Abstract
Increasingly, caregiving to senior citizens and patients requires monitoring of vital signs like heartbeat, respiration, and blood pressure for an extended period. In this paper, we propose a multimodal synchronized biological signal analysis using a deep neural network-based model that may learn to classify different anomalous patterns. The proposed cepstral-based peak fusion technique is designed to model the robust characterization of each biological signal by combining the list of dominant peaks in the input signal and its corresponding cepstrum. This works as an input to the following multimodal anomaly detection process that not only enables accurate identification and localization of aberrant signal patterns but also facilitates the proposed model to adopt an individual’s unique health characteristics over time. In this work, we use Electrocardiogram (ECG), Femoral Pulse, Photoplethysmogram (PPG), and Body Temperature to monitor an individual’s health condition. In both publicly available datasets as well as our lab-based study with 10 participants, the proposed cepstral-based fusion module attains around 7 to \(10\%\) improvement over the baseline of time-domain analysis and the proposed deep learning classifier reports an average accuracy of \(95.5\%\) with 8 classes and \(93\%\) (improvement of \(3\%\)) with 17 classes.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Vital biological Signals, such as heart and respiratory rate, are some of the first-level means to evaluate an individual’s physical health scenario. For example, cardiac motion, which is a primary indicator of an individual’s well-being, is often a unique identifier for each person as no two individuals have the same size, anatomy, or position of heart. While there are scientific tools to estimate basic health conditions from such biological signals, being bulky and hard to use in nature, they are primarily used within a clinical environment, under the supervision of a health professional. Hence, it is typical that these signs are only checked rarely at the annual doctor’s visit or when the patient’s physical health has already drastically deteriorated and symptoms are too prevalent to ignore. The situation becomes more complicated in a COVID-19 like pandemic scenario, where common people around the world, specifically the elderly patients, who are amongst the most vulnerable sections of the community, are trying hard to stay away from the hospitals and clinics to ensure safety. So, the probability of missing the regular health check process is now higher than ever. In fact, to address the criticality, an intensive and expensive medical procedure often turns out to be imperative or unavoidable. However, with early detection and regular monitoring processes in place, such exorbitant events may be circumvented.
Unconstrained means to monitor these vital body signals have rapidly emerged as a popular alternative to the conventional health check process in the last decade [1,2,3,4,5,6]. However, these appliances require frequent charging and are mostly wearable, making the patient uncomfortable (like causing skin irritation), specifically the elderly population. Many times, they also find it awkward due to the devices’ external visibility often compromising the privacy of their personal health information. All these pose severe challenges in continual and accurate data collection. A set of works employ unobtrusive devices, which can be easily installed in frequently used furniture that often appears in closed body contact with the patients [7,8,9,10,11,12]. However, the quality of the signal recorded using such devices often may relies on the frequency of the direct contact between the device surface and the patient’s body.
As such, an obvious way to inconspicuously monitor a person’s physical health is to embed sensors into objects most frequented by an individual. Studies have shown that people usually spend most of their day doing activities that require the person to be seated such as sitting while attending meetings, sitting while eating, sitting in cars, sitting while watching television [13]. A set of recent works [14,15,16,17,18,19,20] which mainly focus on building a hardware system like a chair, often rely on measurements obtained from only one type of signal and thereby have an access to only a limited amount of user’s health information. Additionally, frequently their prediction models derive aggregated decisions on the user’s health condition without allowing enough personalization. On the other hand, works [21,22,23,24,25] focusing primarily on its recognition sub-task tend to fail in a real-life problem setting, where signals collected from the patients are often too noisy for these machine learning-based systems to make an accurate prediction. Toward this, we aim to develop a generic signal processing and anomaly detection framework that may be deployed both in obtrusive and unobtrusive environments to measure and analyze vital signals of humans in real-time. While the proposed algorithm is invariant to its deployment environment, the proposed system has been installed within a SmartChair for real-life evaluation, as such a chair-like setting is known to be a complex application setting in this problem scenario. The extensive set of experiments demonstrate the effectiveness of the proposed cepstral-based peak fusion module by reporting 7 to \(10\%\) improvement over the baseline of a time-domain analysis. Furthermore, the proposed deep anomaly detection reports an average accuracy of 95.3% with 8 classes and \(93\%\) (improvement of \(3\%\)) with 17 classes.
An overview of the proposed method is illustrated in Fig. 1. In our experiments, we have used three vital body signals: respiratory rate, heart rate, and femoral pulse [10, 18, 26,27,28]. The primary contributions of the proposed system include:
-
1.
Generic Machine Learning Based Framework that may analyze both uni- and multi-modal signals within an integrated noise-tolerant framework. The proposed algorithm develops a robust peak detection module by fusing peaks in time and cepstral domain to identify an exhaustive and accurate peak list, which works as an input to the proposed deep learning-based prediction model to predict an individual’s personalized health pattern in an automated manner.
-
2.
Deep Anomaly Detection Strategy, which enables a continual deep learning-based monitoring process to precisely localize the anomalies in the time domain.
-
3.
Real-life Demonstration in an Unobtrusive Experiment Setting, wherein the proposed multimodal signal processing and analysis framework is deployed with a SmartChair health monitoring system that may simultaneously capture different types of vital signals from different parts of the seat occupant’s body (over a wide range of ages) without forcing an interruption in their daily work schedule.
-
4.
Extensive Evaluation and Comparative Study demonstrates an improved performance both in the publicly available datasets as well as our real-life lab experimental settings.
The rest of the paper is organized as follows: Sect. 2 briefly describes related works. The proposed method is explained in Sect. 3. Section 4 and 5 respectively present the experimental results and conclusion.
