Abstract
Crop genomics and genetics have been radically progressed in this past decade, as a whole genome-wide sequencing era. Notwithstanding, it is still hard to exploit the genome sequence information in most of the highly polyploid crops, including hexaploid persimmon (Diospyros kaki). In this chapter, we focus on the genome sequences of a diploid persimmon, D. lotus, which is a close wild relative of D. kaki, and discuss their applications and contribution to evolutionary aspects. The whole-genome sequences of a male D. lotus were drafted recently with one of the next generation sequencing (NGS) technologies, PacBio sequencing, to be defined with 15 pseudomolecules consistent with the basic chromosome numbers of the genus Diospyros. As one of the representative characteristics in plant genomes, the Diospyros genome also underwent at least two genome-wide duplication events. One of the two, named Dd-α, is a lineage-specific duplication that occurred at approx. 60–70 million years ago. This duplication is thought to drive some lineage-specific neofunctionalization in both protein (trans-acting) and expression (cis-regulatory) functions, including an important transition into separated sexualities. Furthermore, the D. lotus genome sequences provided critical insights into sex chromosome evolution. The draft genome sequences have been already widely applied as an alternative reference of the D. kaki genome, in transcriptomic, genomic, and epigenomic analyses. Further exploration in the frontiers of various Diospyros genomes for comparative approaches would expand the possibilities of genomic analysis in persimmon, and shed light on more for the evolution of the genus Diospyros.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
5.1 Potential of Genome Information in the Wild Relatives of Persimmon
Recent progress in biotechnology, especially molecular biological techniques, has allowed various genetic and genomic approaches in non-model plant species, including tree crops. In tree crops, long juvenile phases generally prevent rapid selection of the bred lines, to which the marker-assisted selection (MAS) based on their genomic information has been proposed as a good solution since the 1990s (Ribaut and Hoisington 1998; Collard and Mackill 2008). In contrast to mostly diploid lineages, such as tree crops in the genus Citrus or the family Rosaceae, polyploid species have been thought to be hard to be studied with genetic or genomic approaches, due to its nature of genetic complexity and difficulty in genome sequencing. Persimmon, as a highly polyploid species (2n = 6X or 9X = 90 or 135), would also be an annoying target of genetic/genomic analysis. Notwithstanding, previous studies have managed to develop genetic markers mainly for the understanding of cultivar/variety differentiations (Luo et al. 1995; Badenes et al. 2003; Yonemori et al. 2008; Parfitt et al. 2015) or selection of specific traits, such as fruit astringency derived from proanthocyanidin accumulation (Kanzaki et al. 2001, 2010). While, in hexaploid species, it would be still quite hard to construct a genome-wide sequence database, at least with the current sequencing technology (although further next-generation techniques may easily enable that, considering the progress in these 10 years). For genome-wide analysis in hexaploid persimmon, one feasible approach would be the utilization of genomes in diploid close relatives. This concept would not be limited to persimmon, but has been acceptable to other polyploid crop species, such as strawberry (Shulaev et al. 2011) or sweet potato (Wu et al. 2018). Previous phylogenetic analyzes suggested that diploid Diospyros lotus and Diospyros oleifera would be candidates for the alternative whole-genome sequences resource (Yonemori et al. 2008). The use of D. lotus genome information indeed successfully resulted in the identification of genetic markers linked to the sex determination locus (Akagi et al. 2014) or of the region including the ASTRINGENCY locus (Nishiyama et al. 2018). Although we should be careful about the phenotypic differences in these wild relatives and persimmon (or D. kaki), alternative use of their genomes would often allow various objectives involving genetic research or actual molecular breeding in hexaploid persimmon.
