Abstract
The CODO ontology was designed to capture data about the Covid-19 pandemic. The goal of the ontology was to collect epidemiological data about the pandemic so that medical professionals could perform contact tracing and answer questions about infection paths based on information about relations between patients, geography, time, etc. We took information from various spreadsheets and integrated it into one consistent knowledge graph that could be queried with SPARQL and visualized with the Gruff tool in AllegroGraph. The ontology is published on Bioportal and has been used by two projects to date. This paper describes the process used to design the initial ontology and to develop transformations to incorporate data from the Indian government about the pandemic. We went from an ontology to a large knowledge graph with approximately 5M triples in a few months. Our experience demonstrates some common principles that apply to the process of scaling up from an ontology model to a knowledge graph with real-world data.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Ontology
- Knowledge graph
- Healthcare
- Covid-19
- Agile methods
- Software Development Life-Cycle (SDLC)
- OWL
- SPARQL
- Transformations
1 Introduction
At the beginning of the Covid-19 pandemic (March 2020) we began to develop an ontology called CODO, an Ontology for collection and analysis of COVID-19 data. The ontology followed the FAIR model for representing data and incorporated classes and properties from standard vocabularies such as FOAF, Dublin Core, Schema.org, and SNOMED CT. While other Covid ontologies, such as CIDO, VIDO, CoVoc, etc. (more detail provided in Sect. 1.1) focus on analyzing the virus, CODO focuses on epidemiological issues, such as tracking how the virus was spread based on data about relationships, geography, temporal relations, etc. For more details on the FAIR principles and the general structure of the CODO ontology see [1]. We evolved what started as a small ontology in Protégé to a large knowledge graph (KG) in the AllegroGraph triplestore product from Franz IncFootnote 1. We use the term ontology to refer to the CODO Web Ontology Language (OWL) model with only basic example test data. We use the term knowledge graph to refer to the ontology populated with large amounts of real-world data.
1.1 Relation to Other Work
There has been extensive work in the Semantic Web community to add value to the vast amount of data produced by the pandemic. The existing Covid-19 ontologies can broadly be classified into three categories:
-
1.
High level statistics that illustrate the number of patients infected and the number of deaths per region for various time intervals.
-
2.
Modeling of concepts required to analyze the virus in order to develop treatments and vaccines. This includes modeling the Covid-19 virus and how it is similar and different from related viruses such as SARS and modeling drugs used to treat and develop vaccines for viruses and other illnesses similar to Covid-19.
-
3.
Modeling the space of scientific articles on topics related to Covid-19 in order to provide semantic search capabilities for researchers developing treatments and vaccines.
Examples of the first category include the Johns Hopkins [2] and NYTimes [3] knowledge graphs. Examples of the second category include CIDO [4], IDO-COVID-19Footnote 2, COVID-19 Surveillance OntologyFootnote 3, and CoVocFootnote 4. These ontologies all extend the Infectious Disease Ontology (IDO) [5]. Examples of the third category include the Covid-19 Knowledge Graph [6].
CODO fills a specific niche that is different from these categories. It focuses on modeling epidemiology and the various ways that the virus has spread throughout the population, with a case study of India. For example, the demographics of the patients who were infected by the virus (age, sex, family and social relations, geographic home, travel history) and contact tracing from one patient to another. The graphical features of the AllegroGraph Gruff tool are especially useful for this type of analysis. Information such as the graph of which patient infected which other patients can be generated automatically with Gruff (see Figs. 6 and 7 below). Information that is implicit in the data but difficult to understand without a knowledge graph model can be made explicit and obvious with a knowledge graph and visualization tools. This allows medical professionals to conduct contact tracing and perform epidemiological research. Although, the focus of the current work has been on the pandemic in India, CODO can be applied to any location and indeed to the spread of any infectious disease.
In addition, the CODO team has been taking part in a harmonization process with many of the designers of the ontologies described above [7]. As part of the harmonization process, we have altered the design of the ontology to be more consistent with and more easily integrate with other ontologies that deal with different aspects of the Covid-19 pandemic such as the CIDO ontology.
The main contributions of this paper are:
-
1.
Details the CODO Agile knowledge graph development processes.
-
2.
Describes the issues related to the real world COVID-19 data we incorporated and its transformation to a knowledge graph.
-
3.
Demonstrates some of the visualization capabilities provided by the CODO knowledge graph.
