Abstract
Identifying cells in microscopic images is a crucial step toward studying image-based cell biology research. Cell instance segmentation provides an opportunity to study the shape, structure, form, and size of cells. Deep learning approaches for cell instance segmentation rely on the instance segmentation mask for each cell, which is a labor-intensive and expensive task. An ample amount of unlabeled microscopic data is available in the cell biology domain, but due to the tedious and exorbitant nature of the annotations needed for the cell instance segmentation approaches, the full potential of the data is not explored. This paper presents a weakly supervised approach, which can perform cell instance segmentation by using only point and bounding box-based annotation. This enormously reduces the annotation efforts. The proposed approach is evaluated on a benchmark dataset i.e., LIVECell, whereby only using a bounding box and randomly generated points on each cell, it achieved the mean average precision score of 43.53% which is as good as the full supervised segmentation method trained with complete segmentation mask. In addition, it is 3.71 times faster to annotate with a bounding box and point in comparison to full mask annotation.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
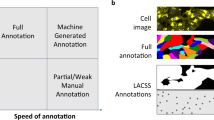
Point2Mask-based instance annotation combines object bounding boxes with points that are sampled randomly inside each box and annotated as the cell (blue) or background (red). We demonstrate that 6 annotated points per instance are faster to collect than the standard cell masks and such groundtruth is sufficient to train the proposed pipeline to achieve 99.2% of its fully supervised performance on the LIVECell dataset. (Color figure online)
Cell segmentation is regarded as the cornerstone of image-based cellular research. Studying cell migration, cell count, cell proliferation, cell morphology, cellular interactions, and cellular events like cell death are all possible with adequate cell segmentation. Deep learning approaches for instance cell segmentation [3, 7, 8, 16, 17, 19, 20] are showing promising results, but they rely heavily on precise full mask supervision for training. Manually annotating a groundtruth mask for each cell is a very labor-intensive, expensive, complex, and time-consuming task. For the natural object dataset like COCO [10], it takes on average 79.2 s per instance to create a polygon-based object mask. The bounding box for the objects is approximately 11 times faster i.e., 7 s [13]. When it comes to image-based cellular research, the LIVECell dataset [3] is the largest dataset of its kind to date. LIVECell is composed of more than 1.6 million cells. On average it contains more than 313 cells per image, which is way more than any other label-free cell segmentation dataset [17, 19]. Some images in the LIVECell dataset contain more than 3,000 cell instances, which can be overly complex, time-consuming, and labor-intensive to manually annotate each cell in a high cell density environment with overlapping cells. Annotating cells in microscopic images is more challenging than the objects in natural images [10] because cells overlap, and the cell boundaries are also very difficult to identify in crowded images. When preparing LIVECell, it took 46 s on average to create segmentation masks, which if we consider the total number of cells in the training data for the LIVECell dataset is more than 13,213 h spent on annotating the masks.
It is important to mention that LIVECell dataset (which is the largest annotated microscopic cell dataset) contains only 8 type of cells which is only a fraction of more then 200 different cells types found in human body [14]. This means that an ample amount of unlabeled image-based cellular data is available in the cell biology domain, but due to the tedious and exorbitant nature of annotations required for the cell instance segmentation approaches, the data is not being used to its full potential. To boost the research in cell biology, it is pivotal to have high-performing systems, which can accurately segment cells and for these methods, it is necessary to have a large number of labeled datasets, which are unfortunately labor-intensive. To tackle that issue, we have proposed a pipeline for weakly supervised cell segmentation, Point2Mask, which considers the bounding box for each cell and the point labels instead of the full mask. The point labels are sampled randomly inside each bounding box as shown in Fig. 1. The annotation required for the proposed Point2Mask can be divided into three steps. First, the bounding boxes need to be drawn, which takes \(\sim \)7 s per cell. After that, random point annotations inside each bounding box are automatically generated. As the last step, random points generated inside each bounding box are classified by an annotator as belonging to the foreground (cell) or background, which takes around \(\sim 0.9\) seconds per point. Table 1 provides insights into the annotation time required for different supervision types. If we only consider a single point for each cell and the bounding box for training, it takes 17.17% of the total time spent on the full mask annotation for all the cells in the LIVECell dataset and is 5.82x faster. For 6 points per cell type, it takes 26.96% of the fully supervised annotation time. The main contributions of this study are as follows:
-
1.