Another importance of the genome information of diploid Diospyros species would be understanding the evolutionary scenario for persimmon. Recent draft genome sequencing in diversified crops has unveiled an “unexpected” (and “undesirable,” as well) possibility for the conservation of gene functions amongst the lineages. In contrast to animal taxa, plants have undergone frequent whole-genome duplication events (WGD) in lineage-specific manner (Van de Peer et al. 2017), which are thought to have provided opportunities for the appearance of new traits representing each species. For instance, in horticultural crops, functional differentiation between paralogs, which had been derived from WGD, resulted in the establishment of specific ripening characteristics in tomato fruits (The Tomato Genome Consortium 2012), specific oil composition in olive (Unver et al. 2017), and specific sex-determination system in kiwifruit (Huang et al. 2013; Akagi et al. 2018, 2019). These are also consistent with an evolutionary theoretical framework that the gene redundancy provided by the presence of duplicate copies allows one copy to be neofunctionalized without loss of the original function (Flagel and Wendel, 2009). In other words, proper (or reliable) draft genome sequences of each crop or its relatives would be indispensable for understanding their lineage-specific traits, which would often involve their commercial importance.
5.2 Draft Genome Sequences in D. Lotus
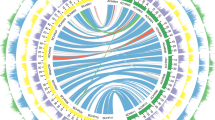
So far, a few of the genome sequences were drafted in diploid Diospyros, such as D. oleifera (introduced in Chap. 6; Zhu et al. 2019; Suo et al. 2020) or D. lotus. Here, I introduce the characteristics and evolution of D. lotus genome, often associating the sex determination systems (as related to Chaps. 9 and 10). The current version (on 31st Aug 2021) of the D. lotus cv. Kunsenshi-male (male, 2A + XY) draft genome (http://persimmon.kazusa.or.jp/), of which the expected haploid genome size is 907 Mb from flow cytometry (Tamura et al. 1998) or 877.7 Mb from kmer analysis in PacBio long-read data, are constituted of 3073 primary contigs totaling 746.1 Mb, and 5901 “secondary” contigs, which are putative allelic contigs to the primary contigs. With the segregating F1 populations (N = 314 and 119, Akagi et al. 2013), a total of 5,959 markers derived from GBS/ddRAD sequencing allowed anchoring of the ca 61.8% scaffolds into 15 pseudomolecules, which is consistent with the basic chromosome numbers in the Diospyros (Akagi et al. 2020). The draft genome (or primary contigs) includes 40,532 predicted gene locations, of which the numbers would be comparable to those in other asterid plant species, such as tomato (N = 34,879) (The Tomato Genome Consortium 2012) or kiwifruit (N = 39,040) (Huang et al. 2013).
Potential whole genome-wide duplications would be one of the important characteristics which would be detectable from mainly two indexes; (i) long syntenic relationships within the genomic regions, and (ii) distribution of silent divergence [or four-fold synonymous (degenerative) third-codon transversion (4DTv), to be more strict] in the duplicated paralogous gene pairs. Previous characterization of the genetic diversities and the distribution of silent divergence (Ks or dS) values in limited numbers of the genes in the genera Actinidia, Diospyros, and Camellia suggested lineage-specific genome-wide duplications in the genus Actinidia and Diospyros (Shi et al. 2010). Consistent with this assumption, in the D. lotus draft genome, the described two indexes suggested at least two genome-wide duplication events, which would correspond to the hexaploidization γ (Hex-γ) (Jaillon et al. 2007) common in the eudicot taxa, and a novel one, named Dd-α (Akagi et al. 2020) (Fig. 5.1). This result was also supported by comparative genomics in the genome drafting of tea (Camellia sinensis), which is nested into the order Ericale, as well as the genus Diospyros (Wang et al. 2021). While, the timing of the later lineage-specific genome-wide duplication (or Dd-α) might remain to be examined more because the species divergences in the ancestral state of the order Ericale is still ambiguously defined (Akagi et al. 2020; Wang et al. 2021). Importantly, the Dd-α occurred concurrently with previously reported whole-genome duplication events in the asterids (Huang et al. 2013; Iorizzo et al. 2016; Reyes-Chin-Wo et al. 2017), as well as across the angiosperms (Vanneste et al. 2014; Van de Peer et al. 2017), concentrated around the K-Pg (Cretaceous-Paleogene) boundary, which is hypothetically 60–70 million years ago (mya) (Fig. 5.2).