The rest of the paper is organized as follows: Sect. 2 discusses the CODO ontology design process and its lifecycle. Section 3 discusses the five phases of CODO KG development activity. The issues related to the pandemic data and their transformations to the graph are discussed in Sect. 4. Section 5 provides example results. Finally, Sect. 6 concludes with the future plans to enhance CODO and to harmonize it with other ontologies designed for the pandemic, especially those in the OBO foundry.
2 CODO Processing Lifecycle
Our development process was a hybrid of two different methods. We utilized the YAMO process for ontology development [8] and Agile Methods [9] to drive our iterations and overall approach to analysis, design, implementation, and testing. These two are complimentary, they address different aspects of the development process. YAMO addresses the specific details of how to design an ontology rather than say an Object Oriented Programming (OOP) or transactional database system. Agile Methods defines the approach to development issues that apply to all software development processes such as length of iterations and interaction between analysis, design, testing, and implementation. It is possible to practice the YAMO methodology in a Waterfall or Agile process. This is similar to the Rational Unified Process (RUP) which defines the design artifacts and processes to develop an OOP system. Although RUP is typically done in an iterative manner it can be used in a Waterfall manner as well [10].
We have created a hybrid approach that is well suited to knowledge graph development in general. The specific Agile methods that we applied were:
-
Test Driven Development. We had test data from the very first ontology as well as various competency questions (stories in Agile terminology) that we used to practice test-driven development of the ontology. As we began to acquire more data it was clear that simply visually inspecting the ontology to validate it was inadequate. Hence, we developed SPARQL queries and Lisp functionsFootnote 5 to facilitate the testing process.
-
Rapid Iterations. Our iterations were approximately on a weekly basis. To present the path of our development we have abstracted these iterations into monthly phases where we describe the major development done in each phase.
-
Refactoring. Our goal was to deliver technology that could be usable from the very beginning. However, as we scaled up the ontology to support larger data, we needed to refactor the model and the transformations we used to transform tabular data into a knowledge graph. As an example of how we used refactoring of the model (as opposed to refactoring the transformations described below), in our initial model the diagnosis date and the date that the patient was released from the hospital were simply stored on the Patient class. When we decided to take advantage of the temporal reasoning in AllegroGraph we refactored these properties onto the Disease class and made Disease a subclass of the Event class which had the appropriate properties for temporal reasoning such as the start and end time of an Event. We used this information to generate visualizations and summary data such as the average length of hospitalization and temporal relations among patients graphed on a timeline.
-
Bottom Up and Top-Down Design. A key concept of Agile is that design emerges over time rather than being set in stone at the end of an Analysis and Design phase as in the Waterfall model. We designed an initial ontology based on our best understanding of the problem and the existing data but refactored that design as we acquired more data and added new ways to utilize the data.
-
Story driven development. The YAMO methodology is designed around competency questions that the model is meant to answer. These competency questions are essentially the same as stories in Agile development.
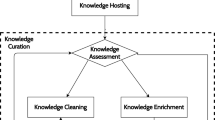
Figure 1 illustrates the complete life cycle as data goes from heterogeneous input formats to a knowledge graph. These processes consist of:
-
1.
The Upload process
-
2.
Data Transformation
-
3.
Reasoning
-
4.
Publication
The Upload process imports data into the initial version of the triplestore. The inputs to this process are various documents and the CODO ontology. The output of this process is an initial triplestore knowledge graph. An additional output of this process are suggested standards fed back to the user communities that recommend canonical formats to standardize future input data in order to make it more amenable to conversion into a knowledge graph. The Data Transformation process transforms text strings from the Upload process into objects and property values. The boundary between the Upload and Transformation process is not rigid. It is possible to do a significant amount of conversion into objects and properties via the initial Upload process using tools such as Cellfie [11]. However, due to the varied nature of our input data we often required the power of a programming language and SPARQL to transform data over those available in tools designed for initial uploading. Thus, we would often simply transfer strings from columns directly to data properties (which we call utility properties) in the initial knowledge graph and then apply more sophisticated transformations to the graph that converted these data properties into objects and object properties. We deleted each utility property string after it had been transformed. The output of the transformation process is the initial semantic knowledge graph model.
