overall gray value, but the difference in image structure cannot be seen.
5 Conclusion
Cochlear implant surgery requires the registration of the pre-operative CT and intra-operative CBCT images to map the preoperatively computed drilling trajectory into the intra-operative space. For robotic surgery, registration speed and precision are especially important. In this paper, we use Elastix to perform intensity-based image registration, which can complete the entire process in one minute. In terms of accuracy, the similarity metrics cannot reflect the geometric difference characteristics, and is easily affected by the gray value. The target registration error of the two corpse head datasets are both below 0.3 mm, whether it is the distance of the screw centroid or feature points. We also find that results of implanted titanium screws and SIFT feature points are very close. So SIFT feature extraction can be used to replace titanium screws implanting during the pre-operation period for registration results evaluation, which will greatly simplify surgical procedure and avoid unnecessary injury. For the 14 human datasets, the similarity metrics are relatively high, and the average point distance is less than one voxel size, which is a reasonable result of image registration. In clinical surgery, when high registration distance accuracy is required, high resolution image should be obtained correspondently.
References
Eshraghi, A.A., Nazarian, R., Telischi, F.F., Rajguru, S.M., Truy, E., Gupta, C.: The cochlear implant: historical aspects and future prospects. Anat. Rec. (Hoboken) 295, 1967–1980 (2012)
Caversaccio, M., et al.: Robotic middle ear access for cochlear implantation: first in man. PLoS ONE 14, e0220543 (2019)
Dong, B., Lu, C., Hu, X., Zhao, Y., He, H., Wang, J.: Towards accurate facial nerve segmentation with decoupling optimization. Phys. Med. Biol. 67, 065007 (2022)
Alam, F., Rahman, S.U., Ullah, S., Gulati, K.: Medical image registration in image guided surgery: issues, challenges and research opportunities. Biocybern. Biomed. Eng. 38, 71–89 (2018)
Xie, Y.Q., Chao, M., Lee, P., Xing, L.: Feature-based rectal contour propagation from planning CT to cone beam CT. Med. Phys. 35, 4450–4459 (2008)
Koutouzi, G., Nasihatkton, B., Danielak-Nowak, M., Leonhardt, H., Falkenberg, M., Kahl, F.: Performance of a feature-based algorithm for 3D–3D registration of CT angiography to cone-beam CT for endovascular repair of complex abdominal aortic aneurysms. BMC Med. Imaging 18, 42 (2018)
Paganelli, C., et al.: Scale invariant feature transform in adaptive radiation therapy: a tool for deformable image registration assessment and re-planning indication. Phys. Med. Biol. 58, 287–299 (2013)
Park, S., Plishker, W., Quon, H., Wong, J., Shekhar, R., Lee, J.: Deformable registration of CT and cone-beam CT with local intensity matching. Phys. Med. Biol. 62, 927–947 (2017)
Xu, P., Yao, D.: A study on medical image registration by mutual information with pyramid data structure. Comput. Biol. Med. 37, 320–327 (2007)
Rohlfing, T.: Image similarity and tissue overlaps as surrogates for image registration accuracy: widely used but unreliable. IEEE Trans. Med. Imaging 31, 153–163 (2012)
Klein, S., Staring, M., Murphy, K., Viergever, M.A., Pluim, J.P.: elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010)
Ishida, T., et al.: Evaluation of performance of pelvic CT-MR deformable image registration using two software programs. J. Radiat. Res. 62, 1076–1082 (2021)
Broggi, S., et al.: A comparative evaluation of 3 different free-form deformable image registration and contour propagation methods for head and neck MRI: the case of parotid changes during radiotherapy. Technol. Cancer Res. Treat. 16, 373–381 (2017)
Styner, M.A., et al.: ACIR: automatic cochlea image registration. In: Medical Imaging 2017: Image Processing (2017)
Chiaruttini, N., Burri, O., Haub, P., Guiet, R., Sordet-Dessimoz, J., Seitz, A.: An open-source whole slide image registration workflow at cellular precision using Fiji, QuPath and Elastix. Front. Comput. Sci. Switz 3, 8 (2022)
Schipper, J., et al.: Navigation as a quality management tool in cochlear implant surgery. J. Laryngol. Otol. 118, 764–770 (2004)
So, R.W.K., Chung, A.C.S.: A novel learning-based dissimilarity metric for rigid and non-rigid medical image registration by using Bhattacharyya Distances. Pattern Recogn. 62, 161–174 (2017)
Wang, M.Y., Maurer, C.R., Fitzpatrick, J.M., Maciunas, R.J.: An automatic technique for finding and localizing externally attached markers in CT and MR volume images of the head. IEEE Trans. Biomed. Eng. 43, 627–637 (1996)
Lowe, D.G.: Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 60(2), 91–110 (2004)
Rister, B., Horowitz, M.A., Rubin, D.L.: Volumetric image registration from invariant keypoints. IEEE Trans. Image Process. 26, 4900–4910 (2017)
De Silva, T., Hotaling, N., Chew, E.Y., Cukras, C.: Feature-based retinal image registration for longitudinal analysis of patients with age-related macular degeneration. In: Proceedings of SPIE (2020)
Acknowledgments
This work was supported in part by the National Key Research and Development Program of China under Grant 2019YFB1311800, in part by the Fundamental Research Funds for the Central Universities under Grant 2021FZZX002-19, in part by the Major Scientific Project of Zhejiang Lab under Grant No. 2020ND8AD01, and in part by the Youth Innovation Team Project of the College of Biomedical Engineering & Instrument Science, Zhejiang University.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Lu, C., Dong, B., Hu, X., Zhao, Y., He, H., Wang, J. (2022). Preoperative CT and Intraoperative CBCT Image Registration and Evaluation in Robotic Cochlear Implant Surgery. In: Yang, G., Aviles-Rivero, A., Roberts, M., Schönlieb, CB. (eds) Medical Image Understanding and Analysis. MIUA 2022. Lecture Notes in Computer Science, vol 13413. Springer, Cham. https://doi.org/10.1007/978-3-031-12053-4_9
Download citation
DOI: https://doi.org/10.1007/978-3-031-12053-4_9
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-12052-7
Online ISBN: 978-3-031-12053-4
eBook Packages: Computer ScienceComputer Science (R0)