Distribution of silent divergences in the genome-wide homologous gene pairs (this panel is cited from Akagi et al. 2020 PLoS Genet 16(2): e1008566). The peaks correspond putative genome-wide (or very large) duplication events, in Vitis (purple), Actinidia (green), Diospyros (orange), and Solanum (red) genomes. The silent divergence would be a proxy age. For the Diospyros genome, the most representative peak, given as the asterisk, was detected almost concurrently with the Solanum-specific (tomato-specific) hexaploidization, named T-tri, which was supposed to occur ca 60–70 mya. The Hex-γ is thought to be a common event to the (eu)dicot species (Jaillon et al. 2007) including the described four species, while recent paleo-ploidizations would tend to mask the genomic traces of Hex-γ
Lineage-specific whole-genome duplication events in representative angiosperm. Putative whole genome-wide duplication events, given in open circles, and their time scales are referred from previous reports (Van de Peer 2009; Huang et al. 2013; Vanneste et al. 2014; Iorizzo et al. 2016; Reyes-Chin-Wo et al. 2017; Harkess et al. 2017; Van de Peer et al. 2017; Akagi et al. 2020). K-Pg, Cretaceous-Paleogene boundary
5.3 New Functions Established via the Lineage-Specific Genome-Wide Duplications
Paleo-genome duplication events, as described, would provide good chances to establish new gene functions, as functional redundancy of the duplicates can facilitate the plasticity, often resulting in subfunctionalization, neofunctionalization, or pseudogenization (Flagel and Wendel, 2009; Van de Peer et al. 2017). Neofunctionalization in expression pattern (or cis-evolution) would be quite simple to define if either of the duplicated pair exhibits novel expression behaviors. The evolution in cis-regulatory elements is thought to be a more rapid and fundamental reaction after duplication events, than that in trans-acting elements (Lynch and Conery 2000; Roulin et al 2013). Consistent with this theoretical framework, in D. lotus genome, the expression patterns of the duplicated pairs derived from the Dd-α showed substantially differentiated, even in focusing only on the expressions between male and female flower buds (Akagi et al. 2020). On the other hand, it might be hard to directly define neofunctionalization in trans-acting elements (or protein function) via duplication events. A potential index to call a novel beneficial function (for surviving) that was triggered by duplication, would be the transition of evolutionary rate (dN/dS values). New beneficial amino-acid mutations are often under positive selections (dN/dS ≪ 1.0), then followed by strong purifying selections (dN/dS ≪ 1.0) to be genetically fixed. Application of the model considering this situation, called “episodic positive selection,” to the D. lotus genome found some candidate genes that acquired a novel function immediately after the Dd-α whole genome-wide duplication event (Fig. 5.3). It is worthy to note that a sex-determining gene, MeGI (see Chap. 8 for sex-determination system in persimmon) derived from its paralogous gene, named Sister of MeGI (SiMeGI), was included in the list of the putatively neofunctionalized genes that underwent episodic positive selection after the Dd-α (Akagi et al. 2020). Importantly, this assumption has been experimentally validated with the transformation of Nicotiana tabacum, where constitutive induction of MeGI resulted in repressed androecium development, or feminization. This feminizing function is not conserved in the paralogous genes, SiMeGI, and its orthologous gene in the other plant species, such as barley (Komatsuda et al. 2007; Sakuma et al. 2013), Arabidopsis (González-Grandío et al. 2017), and maize (Whipple et al. 2011), and also tomato (Lin et al. 2008). Hence, detection of the adaptive evolution of a sex-determining gene, MeGI, by using evolutionary indexes would be a good example for exploitation of the whole-genome information to figure out lineage-specific traits or gene functions. Still, there would be massive untouched gene resources that possibly have evolved Diospyros-specific new functions via lineage-specific duplication events. Considering the potential of this frontier, rapid gene evaluation systems, such as with precocious flowering lines as in the case of kiwifruit nested into the same order Ericale (Varkonyi-Gasic et al. 2019), would be the next breakthrough.