An end-to-end pipeline for weakly supervised point-based cell segmentation using Mask R-CNN [5], Feature pyramid Network with ResNet-50 [6], and bilinear interpolation.
-
2.
Extensive evaluation of the proposed method by increasing point labels for each cell instance to analyze the impact on the performance. Achieved 96.51% to 99.16% of the fully supervised performance using Point2Mask weakly supervised cell segmentation with only 1- to 6-points label per cell instance with a significant reduction in the time required for annotating the data for training.
-
3.
Performed per cell type evaluation to analyze the relationship between the morphological characteristics of different cell cultures like size and the number of point labels required.
2 Related Work
Deep learning-based cell segmentation has evolved drastically in the last decade with the development of the U-net proposed by Ronneberger et al. [16] in 2015. With only 35 images trained U-net model, it outperformed all the other contestants in the 2015 ISBI cell tracking and segmentation challenge. The success of U-net prompted a chain of valuable research in the image-based cellular research with the development of algorithms like DeepCell [22] and Usiigaci [20]. Khalid et al. (2021) [7] proposed a pipeline for cell and nucleus segmentation using the EVICAN dataset [17]. Edlund et al. (2021) proposed anchor-free and anchor-based pipelines for the cell segmentation using the LIVECell dataset [3]. Khalid et al. (2021) [8] proposed a pipeline to perform cell-type aware segmentation in microscopic images using the EVICAN dataset.
Weakly supervised cell segmentation is an active area of research with many different variations of the weak supervision i.e., image tags [12, 25], points [2, 24], missing annotations [4]. Zhou et al. (2018) [25] proposed a promising method for weakly supervised instance segmentation using only class labels of objects appearing in an image. Although this work does not primarily concern itself with cell instance segmentation but object segmentation in general, the approach was also tested on microscopy images and some underlying ideas were developed further to fit the domain [12]. In this method, image regions that produce a particularly high prediction response for a class called class peak responses are backpropagated through a network and mapped to object regions that are high in information. This procedure then allows for full instance masks to be retrieved. Another popular method to make use of weak labels for cell segmentation is using point annotations instead of full pixel-wise mask annotations. Zhao et al. (2020) [24] propose weakly supervised training schemes that only use point annotations to achieve results comparable to those of fully supervised models. In their paper, they propose three distinct methods and compare them to several baseline methods, such as U-Net [16] and the Pyramid-Based fully convolutional network [18]. The first approach, a self-training scheme, updates the output segmentation mask by feeding back the current prediction of the network. For this task, the network is pre-trained using the initial point annotations and a cross-entropy loss, and then a self-training loss is introduced which composes the network’s previous prediction with the previous label and uses it as a new label in a feedback loop. The second approach is a co-training scheme that uses two subsets of the initial dataset and self-trains two networks on them separately. The resulting models then supervise each other’s learning process, guided by a newly defined co-training loss that combines the predictions of both models. A third approach is a hybrid approach, leveraging the advantages of the better-converging self-training approach and the potentially better segmentation results of the co-training scheme. Guerrero-Pena et al. (2019) [4] introduce a method to tackle the frequent problem of missing or incorrect annotations in microscopy images. The method introduced in the paper proposes three key points to improve the effectiveness of deep learning models when trained on incomplete annotation. The first point is to introduce a loss function that helps separate cells by operating in three distinct classes and classifying underrepresented regions. The second point is introducing a weight-aware map model which is especially useful for contour detection and generalization. The third point consists of data augmentation specifically crafted for the weaknesses of a typical microscopy dataset, i.e. strengthening potentially weak signals on edges by adjusting the intensity of regions that contain shared edges of multiple cells.
All these approaches for weakly supervised cell segmentation are trained on small scale datasets like the PHC [11, 21] and Phase100 [23] dataset used in [24], contains 230 and 100 images respectively. This amount of data is too small to enable a trained CNN (Convolutional Neural Network) model to generalize to images beyond its training dataset or for a valid comparison between different supervision approaches. In addition to that, these approaches for weakly supervised cell segmentation are overly complex.