The Reasoning process utilizes OWL and rule-based reasoners. This is required for reasoning about social and family relations as well as other kinds of relations. For example:
-
Reasoning about inverse values. E.g., if a patient X is the father of patient Y then patient Y is the child of patient X.
-
Reasoning about property hierarchies. E.g., if patient X is the father of patient Y then patient X is also the parent of patient Y.
-
Reasoning about transitive place relations. E.g., if city X is contained in state Y and state Y is contained in nation Z then city X is contained in nation Z.
-
SWRL (and later SPARQL) rules to cover reasoning that can’t be done with OWL. E.g., if patient X is the brother of patient Y and patient Z is the daughter of patient Y then patient X is the uncle of patient Z.
For the early iterations of CODO this process consisted of running the Pellet reasoner which included execution of SWRL rules. For later iterations of CODO where Protégé could not support the large number of objects in the knowledge graph we utilized the AllegroGraph triplestore. In these later iterations we utilized the Materializer reasoner in AllegroGraph. In addition, since SWRL is currently not supported in AllegroGraph we replaced SWRL rules with equivalent SPARQL rules.
Finally, the publication process consists of making the knowledge graph available as a SPARQL endpoint. In addition, we publish and update the CODO ontology on Bioportal and the transformation rules and Lisp code on Github.
3 The CODO KG Development Phases
The CODOFootnote 6 project was divided into 5 phases, beginning with a basic ontology in Protégé with only a handful of test data to an AllegroGraph knowledge graph with over 3M triples.
3.1 Phase 1: Protégé Ontology
The initial phase consisted of defining the basic competency questions that we wanted the ontology to answer and building the initial ontology in the desktop version of the Protégé ontology editing tool [12]. Competency questions are a concept from YAMO [8]. Example competency questions are:
-
What is the travel history of patient p (see Fig. 6)?
-
What is the transitive closure for any patient p of all patients who infected and were infected by patient p (see Fig. 7)?
-
Who are the people with any known relationship (family, co-workers, etc.) to patient p?
-
What was the average length of time from infection to recovery for all patients or for patients in a given geographic area or time span?
-
What are summary statistics for the incidents of infection, both globally and in different geographic areas and time periods?
This version of the ontology had no real data. However, as part of our test-driven approach we created representative individuals as example test data. Primarily patients but also test results, cities, etc. These individuals were used to validate the ontology. This version included rules in the Semantic Web Rule Language (SWRL) [13] to define concepts beyond basic OWL Description Logic such as Aunts and Uncles.
3.2 Phase 2: Cellfie and AllegroGraph
In phase 2 we began to use the Cellfie plugin for Protégé to load data from the Indian government about the pandemic. As we loaded our initial data it soon became clear that we required a true database to get acceptable performance. Protégé is a modeling tool and is not designed to accommodate large data sets. We chose the free version of the AllegroGraph triplestore from Franz Inc. As a result, we refactored our SWRL rules into SPARQL as SWRL is not currently supported by AllegroGraph.
We also began to use Web Protégé to store the CODO ontology. This made collaboration much easier. Prior to using Web Protégé we had issues with consistency between the various changes we each made to the ontology. Web Protégé eliminated these issues and also allowed further collaboration capabilities such as having threaded discussions about various entities stored with the ontology. However, there are also capabilities that are currently only supported in the desktop version of Protégé, most significantly the ability to run a reasoner to validate the model. Thus, at regular intervals we would download the ontology into the desktop version of Protégé to run the Pellet reasoner and make other changes not currently supported in Web Protégé.
3.3 Phase 3: SPARQL Transformations
In phase 3 we began to use SPARQL to transform the strings that were too complex for Cellfie to process. This is discussed in more detail in the next section.
3.4 Phase 4: Pattern Matching and LISP
Although we performed pattern matching in phase 3, the majority of our early SPARQL transformations were specific (ad hoc, discussed below). In phase 4 we eliminated most of these transformations with fewer pattern matching transformations in SPARQL and Lisp. In this phase we also wrapped all of our SPARQL transformations in Lisp code. This eliminated the tedious and potentially error filled task of manually running each SPARQL transformation in AllegroGraph’s Gruff editor [14]. Instead, we could run Lisp functions which executed several SPARQL transformations automatically.
We also began to use Lisp to do more complex transformations that were too difficult to do in SPARQL such as iterating through a sequence of patient IDs. We created Lisp functions that utilized the regex extensions in Franz’s version of Common Lisp and directly manipulated the knowledge graph.