Evolutionary model to detect episodic positive selections driven imeediately after the Dd-α. We targeted strong positive selection on either of the duplicates derived from the Dd-α (ω1 ≫ 1, on the thick pink branch), which was not undergone in the background branches and the counter part paralog (ω0 < 1). After establishment of an adaptive new function, the evolutionary rate should be reduced to be under purifying selection, with distinct selective patterns from the original orthologs (ω2 < 1, ω2 ≠ ω0, green triangle)
5.4 Evolution of the Sex Chromosome
The first finding of the sex chromosome in flowering plants was made independently in white champion (Silene latifolia, and Silene spp.), sorrel (Rumex acetosa), hop (Humulus lupulus), and so on, in 1923 (Kihara and Ono 1923; Winge 1923; Blackburn 1923). After that, sex chromosomes have been researched over a century, while yet their nucleotide sequence contexts and evolution of the male(or female in ZW system)-specific regions have been little assessed (Renner and Müller 2021). The genus Diospyros is mostly dioecious, except potentially polygamous polyploids or minorities, which are determined by an XY (or heterogametic male) system (see Chaps. 8 and 9). So far, no reports suggested heteromorphic sex (or XY) chromosomes in Diospyros, and thus, their male-specific region of the Y-chromosome (MSY) is thought to be very small (Akagi et al. 2014). The draft genome of D. lotus successfully anchored some pseudo-autosomal sex-linked scaffolds to chromosome 15 (Akagi et al. 2020). On the other hand, the MSY, including OGI, has not been anchored using genetic markers, presumably due to the large structural variation between the X and Y chromosome and the resultant Y-specific hemizygosity surrounding the OGI gene. Instead, with bacterial artificial chromosomes (BAC) walking started from the seed of OGI (Akagi et al. 2014), Y-chromosomal supercontigs covering most of the MSY were physically anchored (Akagi et al. 2020). The regions surrounding the OGI are male-specific (or hemizygous) and hyper-repetitive, often including palindrome-like structures. These sequence contexts are consistent with those of sex chromosomes in animal taxa (Bachtrog et al. 2014). In the outer regions of the MSY, putative pseudo-autosomal regions (PAR), which include both X- and Y-allelic genes, are partially scattered into the MSY, and dominantly appear only 200–300 kb apart from the single-sex determinant, OGI. Considering the relatively long history of the Y chromosome (or the sex determinant; ca 20–30 million years estimated from the putative silent divergence between MeGI and OGI) in Diospyros, such small MSY (up to 600–1000 kb) might be inconsistent with the conventional hypothesis for sex chromosome evolution in plants, where distinct MSY can form in only a few million years (Ming et al. 2011). On the other hand, recent characterization of various plant sex chromosomes is uncovering their evolutionary diversities, providing some examples for small MSY, such as in Populus, Vitis, and Actinidia, which would be consistent with the sex chromosome evolution in Diospyros (Renner and Müller 2021).
In the MSY of D. lotus, although the boundary to the PAR would be quite ambiguous, some genes were predicted in the hemizygous Y-allelic regions, while they are thought to be mostly the components of transposable elements, which is consistent with the MSY evolution in animal taxa. In the PAR-like sequences, which still include male-specific hemizygous islands, the silent divergence rate between the putative X and Y alleles of the genes is decreased, in inverse proportion to the distance from OGI. This situation is expectable, which reflects historical recombination between the perfectly conserved Y factor, OGI, and the surrounding regions, often forming evolutionary strata (Bergero et al. 2007). The actual dS rate in 1-Mb from OGI, ranges from approx. 0.2 (at OGI) to 0.07, which is comparable to the interspecific dS rate between D. lotus and distant Diospyros species (such as in D. mespiliformis, dS = ca 0.07 against D. lotus), implying suppressed recombination in the regions flanking OGI before the divergence of some Diospyros species. Although OGI is thought to arise via duplications from the ancestral MeGI (Akagi et al. 2014), long syntenic collinearity among the regions surrounding them is not conserved. This means that a local segmental duplication (or simple gene duplication) event followed by an inversion, triggered the establishment of the dioecious sex-determination system of this genus.