3 Point2Mask: The Proposed Approach
Figure 2 provides a system overview of our Point2Mask weakly supervised cell segmentation approach. The proposed pipeline is composed of Feature Pyramid Network [9] with ResNet-50 [6], Region Proposal Network, and Mask R-CNN [5] as the prediction head, which is detailed below.
3.1 Backbone Network for Feature Extraction
The purpose of this block is to extract feature maps from the input image at different scales. The feature extraction module of the proposed methodology is composed of Feature Pyramid Network [9] along with ResNet-50 [6]. Feature Pyramid Network (FPN) extracts features from the images using a pyramid scheme. It utilizes deep convolutional networks (CNNs) for computing features. FPN combines low resolution, semantically strong features with high resolution, semantically weak features. It takes a single-scale image as an input and outputs feature maps of proportional size at multiple levels by operating on a bottom-up pathway, top-down pathway, and lateral connections. The bottom-up pathway uses a normal feed-forward CNN architecture to compute a hierarchy of features consisting of feature maps at various scales. The output of each CNN layer is used later in the top-down pathway via lateral connections. The output of each convolution layer of ResNet-50 is used in the top-down pathway which constructs higher resolution layers from the semantic rich layer. As the final task, the FPN applies a 3\(\,\times \,\)3 convolution operation on each merged map to overcome the aliasing effect after the upsampling to generate the final feature map.
3.2 Region Proposal Network for Cell Region Detection and Groundtruth Matching
Following the extraction of multi-scale features from the backbone network, these features are then passed onto a Regional Proposal Network (RPN) [15]. The primary focus of RPN is to detect regions that contain objects and match them to the groundtruth. This process is performed by generating anchor boxes on the input image which are then matched to the groundtruth by taking Intersection over Union (IoU) between anchors and groundtruth. If IoU is larger than the defined threshold of 0.7, the anchor is linked to one of the groundtruth boxes and assigned to the foreground. If the IoU is greater than 0.3 and smaller than 0.7, it is considered background and otherwise ignored. The anchor strides and aspect ratio parameter used to detect and segment objects in MS-COCO [10] dataset overlooks most of the small cell instances when transferred to this task. Unlike MS-COCO [10] and other commonly used image datasets, the area of some cells especially BV-2 cell culture in the LIVECell [3] dataset is exceedingly small. After extensive experimentation, the anchor sizes and anchor aspect ratios were selected that fit adequately for the task. The details about the anchor parameters are given in Sect. 6. Now that we have the anchor boxes which are assigned to the foreground having shapes like the groundtruth boxes, the next step is anchor deltas calculation which is the distance between groundtruth and anchors. At the final stage of RPN, we choose 3,000 region proposal boxes from the predicted boxes by using non-maximum suppression [1].
3.3 Prediction Head
After the successful generation of proposals, the next block in our pipeline is the prediction head. At the prediction head, we have groundtruth boxes, proposal boxes from RPN, and feature maps from FPN. The job of the prediction head is to predict the class, bounding box, and binary mask for each region of interest. We are using Mask R-CNN [5] as the prediction head, which is an extension of Faster R-CNN [15] by adding a mask branch. Faster R-CNN gives two outputs for each object in an image, classification of the object in an image, and a bounding box around the object. In Mask R-CNN, a third branch is added that outputs an object mask in addition to the other two outputs. The extra branch is composed of Fully Convolutional Network (FCN) which predicts the mask for each RoI in a pixel-to-pixel manner.
In a fully supervised training setting with a mask available for each cell, Mask R-CNN is trained by extracting a matching regular grid of labels from groundtruth masks. On the contrary, for point supervision, predictions are approximated in the locations of groundtruth points from the prediction on the grid using bilinear interpolation (see Fig. 3). Bilinear interpolation is a resampling method that estimates a new pixel value by using the distance weighted average of the four nearest pixel values. When we have prediction and the groundtruth labels at the same points, similar loss as with full supervision can be applied and its gradient will be propagated with bilinear interpolation. Once we have predictions and groundtruth labels at the same points, a loss can be applied in the same way as with full supervision and its gradients will be propagated through bilinear interpolation. In our experiments, we use cross-entropy loss on points.