3.5 Phase 5: Test Harness and Additional Refactoring of Transformations
One issue we identified when testing the transformations developed in the previous phase was that in some cases a general pattern matching transformation might make an inappropriate transformation to a string it wasn’t designed to match. In order to facilitate testing we developed a test harness in Lisp. In testing mode when we deleted a utility property, we would copy it to another test utility property. Thus, we could still take advantage of our strategy of working from specific to more general transformations (see Fig. 4) which required deleting utility property values once they were processed but we could retain an audit trail so that we could inspect objects to ensure that the processed strings were appropriately transformed.
In addition, we further refactored our SPARQL and Lisp transformations to eliminate multiple specific transforms with individual pattern matching transforms. We also imported data on longitude and latitude for the various places (cities, states, nations, etc.) in the CODO ontology. A significant part of our data involves geographic information such as where patients were infected, where they live, travel, etc. We added various SPARQL queries that could provide statistical information and connectivity among patients which could be visualized via the Gruff graph layout tool. Finally, we took advantage of the temporal reasoning capabilities in AllegroGraph. Allegro has a temporal reasoning model based on the well-known Allen model for reasoning about time [15]. We added the required properties from the Allegro model to the CODO ontology so that we could take advantage of the capabilities in Gruff for displaying graphs along a timeline and could also use SPARQL queries to create additional summary data and visualizations about the spread of the pandemic over time.
4 Transforming Strings to Objects
The most difficult part of transforming data from documents, spreadsheets, and relational databases into a knowledge graph is transforming data represented by strings and tables into objects and property values [16]. This is because much of the information required for a useful knowledge graph is implicit in the context of a document. One of the main benefits added by a knowledge graph is to take this implicit context information that users apply when reading the document and make it explicit in the knowledge graph.
For example, one of the columns in the spreadsheets that we used as a data source had the heading Reason. This was meant to be the reason that the patient in that row contracted the virus. Examples of values were:
-
1.
“Travel to Bangalore”
-
2.
“Contact with P134-P135-P136-P137 and P138”
-
3.
“Father”
-
4.
“Policeman on duty”
The meaning for these strings is implicit but easy for humans to understand. Example 1 means that the patient travelled to Bangalore and caught the virus as a result of this trip (information about travel companions was captured in another column). Example 2 means that the patient had contact with a certain group of other patients and caught the virus from one of them. Example 3 means the patient caught the virus from their father (family relations were captured in another column). Example 4 means that the patient was a policeman on duty and caught the virus in the course of their duties. In the CODO ontology this kind of information results in creation of several different objects and property values. In addition to transforming strings in each column our transformations needed to integrate relevant information from other columns.
As an example of the kinds of transformations we developed, ExposureToCOVID-19 is a class with several sub-classes for the different kinds of potential exposures to the virus. Figure 2 shows a partially expanded view of the subclasses of this class in Protégé. For each string in the Reason column, we need to create an instance of the appropriate subclass of ExposureToCOVID-19 and then make that new individual the value of the suspectedReasonOfCatchingCovid-19 property for the patient. We also need to integrate information such as family relations and travel companions from other columns.
Relating back to the examples above, Example 1 should result in an instance of InfectedCo-Passenger. Example 2 should result in an instance of CloseContact. Example 3 should result in an instance of InfectedFather (a subclass of InfectedFamilyMember). Example 4 should result in an instance of InfectedViaPoliceWork.
In addition, depending on the specific instance, other objects or property values may need to get instantiated. For example, for the InfectedCo-Passenger class there is a property to define the place that was the travel destination, in this case the city Bangalore India. For strings such as Example 2 the contractedVirusFrom property on the Patient needs to have values for each Patient referenced in the string as a value. The difficulty with processing these types of strings is that the input data does not adhere to standardized patterns. In some cases, someone may simply enter “Bangalore” in other cases “Travel to Bangalore”. Similarly, when the reason is contact with other patients there are many different patterns used to enter the data. Example 2 can also be entered as: “Contact with P134-P138”, “Contact with P134, 135, 136, 137, and 138”, “Contact with P134-138”, and other formats.