5.5 D. Lotus Genome Shed Light on the Insights into the Transition into Dioecy Associated with Duplication Events
Summarizing so far, the D. lotus genome information would raise the following hypothesis for the transition into dioecy in Diospyros. A lineage-specific WGD event, Dd-α contributed to derive the first twin, proto-MeGI, and SiMeGI, and the proto-MeGI specifically underwent positive selection to be neofunctionalized to act as a feminizing factor to dominantly suppress androecium development. This was followed by a segmental duplication to derive MeGI and a Y-encoded OGI, which dominantly suppresses the expression of MeGI (Akagi et al. 2014, 2020). It would be worthy to note that this type of evolutionary pattern would be reminiscent of potential generality or commonality for the establishment of dioecy in other plant species. In the establishment of dioecy in garden asparagus, the Y-encoded sex-determining gene, SOFF, is thought to have originated from an Asparagus-specific segmental or whole genome-wide duplication, which was followed by the acquisition of its function as a dominant suppressor of feminization (SuF) (Harkess et al. 2017). One of the two Y-encoded sex determinants in kiwifruit (Actinidia spp.), Shy Girl, which is also a dominant suppressor of feminization (Akagi et al. 2018, Varkonyi-Gasic et al. 2021), arose via an Actinidia-specific duplication event (Akagi et al. 2018, 2019), potentially involving one of the two Actinidia-specific WGD events, Ad-α (Huang et al. 2013). Furthermore, in the family Salicaceae, multiple Y-encoded sex determinants, which are mostly small-RNAs acting as dominant suppressors of the “master sex regulator” of ARR17, are independently derived from lineage-specific duplication events (Müller et al. 2020). These parallel paths towards the independent evolution of these sex determinants are probably not coincidental, but consistent with the theoretical framework described above. In flowering plants, transition into separated sexuality would require the appearance and selection of a gain-of-function event in order to acquire a dominant suppressor(s) (Charlesworth and Charlesworth 1978a, 1978b). Although a dominant suppressor would be hard to generate from a non-redundant (or single) gene due to the importance of its original function, while whole-genome duplication events provide good opportunities to relax purifying selective pressure for abundant genes to be neofunctionalized into dominant suppressors.
5.6 Application of the D. Lotus Genome Information and Future Prospects
One of the simplest usages of the D. lotus genome is as a reference for transcripts or genome mapping from D. kaki. As in many cases of the transcriptomic or whole-genomic assessments in polyploid species, alternative use of a diploid close relative for mapping would be an effective way to reduce the issues derived from their genomic/genetic complexities. Although the D. lotus genome information has not been opened until recently, some genome-wide studies already utilized that for the database in D. kaki. Transcriptomic analyses in D. kaki focusing on unlocked male flower production (Masuda et al. 2020a), fruit shape diversity among the cultivars (Maeda et al. 2019), or dwarfism involving gibberellin metabolism (Dong et al. 2021), applied the D. lotus whole-gene sets, and successfully found the candidate genes or molecular mechanisms. Considering the conservation of the genes and their homologies between D. lotus and D. kaki, transcriptomic approaches should have no serious issues. As a genomic reference map, Masuda et al. (2020b) mapped captured gDNA Illumina reads (ddRAD-Seq data, more specifically) from D. kaki segregated population to the D. lotus pseudomolecule and scaffold contigs, to identify genetic regions contributing to the bias in the female/male flower ratio. Importantly, the Y-chromosome (or the Y-encoded sex determinant, OGI) itself and/or its allelic dosage had a substantial association with the male flower ratio. The D. lotus genome sequences were derived from a male (2A + XY), and the Y-specific regions have been well examined as described, so that it would be especially suitable for the studies involving the persimmon sexualities. Epigenetic studies in D. kaki are also acceptable with the D. lotus genome as a reference. Fluctuation of the genome-wide DNA methylation levels, which is potentially associated with male flower production in a genetically female cultivar, were successfully characterized with D. lotus genome as the reference (Masuda et al. 2020c). On the other hand, if we target more “specific” sequences, such as short cis-regulatory elements, in D. kaki, the D. lotus genome might not be applicable, considering the evolutionary distance between them. The genome sequences of D. oleifera, which showed a little bit closer relationship to D. kaki than D. lotus (Yonemori et al. 2008), would be an alternative option, as described in Chap. 6. Still, we should be careful about the hexaploidy in D. kaki, which exhibited substantial sequence variations among the hexaplex alleles. Also, the evolutionary distances (or sequences homologies) between two species would be very flexible among the genomic regions, especially in tree crops, where it would be hard to define uniform distance per genome, mainly due to frequent introgressions (Cornille et al. 2012; Numaguchi et al. 2020). Case-by-case selection of a reference genome, depending on the research purposes, would be key. With the current technologies, construction of a draft genome with nearly perfect quality, such as chromosome-scale assembly, would be becoming quite easier for very cheaper costs. We suppose that it would be reasonable not to persist with the limited genomes information, but to explore the frontiers of other Diospyros genomes for various objectives. Comparative genomic or population genetic approaches with diversified Diospyros species would shed light on more evolutionary aspects, such as the domestication path of D. kaki.