Point2Mask supervision illustration. For a 6\(\,\times \,\)6 prediction mask on the regular grid (green color indicates foreground prediction i.e., cell), the predictions are obtained at the exact location of the groundtruth points (Cell and the background groundtruth points are indicated by red and blue respectively) with bilinear interpolation. The cell contour line is only for illustration purposes. (Color figure online)
4 Dataset
LIVECell dataset [3] has been used in this study, which is the largest fully annotated dataset in image-based cellular research. It contains more than 1.6 million cells in 5,239 images. The images in the dataset are from eight morphologically distinct cell cultures. On average, the LIVECell dataset contains 313 cells per image which is exceedingly high as compared to the EVICAN dataset [17], which contains an average of 5.7 cells per image. That is the reason we opted for the LIVECell dataset for this study. LIVECell train set contains 3,188 images with over 1.03 million cell instances. The validation and the test data contain 539 and 1,512 images with 1,84,371 and 4,67,874 instances, respectively.
For fully supervised training, original LIVECell data with full masks are used for training. For Point2Mask, the mask from the LIVECell dataset is discarded and six different point labels (1, 2, 4, 6, 8, 10) are generated automatically and randomly for each cell of the training data. The point can either be on the cell or anywhere inside or on the edge of the bounding box. If the point annotation is on the cell, it is assigned a point label of 1, and otherwise 0.
5 Evaluation Metrics
To evaluate the performance of the proposed pipeline we are following the standard COCO evaluation protocol [10] with some modifications as reported in [3] for the area ranges. Average Precision (AP) is the precision averaged across all unique recall levels. Mean Average Precision (mAP) is the mean of average precision across all N classes. For the evaluation, we have reported mean average precision for both object detection and segmentation tasks at different IoU thresholds of 0.5 (mAP50), 0.75 (mAP75), and 0.5:0.95 in the steps of 0.05 (mAP). To identify the performance of the model on objects of varied sizes, we have also included mAP for different area ranges. Objects with area less than 320 \(\upmu \mathrm{m}^2\) (corresponding to 500 pixels) belong to APs (small). APm (medium) is for the objects in area ranges of 320 \(\upmu \mathrm{m}^2\) to 970 \(\upmu \mathrm{m}^2\) (corresponding to 1500 pixels) and APl (large) is for objects with area larger than 970 \(\upmu \mathrm{m}^2\).
6 Experimental Setup
We have designed two different experimental settings to evaluate the performance of the proposed pipeline for the point-supervised weak cell segmentation. In the first experimental setting, namely point2Mask vs fully supervised method and impact of validated annotated points, we have performed several experiments with different annotation supervisions using the LIVECell dataset. In the second experimental setting, namely impact of validated annotated points on different cell cultures, the models trained in the first experimental setting under different annotation supervisions are evaluated on test sets of individual cell cultures to analyze the performance of the different numbers of point annotations for each cell culture.
Training for all the experiments used a stochastic gradient descent-based solver with a base learning rate of 0.02 and momentum of 0.9. The anchor sizes and aspect ratios for all settings were set after careful consideration of the cell’s pixel area in the images. Anchor sizes and aspect ratios were set to 8, 16, 32, 64, 128, and 0.5, 1, 2, 3, 4 for all the settings, respectively. The checkpoints for evaluation were chosen based on the higher validation average precision.
The pixel means and pixel standard deviation for the dataset were calculated as 128 and 11.58, respectively. For data augmentation, images are flipped horizontally on a random basis to reduce the risk of over-fitting. All training used multi-scale data augmentation, meaning that image sizes were randomly changed from the original 520\(\times \)704 pixels to size with the same ratios, but the shortest side was set to one of (440, 480, 520, 580, 620) pixels.
6.1 Point2Mask vs Fully Supervised Method and Impact of Validated Annotated Points
In this experimental setting, the objective is to perform weakly supervised cell segmentation for different point annotations as well as fully supervised cell segmentation with a full mask for each cell. All the experiments are performed under the same settings. For point-supervised cell segmentation, six different training experiments are performed with 1-,2-,4-,6-,8-, and 10-point labels per cell instance instead of a full mask.