2 Related Works
In this section, we will briefly describe a set of related research, which can be categorized in parts: (1) Methods focusing on building an intelligent software system, wherein authors assume that a good quality annotated data collection is always available for training a sophisticated machine learning model and the quality of the signals captured during test time may also be considered to be reasonably noise-free, and (2) Methods aiming to build a hardware system that will collect the streaming data for further analysis using machine learning-based methods.
2.1 Health Anomalous Pattern Recognition
Traditional machine learning methods, like Multi-Layer Perceptron (MLP) [29,30,31], Support Vector Machine (SVM) [32,33,34,35], and K nearest neighbors (KNN) [30, 31, 36, 37] have already been used extensively to analyze vital health signals. A set of recent works introduce deep learning models [38,39,40,41,42,43] for improved performance. Deshmane and Madhe [44] have shown some impressive results on ECG Based biometric human identification using the Convolutional Neural Network model. In contrast to the traditional neural networks, Recurrent Neural Network (RNN) can be used for processing sequential data (e.g., cardiac signal) due to their internal state of memory and connection between the nodes. To explore the temporal granular details, RNN and its variant Long Short Term Memory (LSTM) [45,46,47,48] based models have also been introduced for the task. Given the prior knowledge of adjustment between input and output, it can map various sequences with sequences. However, in a practical scenario, specifically in a home-based computationally constrained setting, it is challenging to apply RNN-based methods for continual monitoring tasks, due to its scalability issues. In contrast to these methods, to ensure computational tractability, we use a small set of hand-crafted features to compute a compact feature descriptor that is passed as an input to the subsequent neural network-based prediction module. This not only helps attain a scalable prediction module but also ensures easy adaptability to an individual’s personalized health signal patterns.
2.2 Vital Signal Sensing Modality
Vital signals like ECG have been widely used to determine the health condition in many works [42, 49,50,51]. Kim et al. [52] studies of ECG measurement on the toilet seat for ubiquitous health care. Wu et al. [53] use a capacitive coupling ECG sensor to obtain the signal. While the signal capturing module for many of these methods is unobtrusive in nature, they may still demand the seat occupant’s attention to ensure a perfect connection between the body and the sensor and accurate angular arrangement, which may cause interruption to the seat occupant’s daily routine. A set of recent works have installed multi-channel ECG signals in a chair-based acquisition system to identify the motion artifacts [54]. Important to note that the system either requires the physiological activity in the same fashion as the enrollment stage or periodical resampling of the training dataset [55]. Therefore, the signal capturing process fails to be sustainable enough to ensure long-term usage.
Another set of works design the radio frequency (RF) methods [56] based on the signal reflection, require an off-body reader with the antenna in the far fields, while making the signal acquisition process for a single individual from multiple points, more challenging. A few research have utilized femoral pulse as a component of an active near-field coherent sensing (NCS) system [26, 57,58,59]. However, the arterial blood pressure is dependent on individual’s personal characteristics (e.g., age, height, gender), health conditions, and the administration of vasoactive drugs on the patient. Therefore, it is important to have a personalized prediction model that may effectively utilize the femoral pulse as a vital health signal to evaluate a patient’s health condition. In this paper, we use the ECG, PPG, and SCG vital signals to evaluate the subject’s physical condition. These specific vital signals are chosen as other possible signal resources such as Phonocardiography and Echocardiography are either obtrusive or require high expenses and are difficult to install on chairs.
3 Methodology
We design a generic machine learning-based classification model that performs a comprehensive and synchronized vital health signal analysis both in a uni-modal or multi-modal environment, wherein each mode may represent a signal generated from a unique body part of the participant and make a comprehensive prediction on the health condition of the individual. More specifically, given an annotated data collection \(\mathcal D=\{(s^j,y^j)\}_{j}\), where each vital body signal \(s^j\) is described using a m-mode (\(m\ge 1\)) representation, i.e., \(s^j=\{x^j_l\}^m_{l=1}\) and the corresponding label \(y^j\) is the label for the signal \(s^j\). As shown in Fig. 1, each mode-specific signal is pre-processed via the proposed signal processing module in parallel and later may get combined through feature fusion. In this section, we will describe the process in detail.
3.1 Signal Preprocessing
Noise Filtering. In the real-life setting, the signals received via the sensors are often noisy, due to the individual’s movements or shifts during measurements, dampening and noise from clothing, and noise introduced by the sensor itself. Therefore, any raw input signal is somewhat noisy. To address this challenge, we perform an initial noise filtering using Butterworth filter [60] to process each incoming signal to ensure an accurate prediction performance.
The Butterworth Filter is a filter that separates the high-frequency noise from the signal, such that frequency values within the range of the frequency boundaries are reflected in the signal without a significant amount of change. Also, the impact of higher frequencies is reduced by a significant factor, which is dependent on the filter order, in the filtered out signal. The sharpness of the transition from stopband to passband is controlled by the order, a predefined constant in our experiments. The low-pass Butterworth filter is designed as a rational function, defined as follows:
where \(H_0=1\) the maximum passband gain and \(\omega _0=1\) rad/sec. In our experiments, we have filtered the signal with a cutoff at 2.5 Hz and a fifth-order Butterworth filter, i.e. we have \(n=5\). The filtered signal x is treated as an input for further analysis. Unless mentioned otherwise, any reference of the signal x in the latter part of the paper will assume it as a filtered signal. We use the Scipy Python library [61] to implement the Butterworth filter.