References
Akagi T, Henry IM, Tao R, Comai L (2014) A Y-chromosome–encoded small RNA acts as a sex determinant in persimmons. Science 346(6209):646–650
Akagi T, Pilkington SM, Varkonyi-Gasic E, Henry IM et al (2019) Two Y-chromosome-encoded genes determine sex in kiwifruit. Nat Plants 5(8):801–809
Akagi T, Shirasawa K, Nagasaki H, Hirakawa H, Tao R, Comai L, Henry IM (2020) The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet 16(2):e1008566
Akagi T, Kajita K, Kibe T, Morimura H, Tsujimoto T, Nishiyama S, Kawai T, Tao, R (2013) Development of molecular markers associated with sexuality in Diospyros lotus L. and their Application in D. kaki Thunb J Jpn Soc Hort Sci, CH-109
Akagi T, Henry IM, Ohtani H, Morimoto T, Beppu K, Kataoka I, Tao R (2018) A Y-encoded suppressor of feminization arose via lineage-specific duplication of a cytokinin response regulator in kiwifruit. Plant Cell 30(4):780–795
Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, Perrin N, Ross L, Vamosi, Tree of Sex Consortium (2014) Sex determination: why so many ways of doing it?. PLoS Biol 12(7):e1001899
Badenes M, Garcés A, Romero C, Romero M, Clavé J, Rovira M, Llácer G (2003) Genetic diversity of introduced and local Spanish persimmon cultivars revealed by RAPD markers. Genet Resour Crop Evol 50(6):579–585
Bergero R, Forrest A, Kamau E, Charlesworth D (2007) Evolutionary strata on the X chromosomes of the dioecious plant Silene latifolia: evidence from new sex-linked genes. Genetics 175(4):1945–1954
Blackburn KB (1923) Sex chromosomes in plants. Nature 112(2819):687–688
Charlesworth B, Charlesworth D (1978a) A model for the evolution of dioecy and gynodioecy. Am Nat 112(988):975–997
Charlesworth D, Charlesworth B (1978b) Population genetics of partial male-sterility and the evolution of monoecy and dioecy. Heredity 41(2):137–153
Collard BC, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond B Biol Sci 363(1491):557–572
Cornille A, Gladieux P, Smulders MJ, Roldan-Ruiz I, Laurens F, Le Cam B, Nersesyan A, Clavel J, Olonova M, Feugey L, Gabrielyan I, Zhang XG, Tenalillon M, Giraud T (2012) New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet 8(5):e1002703
Dong Y, Ye X, Xiong A, Zhu N, Jiang L, Qu S (2021) The regulatory role of gibberellin related genes DKGA2ox1 and MIR171f_3 in persimmon dwarfism. Plant Sci 310:110958
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183(3):557–564
González-Grandío E, Pajoro A, Franco-Zorrilla JM, Tarancón C, Immink RG, Cubas P (2017) Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc Natl Acad Sci USA 114(2):E245–E254
Harkess A, Zhou J, Xu C, Bowers JE et al (2017) The asparagus genome sheds light on the origin and evolution of a young Y chromosome. Nat Commun 8(1):1–10
Huang S, Ding J, Deng D, Tang W et al (2013) Draft genome of the kiwifruit Actinidia chinensis. Nat Commun 4(1):1–9
Iorizzo M, Ellison S, Senalik D, Zeng P et al (2016) A high-quality carrot genome assembly provides new insights into carotenoid accumulation and asterid genome evolution. Nature Genet 48(6):657–666
Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Cassagrande A, Wincker P (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449(7161):463–467
Kanzaki S, Yonemori K, Sugiura A, Sato A, Yamada M (2001) Identification of molecular markers linked to the trait of natural astringency loss of Japanese persimmon (Diospyros kaki) fruit. J Am Soc Hortic Sci 126(1):51–55
