Abstract
Supervised deep learning on medical imaging requires massive manual annotations, which are expertise-needed and time-consuming to perform. Active learning aims at reducing annotation efforts by adaptively selecting the most informative samples for labeling. We propose in this paper a novel deep active learning approach for dual-view mammogram analysis, especially for breast mass segmentation and detection, where the necessity of labeling is estimated by exploiting the consistency of predictions arising from craniocaudal (CC) and mediolateral-oblique (MLO) views. Intuitively, if mass segmentation or detection is robustly performed, prediction results achieved on CC and MLO views should be consistent. Exploiting the inter-view consistency is hence a good way to guide the sampling mechanism which iteratively selects the next image pairs to be labeled by an oracle. Experiments on public DDSM-CBIS and INbreast datasets demonstrate that comparable performance with respect to fully-supervised models can be reached using only 6.83% (9.56%) of labeled data for segmentation (detection). This suggests that combining dual-view mammogram analysis and active learning can strongly contribute to the development of computer-aided diagnosis systems.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Breast cancer
- Mass segmentation
- Mass detection
- Dual-view mammogram analysis
- Active learning
- Computer-aided diagnosis
1 Introduction
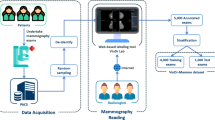
Breast cancer is ranked as the leading cause of global cancer incidence among women in 2020, with an estimated 2.3 million new cases, representing about 25% of all cancers in women [1]. Digital X-ray mammography plays an essential role in diagnosing breast cancer at an early stage. In particular, masses are one of the most common and important type of targeted breast abnormalities. Conventional computer-aided diagnosis (CAD) systems usually use hand-crafted features tailored for mass recognition. Recently, the rise of deep learning made the analysis of mammograms more automatic and accurate thanks to effective training methods, advances in hardware, and most importantly, large amounts of annotated training data [2]. Based on supervised learning using convolutional neural networks (CNN), recent studies have achieved impressive performance regarding mass segmentation [3,4,5] or detection [2, 4, 6,7,8]. Despite such success, supervised deep learning still faces obstacles, including data acquisition and high-quality manual annotations, which are expertise-needed and time-consuming.
Mammography screening involves two standard views acquired for left and right breasts: craniocaudal (CC) and mediolateral-oblique (MLO). In clinical routine, radiologists usually confirm the diagnosis through cross information arising from both views. Examining the CC/MLO correspondence and consistency between suspicious findings thus allows to improve clinical interpretations and subsequent decisions [9]. Computational analysis of dual-view mammograms [10,11,12,13,14] has been validated as an effective way to reduce false-positive cases and improves screening performance. Nevertheless, the labeling workload of radiologists is further increased. Therefore, it is greatly needed to develop an effective annotation suggestion algorithm to alleviate this issue.
Extensively studied in various fields, active learning (AL) aims at reducing human annotation efforts by adaptively selecting the most informative samples for labeling. As for medical imaging, AL has shown high potential in reducing the annotation cost [15]. Recent studies [16, 17] proposed AL frameworks for breast cancer segmentation respectively on immunohistochemistry and biomedical images. However, AL methods have not been widely exploited in X-ray mammography analysis. Zhao et al. [18] first introduced AL into a mammography classification system based on a support vector machine (SVM) classifier. Shen et al. [19] proposed a mass detection framework that incorporates AL and self-paced learning (SPL) to improve the model generalization ability. These studies demonstrate great potential of AL in mammogram analysis. Contrary to existing studies based on the uncertainty and diversity of a single image, our goal is to score the dual-view mammograms according to their prediction consistency. Our work can be seen as a complement to existing methods, and proves that combining inter-view information can bring further improvements.
This paper provides the following contributions. First, we propose a novel approach of deep AL for dual-view mammogram analysis (including breast mass segmentation and detection), where the dual-view prediction consistency is integrated as selection criterion. Second, two task-specific neural networks are carefully designed for more effective mammogram mass segmentation and detection. Third, extensive experiments are conducted to reveal the relationship between dual-view consistency and mammogram informativeness.
2 Methods
To reduce the labeling efforts dealing with breast masses in mammograms, we propose a novel approach of deep active learning for dual-view mammogram analysis. Specifically, we consider two scenarios: mass segmentation and detection. The key insight of our method is to use the consistency of mass segmentation or detection results arising from CC/MLO view-points as active learning criteria.