Peak Identification. In order to analyze the signal characteristic, the first objective of this paper is to propose a robust peak detection scheme that may identify an exhaustive peak list within a signal against the different types of noise resources (e.g. non-stationary effects, low SNR, or several environmental settings of the patient like high heart rate exhibited after exercise) with minimum false positives. In this work, we compute a moving average based on a one-sided window proportional to the sampling frequency, where the proportionality constant is constant and user-defined. In all our experiments, we have chosen the proportionality factor as 0.75 and the sampling frequency as 100. Within each window, any heart rate lying above moving average (where the signal demonstrates a sharp change in gradient) is considered as a peak. While this approach works well in an ideal signal, in presence of a low SNR ratio, the precision performance may still deteriorate significantly resulting in the generation of some false peaks or end up losing some significant peaks in the input signal. An intuitive approach to mitigate the risk of false peak identification is to raise the moving average threshold. However, selecting a universal threshold that would work for all possible noisy signal settings, is difficult and may not be chosen automatically. Therefore, we employ an adaptive approach to dynamically set the threshold by computing the standard deviation of RR intervals [62, 63]. In general, the standard deviation of RR intervals is not large. Marking an extra peak or misplacing a R peak may increase the standard deviation significantly, which indicates the possibility of some false peak identification. Therefore, minimizing RRSD will be key to finding a threshold that finds the most accurate number of peaks. However, if RRSD is zero, then there can be two possibilities: either we have a perfect signal or we are seeing the consequences of undetected noises. So, to provide for the best solution, we choose a threshold from a predefined range that would satisfy both \(min(RRSD)>1\) and \(RRSD>1\). We use Heartpy [64] Python library function for the implementation task.
Peak Identification in the Cepstral Domain. Note that the peak detection in the input signal x is similar to detecting pitch from an audio signal. However, identifying peaks from x directly may not be sufficient in isolation, due to having the chance of missing some important peaks. In fact, this may in turn impact on deteriorating the following feature extraction task. Toward this end, for an improved peak detection performance, we use Cepstrum of the signal x for a granular-level peak analysis. As such, Cepstrum analysis, which is a nonlinear signal processing technique, is typically used for pitch detection (similar in some aspects to peak detection) in audio and speech. The real cepstrum of a signal x [65] is calculated as follows:
where \(X(\omega )\) is the Fourier transform of the sequence x(t). The proposed peak detection scheme (as described in Sect. 3.1) is employed to parallelly capture a set of peaks in the cepstral domain representation \(c_x\) of the input signal x.
Note that in order to ensure an accurate heart rate prediction, we aim to first identify the peaks in an input signal, which will later be used for identifying several key features like beats-per-minute (BPM), Inter-beat-interval (IBI), Root mean square of the successive differences (RMSSD), etc. We will discuss these features more in Sect. 3.2. As such in the cepstral domain, the magnitude of the cepstral coefficient is naturally related to the periodicity of the signal, which is the focus in heart rate estimation and higher values of the cepstrum coefficients reflect increased Signal to Noise Ratio (SNR). A fusion of signal peaks at the cepstrum domain is advantageous to produce a more exhaustive and accurate peak list, which forms the basis of the following feature fusion module (Fig. 2).
Peak Fusion Algorithm. The cepstrum signal \(c_x\) is used as a derived representative for the original unimode input signal x. The proposed method uses both \(c_x\) and x to identify sets of peaks which are fused to obtain a more exhaustive set of peaks in the signal x.
Given \(\mathcal P\) as the set of identified peaks in x, as shown in Fig. 5, for every peak at \(C_i\in \mathcal P\) with co-ordinate \((t_i, x_{(t_i)})\) in the signal x, there is a set of peaks in the corresponding time-domain neighborhood \(N_{t_i}\) around \(t_i\) for the cepstrum signal, \(c_x\). Intuitively multiple such peaks in \(c_x\) within the close neighborhood \(N_{t_i}\)around \(t_i\) do not provide any new peak information. Therefore, while fusing we eliminate all such redundant peaks within \(N_{t_i}\) retaining only the common peak identified at \(C_i\). This is the scenario, which we refer to as Remove Upward. This process is repeated for all peaks in \(\mathcal P\), resulting in retaining only those peaks in \(c_x\), which were not captured within any neighborhood \(N_{t_i}\) for any \(C_i\in \mathcal P\). The set of these remaining peaks in \(c_x\) is denoted as \(\mathcal P_{c_x}\). As illustrated in Fig. 5, to capture these missing peaks within the fused peak list for x, we analyze a close neighborhood \(N_{t_j}\) around every remaining peak in \(\mathcal P_{c_x}\). The time instant \(t_j\) within \(N_{t_j}\) at which the signal magnitude \(c_x[t_j]\) is maximum is mapped down to identify an additional peak \(D_j\) with magnitude \(x[t_j]\) in x. This process is referred to as Add Downward. The process is repeated for all elements of \(\mathcal P_{c_x}\). The combined peak list obtained at the end of a sequence of Remove Upward followed by a sequence of Add Downward is treated as the fused peak list that is used as the input to the following feature extraction module.
3.2 Multimodal Feature Extraction
Given the fused peak list obtained from the processed signal x, we derive several handcrafted signals including RRSD; RMSSD; BPM; IBI; SDNN; SDSD; NN20; NN50; PNN20; and PNN50 to represent the incoming signal in terms of a compact feature descriptor \(f_x\in \mathbb R^d\). RRSD can be computed as the standard deviation between the RR intervals (difference in time between the R-peaks) of a heart signal. RRSD can be computed as the standard deviation between the RR intervals (difference in time between the R-peaks) of a heart signal. RMSSD is defined as the root mean square of successive RR-Intervals and calculated by squaring each RR-interval. Then, the resulting values are averaged before the square root of the total is obtained. BPM can be calculated as the total number of peaks divided by the amount of time passed. IBI, the inter beat interval, can be calculated as the overall average of the RR Intervals. SDNN reflects the changes in heart rate due to cycles longer than 5 min. SDNN can be measured by computing the standard deviation of the time between the consecutive R-peaks. SDSD can be computed as the standard deviation of the successive differences between adjacent RR intervals. NN20 and NN50 can be computed by measuring the number of successive RR intervals that differ by more than 20 and 50 milliseconds respectively. PNN20 and PNN50 can be obtained by dividing NN20 and NN50 by a total number of RR intervals respectively.