While upload tools such as Cellfie can do simple pattern matching these more complex examples are difficult to process with upload tools. As a result, we utilized a two-step process for uploading and transforming data as illustrated in Fig. 1. During the Upload process we would where possible directly transform strings to data types or objects. However, where there were many patterns to the data, we would simply upload those strings into data properties we defined as utility properties. Then in the Transformation process we would use tools such as SPARQL and Lisp to perform more complex pattern matching on the strings uploaded into the utility data properties. The Lisp and SPARQL files can be found at [17].
Our process for transforming these types of strings illustrates our Agile development approach. To begin with when we had a small amount of sample data, we wrote specific SPARQL queries. We call these queries transformations because they don’t just query the data but change it via INSERT and DELETE statements. To begin we had many SPARQL transformations in a text file which we would execute by hand via AllegroGraph’s Gruff tool. These included transformations (which we call ad hoc transformations) that directly match for specific strings via WHERE clauses in SPARQL and then perform the appropriate creation of objects and property values via INSERT clauses.
Figure 3 shows an example of an ad hoc transformation. The FILTER statement exactly matches a specific string, and the INSERT statement adds the appropriate new triples. E.g., it creates a new instance of the CloseContact class shown in Fig. 2 and adds that to the suspectedReasonOfCatchingCovid-19 property for the patient. The DELETE statement removes the utility string value.
Of course this approach was not scalable. As a result, we defined a more scalable approach to transforming our data. That process was as follows:
-
1.
Delete each utility data property value after it has been transformed (in Fig. 3 the reasonString property is a utility property).
-
2.
Apply transformations in an order from the most specific transformations to more general transformations. See Fig. 4.
This process illustrated in Fig. 4 allowed us to write pattern matching transformations that were very general and would not correctly process certain unusual strings. These more specific strings were processed first by less general pattern matching transformations. After processing, the value for the processed utility property was deleted so that the more general pattern matching transformations could be applied without risk of error on the more unusual strings.
Figure 5 shows a pseudo code fragmentFootnote 7 from a Lisp function that does general pattern matching. This function matches strings such as: “Contact of P6135-6139”. The function first executes a SPARQL query to find all the patients with a reasonString that matches the pattern. It then uses the Allegro CL function match-re to extract the sub-string required for the transformation (e.g., “6135-6139”). It then uses the function split-re to extract the two patient ID strings, in this case “6135” and “6139”. It then converts these strings to integers and loops from the first to last integer. Within the loop it performs the appropriate manipulation of the knowledge graph. This requires converting each integer back into a string and performing a lookup of the Patient object that matches the ID. Finally, it calls the function make-close-contact-object which does the equivalent of the part of the SPARQL transformation in Fig. 3 to create an instance of the CloseContact class and fill in appropriate property values. The make-close-contact-object function also deletes the utility property value. If the system is running in test mode it saves a copy of the value on a testing utility property.
The most general pattern matching function for these types of contact strings (a transformation at the bottom of Fig. 4) simply uses the Allegro CL function: (split-re “\(\backslash \backslash \)D+” rs) where rs is bound to the reasonString to extract all the numeric substrings in the string. This function needs to be run after other functions that match patterns for locations, relatives, etc. since those strings may have numbers in them that are not related to patient ID’s. It also needs to run after transformations that have patterns such as “P6135-6139” since it only finds each individual numeric string and would not correctly process the iteration implied in those types of strings. When run at the appropriate time, after the more specific strings have been removed this general pattern matching function processes a great deal of the strings that were previously handled by several more specific transformations. This is an example of how refactoring can help us build a knowledge graph capable of handling increasing amounts of data.
5 Results
The CODO knowledge graph has over 71 thousand patients and approximately 5M triples after running the Materializer reasoner. We implemented 100% of the initial competency questions defined in the original phase of the project via SPARQL queries. These queries can be found at [17]. In addition, we found countless opportunities for providing new ways to visualize the data once it is in one integrated graph format. Just three of these are shown in Figs. 6 and 7 below. These visualizations come from simple SPARQL queries. Typically, (as in these figures) we use Gruff to automatically transform the results of the SPARQL query into a visualization. We have also imported the output of SPARQL queries into Excel to create pie and other charts.