The checkpoints at 3,000 have been chosen for 1-, 10-points, and full mask training settings, and 2,9500 for 4-,6-, and 8-point training settings on the basis of higher validation average precision.
Results. Table 2 shows the overall detection and segmentation average precision scores of the proposed pipeline on the LIVECell dataset. For the full mask supervision setting, we are getting detection and segmentation AP scores of 43.12% and 43.90% respectively. The area ranges scores show that the model is performing best for the cells of larger areas. For the 1-point supervision, we are getting AP scores of 42.67% and 43.27% for detection and segmentation tasks, respectively. 1.01% improvement in performance is seen for 2-point supervision in comparison to the 1-point supervision. Similarly, 1.01% gain in performance is achieved for the 4-point supervision as compared to the 2-point supervision. For the 6-point supervision, we are getting the best results with an AP score of 43.53% for segmentation. For the 8- and 10- point supervision, we are getting a decline in the performance for cell segmentation.
6.2 Impact of Validated Annotated Points on Different Cell Cultures
In this experimental setting, we are mostly concerned with finding the interlink between the morphological properties of the cells and the number of point annotations required for each different cell culture. The models trained in experimental setting 1 are evaluated on the individual test set of each cell culture.
Results. Table 3 gives insights into per class AP scores for different point-supervised training settings. For the cell culture A172 and BT-474, the best performance is achieved by 6-point supervision. When we analyze the area of the A172 cells in the LIVECell dataset, it is observed that more than 50% of the cells belong to the medium area range (320 \(\upmu \mathrm{m}^2\) to 970 \(\upmu \mathrm{m}^2\)). The best performance is achieved by the 10-point supervision for the cell cultures Huh7 and SK-OV-3 because more than 48% and 59% of the cells in these cell cultures respectively have cells in a large area range (larger than 970 \(\upmu \mathrm{m}^2\)). For the cell culture BV-2, the best performance is seen across the 6-point supervision, but the interesting thing to notice is that for the 1-point supervision, we are getting 99.5% of 6-point annotation performance with 6x less time spent on the annotation. From these observations, we can conclude that the morphological characteristics like the size of the cells in the dataset can give insights into how many points are enough to achieve the best performance for each cell culture.
7 Analysis and Discussion
Inference results using the models trained on the different number of point annotations and full mask. The solid yellow line represents the groundtruth mask for each cell and the dotted red line represents the prediction made by the model. The red, green, blue, and purple columns represent the inference results obtained from the models trained on 1,2,6,10-point annotations and full mask respectively. Each row represents the inference result from an image from different cell culture. (Color figure online)
In this section, we discuss the results of the point-supervised weak cell segmentation pipeline for both experimental settings. In experimental setting 1 (Point2Mask vs Fully supervised method and impact of validated annotated points), 6 different points and full mask annotation were used for training. Results in Table 2 show that we have achieved 96.51% to 99.16% of the fully supervised performance by using weakly supervised cell segmentation with only 1- to 6-points label per cell instance with a significant reduction in the time required for annotating the data for training. Figure 4 presents the mask AP scores on the LIVECell test set with a different number of points used for training. For the 1-point supervision (\(\mathcal {P}_{\textit{1}}\)), we have achieved a mask AP score of 42.37%, which is 96.51% of the fully supervised trained model performance under the same settings. Similarly, for 2- (\(\mathcal {P}_{\textit{2}}\)) and 4-point supervisions (\(\mathcal {P}_{\textit{4}}\)), we have achieved 97.63% and 98.34% of the full supervision performance. For the 6-point supervision (\(\mathcal {P}_{\textit{6}}\)), we have achieved the best performance in terms of mask AP with the score of 43.53%, which is 99.16% of the fully supervised performance. For the 8-(\(\mathcal {P}_{\textit{8}}\)) and 10-point(\(\mathcal {P}_{\textit{10}}\)) supervision, the performance starts to decline compared to 6-point(\(\mathcal {P}_{\textit{6}}\)) with mask AP scores of 43.41% and 43.40% respectively.