In a multimodal environment, feature descriptor collection \(\{f^j_{x_l}\}^m_{l}\) representing multiple unimode signals \(s^j=\{x^j_l\}^m_{l=1}\) is transformed into a fused feature \(f=\phi (\{f^j_{x_l}\}^m_{l})\). In this work, we use vector concatenation function [66] as \(\phi \) to produce md dimensional fused feature \(f^j\).
3.3 Anomalous Pattern Recognition
Given an input signal x, we feed the feature vector \(f_x\) (as defined above) into a Neural Network model consisting of 3 fully connected (FC) layers with rectified linear unit (ReLU) activation function. The activation of the last FC layer is fed into a softmax layer to obtain the probabilistic category membership scores for the incoming signal’s anomaly score. While adding more layers makes the network more expressive, it simultaneously becomes harder to train due to increased computational complexity, vanishing gradients, and model over-fitting. The standard backpropagation algorithm is employed to update the fully connected layer weight parameters.Footnote 1 The loss function L is defined as follows:
where \(\mathbf{1}{.}\) is the indicator function, \(\mathbf{W}\) represents the neural network weight parameters and \(log(p(y^{j}=y|s^{j}; \mathbf{W})\) computes the probabilistic score of the sample \(x_i\) for the class \(y\in \mathcal Y\). The learning task is formulated as solving the minimization problem defined as: \(\underset{\mathbf{W}}{min}L(\mathbf{W})\).
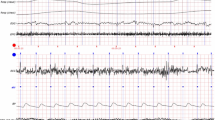
Peak detection performance in the OHSU ECG signal dataset [67] (shown in (a)) and our in-house dataset with 13 participants (shown in (b)).
4 Experiments
The proposed method is evaluated from two different perspectives: 1) accuracy evaluation of the peak detection module and 2) the effectiveness of its two-class neural network based prediction module, where the goal is to precisely identify the ‘anomalous’ signal characteristics of a participant in near real-time. Different datasets are used to evaluate the performance of the model.
4.1 Dataset
To evaluate the performance of our peak detection algorithm, which forms the core of the subsequent prediction module, we use two datasets: the publicly available Oregon Health and Science University (OHSU) ECG signal dataset with 28 participants [67] and our in-house dataset with 13 participants. The OHSU dataset has recorded its signals at a sampling rate of 200 Hz and at an amplitude resolution of 4.88 muV. We have used only the health signals from 26 participants. As for the remaining 2 participants, the ECG signals were missing at several time instants. Therefore, we have not used these 2 participants’ data. In our in-house dataset collected via the prototype VitalChair (which has sensors at different positions for recording signals from the seat occupant and details to follow in Sect. 4.2), the synchronized Femoral pulse (FP), Wrist Pulse (WP), and ECG signals are collected from 13 participants sitting at 7 different positions in a chair for 30 seconds. Among the participants, 4 are high school students, 6 are healthy functioning adults, and 3 are senior adults who have gone through heart surgeries in the past year. The system performs sensor fusion, analyzing the signal patterns to highlight potential anomalous patterns if any. Tests include two scenarios: 1) heart rate of a person at ‘calm’ state, 2) excited state after 30 min of ‘after exercise’.
To evaluate the performance of the proposed neural network model that uses a compact feature descriptor derived from the identified fused peak list as input to predict the participant’s health condition, we use Mendeley ECG 1000 Fragments Dataset [25] and our in-house dataset. The Mendeley ECG 1000 Fragments Dataset [25] is the publicly available dataset that we have used to evaluate our framework. This dataset has data from 45 different patients in different health conditions, which comprise of: 2 types of normal rhythms including a pace-maker rhythm and a normal sinus rhythm; 15 types of cardiac dysfunctions including Atrial premature beat, Atrial flutter, Atrial fibrillation, Supraventricular tachyarrhythmia, Pre-excitation (WPW), Premature ventricular contraction, Ventricular bigeminy, Ventricular trigeminy, and Ventricular tachycardia. All the recorded signals are documented at a sampling rate 360 Hz and a gain of 2200 [adu/mV]. In our experiments we have used the above-mentioned 2 types of normal rhythm signal collection as our ‘normal’ class, which combined together is referred to as Class 8, while all the other classes are treated as a specific type of ‘anomalous’ classes. The class population ratio between two types of classes (i.e. ‘normal’ and ‘anomalous’) are highly skewed and the Class 8 population has size 14, 000. So, we refrain from using any ‘anomalous’ class with samples less than 1, 000. Therefore, in our derived dataset, we have only samples from Class 8 forming the ‘normal’ class population and 7 different ‘anomalous’ classes. In our binary prediction module, we reiterate the experiments several times. At each session, Class 8 is used as the ‘normal’ class and one of the remaining 7 classes is treated as the ‘anomalous’ class. Also to note that the signals in this collection are typically high-sampled and the ratio of the anomaly to non-anomaly classes is still very low. Therefore, to further balance the class population at every experimental session, 50 randomly selected sub-sampled signals (of length 500) from the entire signal comprising of nearly 3600 samples, are randomly selected to form the larger training collection. To maintain the balance we just randomly select an equal-sized subset of sub-sampled normal signals to represent the Class 8 population.