5.1 Evaluation of CODO
The most significant failure of the team to follow an Agile process is that as much as we tried, it was very difficult to find medical professionals to define requirements and to utilize the ontology and give us feedback. The reason is analogous to the classic Knowledge Acquisition Bottleneck problem. Healthcare professionals in India were so overwhelmed with simply dealing with the pandemic that they were unable to participate in research. As part of the Covid harmonization process [7], Dr. Sivaram Arabandi MD reviewed the ontology. CODO was also used as a test ontology for the OOPS! Ontology evaluation tool [18]. Design changes were made as a result of both these reviews. We hope that future versions of CODO will be utilized and evaluated by more healthcare professionals as their time is freed from the crisis of the pandemic.
5.2 Privacy Issues
Although the data in CODO was anonymous, legislation such as the US Health Insurance and Portability Act (HIPAA) and the European Union’s Regulation 2016/679 may make utilization of CODO for functions such as contact tracing problematic. However, as [19] points out, Google and Apple have released voluntary contact tracing apps and nations such as Singapore and Ireland have had success in the voluntary usage of these apps by patients and consumers.
6 Conclusions
Most current ontology design methods (e.g., [20, 21], the models surveyed in [22]) emphasize an approach that is essentially a Waterfall model where all the emphasis is on designing the model (essentially the T-Box). For example, in the evaluation of the productivity of upper models in [23] the evaluation criteria were focused only on the model with no consideration of the model’s ability to incorporate and provide value to actual data. These approaches have the same problems for designing ontologies as the waterfall model in general has shown for most software development projects [9]. The emphasis on getting a “perfect” design the first time is doomed to failure. This insight goes back to Boehm’s spiral model [24]. What might be a correct ontology in terms of the actual domain may turn out to be difficult to use because of issues with existing data or other non-functional requirements. In the real-world good software design comes both from the bottom up (from constraints imposed by legacy data, business processes, etc.) as from the top down (by analysis of the problem domain). Our experience with CODO, where only two developers developed a large knowledge graph in a few months, is evidence that the Agile approach provides the same benefits for the design of knowledge graphs as it has demonstrated for many other types of software systems [25]. In addition, our experience demonstrates principles that apply to real-world knowledge graph development in general:
-
Use of pattern matching transformations. In transforming from “strings to things” [16] the basic capabilities of upload tools may not be sufficient and may require transformations that utilize features such as iteration in programming languages.
-
Transforming from specific to general. In developing transformations, the most productive strategy is to begin with the most specific types of patterns and then use more general transformations after ensuring that outliers that would be incorrectly processed by the general transformations have been processed and removed.
In the future we plan to investigate the use of ML and NLP for these transformations.
Notes
- 1.
- 2.
- 3.
- 4.
- 5.
We utilized Lisp because our team was very small (2 people), and the lead developer had the most experience in Lisp and we wanted to work as rapidly as possible to meet the needs of the pandemic. In future versions we will re-implement the functions in Python.
- 6.
The details of the CODO project can be found at: https://github.com/biswanathdutta/CODO. The CODO ontology can be found at: https://bioportal.bioontology.org/ontologies/CODO.
- 7.
See [17] for the actual Lisp and SPARQL code for this and all transformations.
References
Dutta, B., DeBellis, M.: CODO: an ontology for collection and analysis of COVID-19 data. In: Proceedings of the 12th International Joint Conference on Knowledge Discovery, Knowledge Engineering and Knowledge Management (2020)
Gardner, L.: Modeling the spreading risk of 2019-nCov (2020). https://systems.jhu.edu/research/public-health/ncov-model-2/
Sirin, E.: Analyzing COVID-19 data with SPARQL (2020). https://www.stardog.com/labs/blog/analyzing-covid-19-data-with-sparql/