In experimental setting 2 (Impact of validated annotated points on different cell cultures), we aimed to find the connection between the morphological characteristics of the cells and the \(\mathcal {N}\) point supervision required to get the optimal performance. From the analysis of the results in Table 3, it can be seen that for the cultures which contain cells in the small area ranges like BV2, minimal point supervision yields optimal results. For the cells in the medium area ranges like A172, BT-474, and SkBr3, the best performance is achieved with 6-point supervision. Similarly, 10-point supervision outputs the best performance for the cell cultures in large area ranges like Huh7 and SK-OV-3. These findings can help the annotators and the biologists in targeted point annotation according to the morphological characteristics of the different cell cultures.
Figure 5 shows the inference results on some samples using the models trained on the different number of point annotations and full mask. The solid yellow lines are the groundtruth mask for each cell and the dotted red lines are the predictions made by the model. The red, green, blue, and purple columns are the inference results obtained from the models trained on 1-,2-,6-,10-point annotations, and full mask, respectively. Each row shows the qualitative performance of different supervisions on the identical image from different cell culture for comparison. AP50 on top of every prediction sub-image is the segmentation average precision score at the IoU threshold of 0.5. For the image in the first row belonging to cell culture A172, the 1-point supervision model performs best with an AP50 score of 97.29%. The best performance for the image in the second row (BV-2) is seen across the model trained with 6-point supervision. For the image in the third row belonging to the SH-SY5Y cell culture, the best performance is recorded against the 10-point supervision model. The last 2 images in the fourth and the fifth row belong to SkBr3 cell culture. The best performance for both the images can be seen against the model trained on 1- and 6-point supervision, respectively.
We have achieved close to the full supervision performance by reducing the time required to annotate the data by a significant amount compared to the full mask annotation. In this study, quality assurance time has not been considered for both the full mask and the point annotations. Quality assurance for point labels in overlapping cells in crowded images can sometimes take more time than drawing the full mask. Even with the 1-point supervision for training, we are getting more than 96% of the fully supervised performance. As explained earlier, annotation of cells in microscopic images is a very labor-intensive and expensive task and requires expert knowledge of the biomedical staff. One single image of the cell culture BV2 can contain up to 3,000 cell instances, which can be very time-consuming and complex to annotate. With the help of the proposed pipeline, we can annotate the data semi-automatically by using the proposed pipeline for weakly supervised cell segmentation to generate a mask for each cell, which can then be improved by the annotators in case of false positive or missed detection. Also, the findings of experimental setting 2 can help us decide how many point annotations are required for specific cell culture according to its morphological properties.
8 Conclusion
In this study, we have proposed a pipeline for weakly supervised cell segmentation using point annotations. Point2Mask generates a mask for the cell, providing just the bounding box and the point labels. With the help of the proposed pipeline, we have achieved 99.16% of the fully supervised performance with just 6-point labels instead of drawing a full mask. With only 0.84% loss in the performance compared to the fully supervised setup, significant amount of time required for the fully supervised training can be saved. The performance achieved for a 1-point label per cell instance e, 96.51%, is still adequate and can save an ample amount of time spent on labeling the full mask for each cell. The findings of this paper can help biologists and doctors to save enough time in labeling the data and can expedite the field of medicine and disease diagnosis to a great extent. With the help of the results in this study, we have proved that we can not only reduce the time and the cost required for the full annotation, but we can also reduce the amount of expert knowledge required from the biologists to draw the boundaries of each cell. An abundant amount of unlabeled image-based cellular data is available, which can be semi-automatically annotated using the proposed pipeline for weakly supervised cell segmentation. Furthermore, we have also pointed out the relationship between morphological characteristics of different cell cultures and the number of point annotations required. These findings can help biologists to design the targeted point annotation for specific cell cultures.