Kanzaki S, Akagi T, Masuko T, Kimura M, Yamada M, Sato A, Mitani N, Ustunomiya N, Yonemori K (2010) SCAR markers for practical application of marker-assisted selection in persimmon (Diospyros kaki Thunb.) breeding. J Jpn Soc Hort Sci 79(2):150–155
Kihara H, Ono T (1923) Cytological studies on rumex L. II on the relation of chromosome number and sexes in rumex acetosa L. Bot Mag Tokyo 37:147–149
Komatsuda T, Pourkheirandish M, He C, Azhaguvel P et al (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci USA 104(4):1424–1429
Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D (2008) A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J 55(2):301–310
Luo ZR, Yonemori K, Sugiura A (1995) Evaluation of RAPD analysis for cultivar identification of persimmons. J Jpn Soc Hort Sci 64(3):535–541
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290(5494):1151–1155
Maeda H, Akagi T, Onoue N, Kono A, Tao R (2019) Evolution of lineage-specific gene netorks underlying the considerable fruit shape diversity in persimmon. Plant Cell Physiol 60(11):2464–2477
Masuda K, Akagi T, Esumi T, Tao R (2020a) Epigenetic flexibility underlies somaclonal sex conversions in hexaploid persimmon. Plant Cell Physiol 61(2):393–402
Masuda K, Fujita N, Yang HW, Ushijima K, Kubo Y, Tao R, Akagi T (2020b) Molecular mechanism underlying derepressed male production in hexaploid persimmon. Front Plant Sci 11:567249
Masuda K, Yamamoto E, Shirasawa K, Onoue N, Kono A, Ushijima K, Kubo Y, Tao R, Henry IM, Akagi T (2020c) Genome-wide study on the polysomic genetic factors conferring plasticity of flower sexuality in hexaploid persimmon. DNA Res 27(3):dsaa012
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Annu Rev Plant Biol 62:485–514
Müller NA, Kersten B, Leite Montalvão AP, Mähler N, Bernhardsson C, Bräutigam K, Lorenzo ZC, Hoenicka H, Kumar V, Mader M, Pakull B, Robinson KM, Sabatti M, Vettori C, Ingvarsson PK, Cronk Q, Street NR, Fladung M (2020) A single gene underlies the dynamic evolution of poplar sex determination. Nature Plants 6(6):630–637
Nishiyama S, Onoue N, Kono A, Sato A, Yonemori K, Tao R (2018) Characterization of a gene regulatory network underlying astringency loss in persimmon fruit. Planta 247(3):733–743
Numaguchi K, Akagi T, Kitamura Y, Ishikawa R, Ishii T (2020) Interspecific introgression and natural selection in the evolution of Japanese apricot (Prunus mume). Plant J 104(6):1551–1567
Parfitt DE, Yonemori K, Honsho C, Nozaka M, Kanzaki S, Sato A, Yamada M (2015) Relationships among Asian persimmon cultivars, astringent and non-astringent types. Tree Genet Genomes 11(2):24
Renner SS, Müller NA (2021) Plant sex chromosomes defy evolutionary models of expanding recombination suppression and genetic degeneration. Nat Plants 7(4):392–402
Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A et al (2017) Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat Commun 8(1):1–11
Ribaut JM, Hoisington D (1998) Marker-assisted selection: new tools and strategies. Trends Plant Sci 3(6):236–239
Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA (2013) The fate of duplicated genes in a polyploid plant genome. Plant J 73(1):143–153
Sakuma S, Pourkheirandish M, Hensel G, Kumlehn J, Stein N, Tagiri A, Yamaji N, Ma JF, Sassa H, Koda T, Komatsuda T (2013) Divergence of expression pattern contributed to neofunctionalization of duplicated HD-Z ip I transcription factor in barley. New Phytol 197(3):939–948