Note that in Mendeley Dataset [25], there are 7 anomaly classes and 1 normal class (namely, Class 1: Ventricular Bigeminy, Class 2: Ventricular Trigeminy, Class 3: Supraventricular Tachyarrhythmia, Class 4: Atrial Fibrillation, Class 5: Left Bundle Branch Block Beat, Class 6: Atrial Premature Beat, Class 7: Premature Ventricular Contraction, and Class 8: Normal Sinus Rhythm) and the ratio of the anomalies to the normal classes is exceptionally low. For the training and testing of the neural network, 50 randomly selected segments of 500 samples for every 3600 samples of the ECG signal were randomly selected. This procedure of subsampling was performed to ensure that the neural network produced by training on this data is not biased or overfitted due to the lack of anomaly-class data. Furthermore, this act of subsampling allows the neural network to produce a more fine-grained interval in which the anomalies are prevalent.
4.2 Prototype Implementation: VitalChair
The custom-built circuit used for the Vitalchair used in our experiments for real-life study, consists of several capacitors, resistors, and photodiodesFootnote 2. It includes Arduino UNO; Breadboard; USB Cable; Power supplies; Jumper-wires (M/M, M/F); 1.0 M/4,7M Ohm Resistors; Piezoelectric; DS18B20 1-wire waterproof Temperature Sensors; Heart Rate pulse-sensors; different colored LEDs. To build the software module, I have used Arduino IDE and Python. Multiple biological signals including Electro-Cardiogram (ECG), Photoplethysmogram (PPG) from the wrist, Femoral Pulse (FP) are recorded using its corresponding sensor placed at different parts of the chair as illustrated in Fig. 1. The resulting signal from each sensor is passed onto an Arduino microcontroller attached to the bottom of the chair to collect readings from each sensor. Data was acquired onto a server connected to the Arduino over USB and analyzed using the Arduino software in real-time. The outputs of the sensors at different positions on the SmartChair are collected in a synchronized fashion for a comprehensive understanding of the seat occupant’s overall wellbeing.
4.3 Performance Evaluation
Peak Detection Accuracy Metric. The results of the first type of experiments, evaluating the peak detection module of the proposed method, use Accuracy as the evaluation metric. Given g as the number of hand-picked peaks by an independent evaluator and p is the number of system-identified peaks, we compute the \(Accuracy=1-\frac{|p-g|}{g}\) The quantitative results obtained in the OHSU ECG signal dataset and our in-house dataset are reported in Fig. 3(a) and (b) respectively. As observed in Fig. 3(a), the average accuracy achieved by the proposed prediction module over 28 participants is around \(94.18\%\). Specifically for the participant id 26, the accuracy (approx \(75\%\)) is considerably lower compared to the rest, which is due to the missing data at several time instants that resulted in missing some significant peaks. The deteriorated peak detection performance propagated to influence the performance of the subsequent prediction module.
Peak Detection Performance. In Fig. 3(b), we notice that the accuracy of the ‘calm’ state is usually greater than the after exercise accuracy. This is the case because, after exercise, an individual’s heart rate increases significantly, which causes many additional consecutive peak occurrences. However, the system perceives this extra flow of peaks as noises and thus, some of the peaks are not counted. This results in missing peaks that impact reducing the overall accuracy of the prediction module. However, this high-frequency heart-rate period only lasts for a couple of minutes and the individual (if indeed ‘healthy’) quickly regains their normal heart rate. To mitigate this noise impacted response, we pause the prediction task during the initial minute, so that any alert regarding the participant’s health condition is generated only if it has been more than a minute since their seat occupancy.
As observed in Fig. 4, combining peaks from the processed signal x and the cepstrum signal \(c_x\) have been useful to improve the resulting peak detection performance of the proposed method by an average of \(3\%\). In fact, in several instances (like participants 1, 8, 15, 22, and 23) the improvement reported was around 7–10%.
The performance of the proposed prediction module on the Mendeley dataset, reported using class specific ROC Curves (in (a)) and the classification accuracy (in (b)), wherein Class 1: Ventricular Bigeminy, Class 2: Ventricular Trigeminy, Class 3: Supraventricular Tachyarrhythmia, Class 4: Atrial Fibrillation, Class 5: Left Bundle Branch Block Beat, Class 6: Atrial Premature Beat, and Class 7: Premature Ventricular Contraction with their AUC scores respectively as: 0.98, 0.98, 0.97, 0.95, 0.94, 0.95, and 0.92.
Anomaly Detection Performance Metrics. The Classification Accuracy and Sensitivity score are used as the compact evaluation metrics computed by relating FP (False Positives), FN (False Negatives), TP (True Positives) and TN (True Negatives) and defined as:
where the scores are computed based on N-fold cross-validated test process, In our experiments, we have used \(N=5\). We also report the performance details using Area Under the Receiver Operating Curve, known as AUC score [68]. This metric is significant as the population distribution across different classes varies widely. While Classification_Accuracy (or Sensitivity) may provide an overall performance at a given experimental parameter setting, the AUC metric provides greater insight on the class-specific performances of the proposed method. Also, Sensitivity score is used to report the comparative performance of the proposed method.
Anomaly Detection Performance. Figure. 6 reports the performance of the proposed method using ROC Curves, AUC Scores as the total area under the ROC curve, and the Classification Accuracy as the evaluation metrics [69]. As seen in the figure, note that, the average performance on all seven anomalous classes is around \(95.29\%\). While the performance on Class 5 is approximately \(87.22\%\), it is primarily attributed to the sparse signal (with also missing ECG values) obtained from participants.
4.4 Comparative Study
The performance of the proposed method is compared against that of several methods reported in [25], and the result is reported in Table 1. To attain an equivalent experimental setting, for this experiment, we combine all the 17 cardiac disorders into an anomaly class, while the healthy signals form the second class. As seen by comparing the results reported in the table, the proposed method shows an improved performance by reporting \(3\%\) increased Sensitivity score. An equivalent experiment is also performed using only 8 classes and as shown in the table, the proposed method attains an impressive performance gain of \(2.5\%\) compared to the best result reported on the data-set.