References
Cai, Z., Vasconcelos, N.: Cascade R-CNN: delving into high quality object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2018)
Chen, Z., et al.: Weakly supervised histopathology image segmentation with sparse point annotations. IEEE J. Biomed. Health Inform. 25, 1673–1685 (2020)
Edlund, C., et al.: Livecell-a large-scale dataset for label-free live cell segmentation. Nat. Methods 18, 1038–1045 (2021)
Guerrero-Peña, F.A., Fernandez, P.D.M., Ren, T.I., Cunha, A.: A weakly supervised method for instance segmentation of biological cells. In: Wang, Q., et al. (eds.) DART/MIL3ID -2019. LNCS, vol. 11795, pp. 216–224. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-33391-1_25
He, K., Gkioxari, G., Dollár, P., Girshick, R.: Mask R-CNN. In: Proceedings of the IEEE International Conference on Computer Vision (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2016)
Khalid, N., et al.: Deepcens: an end-to-end pipeline for cell and nucleus segmentation in microscopic images. In: 2021 International Joint Conference on Neural Networks (IJCNN). IEEE (2021)
Khalid, N., et al.: Deepcis: an end-to-end pipeline for cell-type aware instance segmentation in microscopic images. In: 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI). IEEE (2021)
Lin, T.Y., Dollár, P., Girshick, R., He, K., Hariharan, B., Belongie, S.: Feature pyramid networks for object detection. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2017)
Lin, T.-Y., et al.: Microsoft COCO: common objects in context. In: Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T. (eds.) ECCV 2014. LNCS, vol. 8693, pp. 740–755. Springer, Cham (2014). https://doi.org/10.1007/978-3-319-10602-1_48
Maška, M., et al.: A benchmark for comparison of cell tracking algorithms. Bioinformatics 30, 1609–1617 (2014)
Nishimura, K., Ker, D.F.E., Bise, R.: Weakly supervised cell instance segmentation by propagating from detection response. In: Shen, D., et al. (eds.) MICCAI 2019. LNCS, vol. 11764, pp. 649–657. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-32239-7_72
Papadopoulos, D.P., Uijlings, J.R., Keller, F., Ferrari, V.: Extreme clicking for efficient object annotation. In: Proceedings of the IEEE International Conference on Computer Vision (2017)
Regev, A., et al.: Science forum: the human cell atlas. Elife 6, e27041 (2017)
Ren, S., He, K., Girshick, R., Sun, J.: Faster R-CNN: Towards real-time object detection with region proposal networks. arXiv preprint arXiv:1506.01497 (2015)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). https://doi.org/10.1007/978-3-319-24574-4_28
Schwendy, M., Unger, R.E., Parekh, S.H.: Evican-a balanced dataset for algorithm development in cell and nucleus segmentation. Bioinformatics 36, 3863–3870 (2020)
Seferbekov, S., Iglovikov, V., Buslaev, A., Shvets, A.: Feature pyramid network for multi-class land segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops (2018)
Stringer, C., Wang, T., Michaelos, M., Pachitariu, M.: Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2020)
Tsai, H.F., Gajda, J., Sloan, T.F., Rares, A., Shen, A.Q.: Usiigaci: instance-aware cell tracking in stain-free phase contrast microscopy enabled by machine learning. SoftwareX 9, 230–237 (2019)
Ulman, V., et al.: An objective comparison of cell-tracking algorithms. Nat. Methods 14, 1141–1152 (2017)
Van Valen, D.A., et al.: Deep learning automates the quantitative analysis of individual cells in live-cell imaging experiments. PLoS Comput. Biol. 12, e1005177 (2016)
Zhao, T., Yin, Z.: Pyramid-based fully convolutional networks for cell segmentation. In: Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G. (eds.) MICCAI 2018. LNCS, vol. 11073, pp. 677–685. Springer, Cham (2018). https://doi.org/10.1007/978-3-030-00937-3_77
Zhao, T., Yin, Z.: Weakly supervised cell segmentation by point annotation. IEEE Transa. Med. Imaging 40, 2736–2747 (2020)
Zhou, Y., Zhu, Y., Ye, Q., Qiu, Q., Jiao, J.: Weakly supervised instance segmentation using class peak response. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2018)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this paper
Cite this paper
Khalid, N. et al. (2022). Point2Mask: A Weakly Supervised Approach for Cell Segmentation Using Point Annotation. In: Yang, G., Aviles-Rivero, A., Roberts, M., Schönlieb, CB. (eds) Medical Image Understanding and Analysis. MIUA 2022. Lecture Notes in Computer Science, vol 13413. Springer, Cham. https://doi.org/10.1007/978-3-031-12053-4_11
Download citation
DOI: https://doi.org/10.1007/978-3-031-12053-4_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-12052-7
Online ISBN: 978-3-031-12053-4
eBook Packages: Computer ScienceComputer Science (R0)