Shi T, Huang H, Barker MS (2010) Ancient genome duplications during the evolution of kiwifruit (Actinidia) and related Ericales. Ann Bot 106(3):497–504
Shulaev V, Sargent DJ, Crowhurst RN et al (2011) The genome of woodland strawberry (Fragaria vesca). Nat Genet 43(2):109–116
Suo Y, Sun P, Cheng H, Han W, Diao S, Li H, Mai Y, Zhao X, Fu J (2020) A high-quality chromosomal genome assembly of Diospyros oleifera Cheng. GigaScience 9(1):giz164
Tamura M, Tao R, Yonemori K, Utsunomiya N, Sugiura A (1998) Ploidy level and genome size of several Diospyros species. J Jpn Soc Hort Sci 67(3):306–312
Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485(7400):635
Unver T, Wu Z, Sterck L, Turktas M et al (2017) Genome of wild olive and the evolution of oil biosynthesis. Proc Natl Acad Sci USA 114(44):E9413–E9422
Van de Peer Y, Mizrachi E, Marchal K (2017) The evolutionary significance of polyploidy. Nat Rev Genet 18(7):411–424
Vanneste K, Baele G, Maere S, Van de Peer Y (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res 24(8):1334–1347
Varkonyi-Gasic E, Wang T, Voogd C, Jeon S, Drummond RS, Gleave AP, Allan AC (2019) Mutagenesis of kiwifruit CENTRORADIALIS-like genes transforms a climbing woody perennial with long juvenility and axillary flowering into a compact plant with rapid terminal flowering. Plant Biotechnol J 17(5):869–880
Varkonyi‐Gasic E, Wang T, Cooney J, Jeon S, Voogd C, Douglas MJ, Pilkington SM, Akagi T, Allan AC (2021) Shy Girl, a kiwifruit suppressor of feminization, restricts gynoecium development via regulation of cytokinin metabolism and signalling. New Phytol 230(4):1461–1475
Wang Y, Chen F, Ma Y, Zhang T, Sun P, Lan M, Li F, Fang W (2021) An ancient whole-genome duplication event and its contribution to flavor compounds in the tea plant (Camellia sinensis). Hortic Res 8(1):1–12
Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP (2011) grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci USA 108(33):E506–E512
Winge O (1923) On sex chromosomes, sex determination and preponderance of female in some dioecious plants. Comp Rend Trav Lab Carlsberg 15(5):1–25
Wu S, Lau KH, Cao Q et al (2018) Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement. Nat Commun 9(1):1–12
Yonemori K, Honsho C, Kanzaki S, Ino H, Ikegami A, Kitajima A, Sugiura A, Parfitt DE (2008) Sequence analyses of the ITS regions and the mat K gene for determining phylogenetic relationships of Diospyros kaki (persimmon) with other wild Diospyros (Ebenaceae) species. Tree Genet Genomes 4(2):149–158
Zhu QG, Xu Y, Yang Y, Guan CF, Zhang QY, Huang JW, Grierson D, Chen KS, Gong BC, Yin XR (2019) The persimmon (Diospyros oleifera Cheng) genome provides new insights into the inheritance of astringency and ancestral evolution. Hortic Res 6(1):1–15
Acknowledgements
We thank Dr. Deborah Charlesworth for the extensive discussions on the interpretation of D. lotus genome evolution, especially on the parts involving sex determination, and the many thoughtful pieces of advice provided through this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Akagi, T. (2022). Diospyros Lotus Genome. In: Tao, R., Luo, Z. (eds) The Persimmon Genome. Compendium of Plant Genomes. Springer, Cham. https://doi.org/10.1007/978-3-031-05584-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-031-05584-3_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-05583-6
Online ISBN: 978-3-031-05584-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)