Also in this scenario, it is also important to note that the performance varies from class to class (please refer to Fig. 6(b)). So, such course-level overall performance evaluation may not be sufficiently insightful in terms of getting sufficient insight on the effectiveness of the proposed model. For example, accurate identification of samples from Class 5 samples is harder than that of Class 6. Moreover, the above-mentioned paper obtains the accuracy in an obtrusive manner, which significantly reduces the amount of noise corrupting the signal. Therefore, the applicability of these methods is limited in various real-life environments, where signals can only be received in an unobtrusive manner. In contrast, the proposed method, which is generic and sufficiently robust to handle the noisy signal inputs, and allows for sequential learning by continual signal capturing process, is more effective and efficient. As described earlier, to further investigate the robustness of the method, we perform experiments in a real-life environment by deploying the software in the SmartChair, which collects signals by placing sensors at different parts of the chair. Also, the method is evaluated at different levels of stress and physical activity state of the participants to investigate the efficacy of the signal filtering and feature extraction methods.
5 Conclusion
In this paper, we have presented a framework that is able to accurately classify multiple input signals like ECG, PPG, and Femoral Pulse from a specific individual into two categories: healthy or unhealthy. Having been able to continuously monitor the patient’s vital signals, this system has several life-changing effects, including the ability to identify pathology conditions before they can turn into a serious threat to the human’s life or severe measures are required to cure them such as amputations. To demonstrate our proposed model’s real-life feasibility, we have physically implemented this framework into a chair. However, the proposed method is sufficiently generic to be deployed into other frequently used furniture items like beds, sofas, etc. We plan to extend this work to include other means of extracting pathological information, like vocal signals, and synchronize them all to make a smart home system that will be able to accurately classify the disease-specific pathological condition the individual has using multi-modal information.
References
Mukhopadhyay, S.C.: Wearable sensors for human activity monitoring: a review. IEEE Sensor. J. 15(3), 1321–1330 (2014)
Fletcher, R., Poh, M., Eydgahi, H.: Wearable sensors: opportunities and challenges for low-cost health care. In: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), vol. 1766 (1763)
Niswar, M., Nur, M., Ilham, A.A., Mappangara, I.: A low cost wearable medical device for vital signs monitoring in low-resource settings. Int. J. Electr. Comput. Eng. 9, 2088–8708 (2019)
Huang, M.C., Liu, J.J., Xu, W., Gu, C., Li, C., Sarrafzadeh, M.: A self-calibrating radar sensor system for measuring vital signs. IEEE Trans. Biomed. Circuits Syst. 10(2), 352–363 (2016)
Rathore, A.S., Li, Z., Zhu, W., Jin, Z., Xu, W.: A survey on heart biometrics. ACM Comput. Surv. (CSUR) 53(6), 1–38 (2020)
Patil, O.R., Wang, W., Gao, Y., Xu, W., Jin, Z.: A low-cost, camera-based continuous PPG monitoring system using Laplacian pyramid. Smart Health 9—-10, 2–11 (2018). CHASE 2018 Special Issue
Lim, Y.G., et al.: Monitoring physiological signals using nonintrusive sensors installed in daily life equipment. Biomed. Eng. Lett. 1(1), 11–20 (2011). https://doi.org/10.1007/s13534-011-0012-0
Brüser, C., Antink, C.H., Wartzek, T., Walter, M., Leonhardt, S.: Ambient and unobtrusive cardiorespiratory monitoring techniques. IEEE Rev. Biomed. Eng. 8, 30–43 (2015)
Kang, S., et al.: Sinabro: opportunistic and unobtrusive mobile electrocardiogram monitoring system. In: Proceedings of the 15th Workshop on Mobile Computing Systems and Applications, pp. 1–6 (2014)
Malik, A.R., Pilon, L., Boger, J.: Development of a smart seat cushion for heart rate monitoring using ballistocardiography. In: 2019 IEEE International Conference on Computational Science and Engineering (CSE) and IEEE International Conference on Embedded and Ubiquitous Computing (EUC), pp. 379–383 IEEE (2019)
Yu, X., et al.: A multi-modal sensor for a bed-integrated unobtrusive vital signs sensing array. IEEE Trans. Biomed. Circuits Syst. 13(3), 529–539 (2019)
Wang, S., et al.: Noninvasive monitoring of vital signs based on highly sensitive fiber optic mattress. IEEE Sens. J. 20(11), 6182–6190 (2020)
Pronk, N.: The problem with too much sitting: a workplace conundrum. ACSM’s Health Fit. J. 15(1), 41–43 (2011)
Arnrich, B., Setz, C., La Marca, R., Tröster, G., Ehlert, U.: What does your chair know about your stress level? IEEE Trans. Inf. Technol. Biomed. 14(2), 207–214 (2009)
Zheng, Y., Morrell, J.B.: A vibrotactile feedback approach to posture guidance. In: 2010 IEEE haptics Symposium, pp. 351–358. IEEE (2010)
Baek, H.J., Chung, G.S., Kim, K.K., Park, K.S.: A smart health monitoring chair for nonintrusive measurement of biological signals. IEEE Trans. Inf. Technol. Biomed. 16(1), 150–158 (2011)