He, Y., et al.: CIDO, a community-based ontology for coronavirus disease knowledge and data integration, sharing, and analysis. Sci. Data 7(1), 1–5 (2020)
Cowell, L.G., Smith, B.: Infectious disease ontology. In: Sintchenko, V. (ed.) Infectious Disease Informatics, pp. 373–395. Springer, New York (2010). https://doi.org/10.1007/978-1-4419-1327-2_19
Domingo-Fernández, D., et al.: COVID-19 knowledge graph: a computable, multi-modal, cause-and-effect knowledge model of COVID-19 pathophysiology. Bioinformatics 37(9), 1332–1334 (2021)
Lin, A., et al.: A community effort for COVID-19 ontology harmonization. In: The 12th International Conference on Biomedical Ontologies (2021)
Dutta, B., Chatterjee, U., Madalli, D.P.: YAMO: yet another methodology for large-scale faceted ontology construction. J. Knowl. Manag. 19(1), 6–24 (2015)
Beck, K.: Extreme Programming Explained: Embrace Change. Addison-Wesley Professional, Boston (2000)
Kroll, P., Kruchten, P.: The Rational Unified Process Made Easy: A Practitioner’s Guide to the RUP: A Practitioner’s Guide to the RUP. Addison-Wesley Professional, Boston (2003)
O’Connor, M.J., Halaschek-Wiener, C., Musen, M.A., et al.: Mapping master: a flexible approach for mapping spreadsheets to OWL. In: Patel-Schneider, F. (ed.) ISWC 2010. LNCS, vol. 6497, pp. 194–208. Springer, Heidelberg (2010). https://doi.org/10.1007/978-3-642-17749-1_13
Musen, M.A.: The protégé project: a look back and a look forward. AI Matters 1(4), 4–12 (2015)
SWRL: A semantic web rule language combining OWL and ruleML. W3C Member Submission 21 May 2004 (2016). Accessed Feb 2016
Aasman, J., Cheetham, K.: RDF browser for data discovery and visual query building. In: Proceedings of the Workshop on Visual Interfaces to the Social and Semantic Web (VISSW 2011), Co-located with ACM IUI, p. 53 (2011)
Aasman, J.: Unification of geospatial reasoning, temporal logic, & social network analysis in event-based systems. In: Proceedings of the Second International Conference on Distributed Event-Based Systems, pp. 139–145 (2008)
Singhal, A.: Introducing the knowledge graph: things, not strings. Official Google Blog 5, 16 (2012)
Debellis, M.: Lisp and SPARQL files for CODO ontology (2020). https://github.com/mdebellis/CODO-Lisp
Chansanam, W., Suttipapa, K., Ahmad, A.R.: COVID-19 ontology evaluation. Int. J. Manag. (IJM) 11(8), 47–57 (2020)
Kejriwal, M.: Knowledge graphs and COVID-19: opportunities, challenges, and implementation. Harv. Data Sci. Rev. (2020)
Arp, R., Smith, B., Spear, A.D.: Building Ontologies with Basic Formal Ontology. MIT Press, Cambridge (2015)
de Almeida Falbo, R.: SABiO: systematic approach for building ontologies. In: CEUR Workshop Proceedings, vol. 1301 (2014)
Garcia, A., et al.: Developing ontologies within decentralised settings. In: Chen, H., Wang, Y., Cheung, K.H. (eds.) Semantic e-Science. AOIS, vol. 11, pp. 99–139. Springer, Boston (2010). https://doi.org/10.1007/978-1-4419-5908-9_4
Keet, C.M., et al.: The use of foundational ontologies in ontology development: an empirical assessment. In: Antoniou, G. (ed.) ESWC 2011. LNCS, vol. 6643, pp. 321–335. Springer, Heidelberg (2011). https://doi.org/10.1007/978-3-642-21034-1_22
Boehm, B.W.: A spiral model of software development and enhancement. Computer 21(5), 61–72 (1988)
Pallozzi, D.: The word that took the tech world by storm: returning to the roots of agile (2018). https://www.thoughtworks.com/en-in/perspectives/edition1-agile/article
Acknowledgement
This work has been supported by Indian Statistical Institute, Kolkata. This work was conducted using the Protégé resource, which is supported by grant GM10331601 from the National Institute of General Medical Sciences of the United States National Institutes of Health. Thanks to Franz Inc. (http://www.allegrograph.com) for their help with AllegroGraph and Gruff. Thanks to Dr. Sivaram Arabandi, MD for his feedback on the CODO ontology.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
DeBellis, M., Dutta, B. (2021). The Covid-19 CODO Development Process: an Agile Approach to Knowledge Graph Development. In: Villazón-Terrazas, B., Ortiz-Rodríguez, F., Tiwari, S., Goyal, A., Jabbar, M. (eds) Knowledge Graphs and Semantic Web. KGSWC 2021. Communications in Computer and Information Science, vol 1459. Springer, Cham. https://doi.org/10.1007/978-3-030-91305-2_12
Download citation
DOI: https://doi.org/10.1007/978-3-030-91305-2_12
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-91304-5
Online ISBN: 978-3-030-91305-2
eBook Packages: Computer ScienceComputer Science (R0)