Kumar, R., Bayliff, A., De, D., Evans, A., Das, S.K., Makos, M.: Care-chair: sedentary activities and behavior assessment with smart sensing on chair backrest. In: 2016 IEEE International Conference on Smart Computing (SMARTCOMP), pp. 1–18. IEEE (2016)
Griffiths, E., Saponas, T.S., Brush, A.B.: Health chair: implicitly sensing heart and respiratory rate. In: Proceedings of the 2014 ACM International Joint Conference on Pervasive and Ubiquitous Computing, pp. 661–671 (2014)
Ravichandran, R., Saba, E., Chen, K.Y., Goel, M., Gupta, S., Patel, S.N.: WiBreathe: estimating respiration rate using wireless signals in natural settings in the home. In: 2015 IEEE International Conference on Pervasive Computing and Communications (PerCom), pp. 131–139. IEEE (2015)
Zhang, Y., Chen, Z., Chen, W., Li, H.: Unobtrusive and continuous bcg-based human identification using a microbend fiber sensor. IEEE Access 7, 72518–72527 (2019)
Bazi, Y., Alajlan, N., AlHichri, H., Malek, S.: Domain adaptation methods for ECG classification. In: 2013 International Conference on Computer Medical Applications (ICCMA), pp. 1–4 (2013)
Lin, C.C., Yang, C.M.: Heartbeat classification using normalized RR intervals and wavelet features. In: 2014 International Symposium on Computer. Consumer and Control, pp. 650–653 (2014)
Zhang, Z., Luo, X.: Heartbeat classification using decision level fusion. Biomed. Eng. Lett. 4(4), 388–395 (2014). https://doi.org/10.1007/s13534-014-0158-7
Zhang, Z., Dong, J., Luo, X., Choi, K.S., Wu, X.: Heartbeat classification using disease-specific feature selection. Comput. Biol. Med. 46, 79–89 (2014)
Pławiak, P.: Novel methodology of cardiac health recognition based on ECG signals and evolutionary-neural system. Expert Syst. Appl. 92, 334–349 (2018)
Hui, X., Kan, E.C.: Seat integration of RF vital-sign monitoring. In: 2019 IEEE MTT-S International Microwave Biomedical Conference (IMBioC), vol. 1, pp. 1–3. (2019)
Gui, Q., Ruiz-Blondet, M.V., Laszlo, S., Jin, Z.: A survey on brain biometrics. ACM Comput. Surv. (CSUR) 51(6), 1–38 (2019)
Anttonen, J., Surakka, V.: Emotions and heart rate while sitting on a chair. In: Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, pp. 491–499 (2005)
Pinto, J.R., Cardoso, J.S., Lourenço, A., Carreiras, C.: Towards a continuous biometric system based on ECG signals acquired on the steering wheel. Sensors 17(10), 2228 (2017)
Sidek, K.A., Mai, V., Khalil, I.: Data mining in mobile ECG based biometric identification. J. Netw. Comput. Appl. 44, 83–91 (2014)
Kavsaoğlu, A.R., Polat, K., Bozkurt, M.R.: A novel feature ranking algorithm for biometric recognition with PPG signals. Comput. Biol. Med. 49, 1–14 (2014)
Li, M., Narayanan, S.: Robust ECG biometrics by fusing temporal and cepstral information. In: 2010 20th International Conference on Pattern Recognition, pp. 1326–1329. IEEE (2010)
Da Silva, H.P., Fred, A., Lourenço, A., Jain, A.K.: Finger ECG signal for user authentication: usability and performance. In: 2013 IEEE Sixth International Conference on Biometrics: Theory, Applications and Systems (BTAS), pp. 1–8. IEEE (2013)
Lin, S.L., Chen, C.K., Lin, C.L., Yang, W.C., Chiang, C.T.: Individual identification based on chaotic electrocardiogram signals during muscular exercise. IET Biometrics 3(4), 257–266 (2014)
Lin, F., Song, C., Zhuang, Y., Xu, W., Li, C., Ren, K.: Cardiac scan: a non-contact and continuous heart-based user authentication system. In: Proceedings of the 23rd Annual International Conference on Mobile Computing and Networking, pp. 315–328 (2017)
Gürkan, H., Guz, U., Yarman, B.S.: A novel biometric authentication approach using electrocardiogram signals. In: 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), pp. 4259–4262. IEEE (2013)
Venkatesh, N., Jayaraman, S.: Human electrocardiogram for biometrics using DTW and FLDA. In: 2010 20th International Conference on Pattern Recognition, pp. 3838–3841. IEEE (2010)
Donida Labati, R., Muñoz, E., Piuri, V., Sassi, R., Scotti, F.: Deep-ECG: convolutional neural networks for ECG biometric recognition. Pattern Recogn. Lett. 126, 78–85 (2019)
Tavallali, P., Razavi, M., Pahlevan, N.: Artificial intelligence estimation of carotid-femoral pulse wave velocity using carotid waveform. Sci. Rep. 8, 1–12 (2018)
Ledezma, C.A., Zhou, X., Rodríguez, B., Tan, P.J., Díaz-Zuccarini, V.: A modeling and machine learning approach to ECG feature engineering for the detection of ischemia using pseudo-ECG. PLOS One 14, 1–21 (2019)
da Silva Luz, E.J., Moreira, G.J., Oliveira, L.S., Schwartz, W.R., Menotti, D.: Learning deep off-the-person heart biometrics representations. IEEE Trans. Inf. Forensics Secur. 13(5), 1258–1270 (2017)
Zhang, Q., Zhou, D., Zeng, X.: HeartID: a multiresolution convolutional neural network for ECG-based biometric human identification in smart health applications. Ieee Access 5, 11805–11816 (2017)
Xiang, Y., Luo, J., Zhu, T., Wang, S., Xiang, X., Meng, J.: ECG-based heartbeat classification using two-level convolutional neural network and RR interval difference. IEICE Trans. Inf. Syst. 101(4), 1189–1198 (2018)
Deshmane, M., Madhe, S.: ECG based biometric human identification using convolutional neural network in smart health applications. In: International Conference on Computing Communication Control and Automation (ICCUBEA), pp. 1–6 (2018)
Salloum, R., Kuo, C.C.J.: ECG-based biometrics using recurrent neural networks. In: 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 2062–2066. IEEE (2017)
Everson, L., et al.: BiometricNet: deep learning based biometric identification using wrist-worn PPG. In: 2018 IEEE International Symposium on Circuits and Systems (ISCAS), pp. 1–5. IEEE (2018)
Salloum, R., Kuo, C.J.: ECG-based biometrics using recurrent neural networks. In: 2017 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 2062–2066 (2017)
Dezaki, F.T., et al.: Deep residual recurrent neural networks for characterisation of cardiac cycle phase from echocardiograms. In: Cardoso, M.J., et al. (eds.) DLMIA/ML-CDS-2017. LNCS, vol. 10553, pp. 100–108. Springer, Cham (2017). https://doi.org/10.1007/978-3-319-67558-9_12
Hui, X., Conroy, T.B., Kan, E.C.: Multi-point near-field RF sensing of blood pressures and heartbeat dynamics. IEEE Access 8, 89935–89945 (2020)
Sidikova, M., et al.: Vital sign monitoring in car seats based on electrocardiography, ballistocardiography and seismocardiography: A review. Sensors 20(19), 5699 (2020)
Singh, R.K., Sarkar, A., Anoop, C.S.: A health monitoring system using multiple non-contact ECG sensors for automotive drivers. In: 2016 IEEE International Instrumentation and Measurement Technology Conference Proceedings, pp. 1–6 (2016)
Kim, K.K., Lim, Y.K., Park, K.S.: The electrically noncontacting ECG measurement on the toilet seat using the capacitively-coupled insulated electrodes. In: The 26th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, vol. 1, pp. 2375–2378. IEEE (2004)
Wu, K.F., Chan, C.H., Zhang, Y.t.: Contactless and cuffless monitoring of blood pressure on a chair using e-textile materials. In: 2006 3rd IEEE/EMBS International Summer School on Medical Devices and Biosensors, pp. 98–100. IEEE (2006)
Choi, M., Jeong, J., Kim, S., Kim, S.: Reduction of motion artifacts and improvement of R peak detecting accuracy using adjacent non-intrusive ECG sensors. Sensors 16, 715 (2016)
Coutinho, D.P., Silva, H., Gamboa, H., Fred, A., Figueiredo, M.: Novel fiducial and non-fiducial approaches to electrocardiogram-based biometric systems. IET Biometrics 2(2), 64–75 (2013)
Adib, F., Mao, H., Kabelac, Z., Katabi, D., Miller, R.C.: Smart homes that monitor breathing and heart rate. In: Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems, pp. 837–846. Association for Computing Machinery, New York (2015)
Welkowitz, W., Cui, Q., Qi, Y., Kostis, J.B.: Noninvasive estimation of cardiac output. IEEE Trans. Biomed. Eng. 38(11), 1100–1105 (1991)
Smith, J., Camporota, L., Beale, R.: Monitoring arterial blood pressure and cardiac output using central or peripheral arterial pressure waveforms. In: Vincent, J.L. (ed.) Intensive Care Medicine, pp. 285–296. Springer, New York (2009). https://doi.org/10.1007/978-0-387-92278-2_27
Chen, J., et al.: High durable, biocompatible, and flexible piezoelectric pulse sensor using single-crystalline III-N thin film. Adv. Funct. Mater. 29, 1903162 (2019)
Winder, S.: Analog and digital filter design. Elsevier, Amsterdam (2002)
Virtanen, P., et al.: SciPy 1.0: Fundamental algorithms for scientific computing in python. Nat. Methods 17(3), 261–272 (2020)
O’Haver, T.: Pragmatic introduction to signal processing (2018)
Richig, J.W., Sleeper, M.M.: Electrocardiography of Laboratory Animals. Academic Press, San Diego (2018)
van Gent, P., Farah, H., Nes, N., Arem, B.: HeartPy: a novel heart rate algorithm for the analysis of noisy signals. Transp. Res. Part F Traffic Psychol. Behav. 66, 368–378 (2019)
Pei, S.C., Lin, H.S.: Minimum-phase FIR filter design using real cepstrum. IEEE Trans. Circuits Syst. II Express Briefs 53(10), 1113–1117 (2006)
Bretscher, O.: Linear Algebra with Applications. Prentice Hall, Eaglewood Cliffs (1997)
Rogovoy, N., et al.: The dialysis procedure triggers autonomic imbalance and cardiac arrhythmias: insights from continuous 14-day ECG monitoring, April 2019
Fan, J., Upadhye, S., Worster, A.: Understanding receiver operating characteristic (roc) curves. Can. J. Emerg. Med. 8(1), 19–20 (2006)
Pedregosa, F., et al.: Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011)
Ye, C., Kumar, B.V., Coimbra, M.T.: Heartbeat classification using morphological and dynamic features of ECG signals. IEEE Trans. Biomed. Eng. 59(10), 2930–2941 (2012)
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 ICST Institute for Computer Sciences, Social Informatics and Telecommunications Engineering
About this paper
Cite this paper
Bhattacharjee, S., Li, H., Xu, W. (2022). Anomalous Pattern Recognition in Vital Health Signals via Multimodal Fusion. In: Ur Rehman, M., Zoha, A. (eds) Body Area Networks. Smart IoT and Big Data for Intelligent Health Management. BODYNETS 2021. Lecture Notes of the Institute for Computer Sciences, Social Informatics and Telecommunications Engineering, vol 420. Springer, Cham. https://doi.org/10.1007/978-3-030-95593-9_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-95593-9_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-95592-2
Online ISBN: 978-3-030-95593-9
eBook Packages: Computer ScienceComputer Science (R0)