The proposed AL process starts by pre-training the model on a small labeled subset \(D_l\). Then, we perform model inference on the unlabeled dataset \(D_u\) to select the most informative mammogram pairs according to the calculated dual-view prediction consistency. These selected pairs are then sent to radiologists for annotation and appended to \(D_l\), where the model is consequently fine-tuned on. Such AL cycle is repeated several times to gradually improve the model performance, until the annotation budget is exhausted. The key feature of AL is the query algorithm for the informativeness ranking of unlabeled images, which in our work is the scoring function of the dual-view prediction consistency.
2.1 Proposed Network Architectures
Breast mass segmentation and detection are two main tasks in mammogram analysis. We take inspiration from recent advances of deep neural networks [20,21,22], and design simple and efficient networks for each of these tasks (Fig. 2).
Mass Segmentation Network (MSN). The architecture is composed of an encoder for feature extraction, a decoder for spatial detail reconstruction and several skip-connections between both branches to recover spatial information. Instead of using a standard symmetric encoder-decoder architecture [21, 23], we apply an alternative asymmetric architecture where residual blocks are integrated into the encoder and \(1\times 1\) convolution layers are part of the decoder (Fig. 2(a)). The network complexity is greatly reduced while the performance stays unchanged. The optimization is supervised by the combination of binary cross-entropy (\(L_{bce}\)) and Dice (\(L_{dice}\)) losses following \(L_{seg} = L_{dice} + \lambda _1 L_{bce}\) with:
where p and y represent the prediction mask and the ground truth mask respectively, |.| and \(\circ \) the pixel-wise sum and multiplication operations. The empirical factor \(\lambda _1\) is set to 0.5 to prevent the combined loss from degenerating into \(L_{bce}\).
Mass Detection Network (MDN). We designed a single-stage mass detection network where a multi-scale prediction strategy is applied to detect masses of different scales. Three detection branches with different scales \(\{64\times 32,32\times 16, 16\times 8\}\) are attached to a regular feature extraction network (Fig. 2(b)) consisting of 3 residual blocks. The multi-scale architecture allows the network to be more robust to lesions of different sizes, i.e. larger scale for smaller masses and vise-versa. Each branch consists of a localization module and a classification module, where the former is in charge of regressing the spatial transformation (4 coordinates offset) from predefined anchor boxes to ground truth boxes, and the latter predicts the mass presence probability for each anchor box. We use the focal loss (\(L_{focal}\)) to supervise classification modules and the balanced L1 loss (\(L_{bl1}\)) to supervise localization modules, following \(L_{det} = L_{focal} + \lambda _2 L_{bl1}\) with:
We use the default parameters of \(L_{focal}\) and \(L_{bl1}\) as respectively introduced in [22] and [24]: \(\alpha _1 = 0.25, \gamma _1=2.0\) for \(L_{focal}\), \(\alpha _2 = 0.5, \gamma _2=1.5, \beta =1.0\) for \(L_{bl1}\). The final detection loss is the combination of \(L_{focal}\) and \(L_{bl1}\) with \(\lambda _2=1\).
2.2 Dual-View Consistency
At the selection stage of each AL cycle, we aim at filtering the most informative mammograms in \(D_u\) through the analysis of dual-view consistency. Theoretically, given a pair of mammograms \(\{I_{CC},I_{MLO}\}\) from the same breast, the analysis results should be coherent. Many latent relationships can potentially be exploited as query factors, such as the number of masses detected on both views, or the mass size, position, shape, texture... In our work, we consider the first two factors as consistency criteria since their correlation is more obvious. In particular, the number of identified masses from both views \(\{N_{CC},N_{MLO}\}\) should be identical and their sizes \(\{S_{CC},S_{MLO}\}\) (i.e. number of pixels) should be similar. We define two scores (\(S_{num}\) and \(S_{size}\)) as the measurements of the following factors:
where \(S_{num}\) and \(S_{size}\) varies from 0 (low consistency) and 1 (high consistency). Correct predictions should meet the above two conditions simultaneously, thus the final combined score is calculated as the minimum of \(S_{num}\) and \(S_{size}\):
The proposed consistency score S provides a rough estimation of the mass segmentation/detection prediction quality: mammogram pairs with higher S values are regarded as easy samples and vise-versa. Figure 3 shows mammogram pairs with different S values for both segmentation and detection tasks. When S is low, the prediction on at least one mammogram appears inaccurate. Considering the existence of labeling errors, verifying the number of found lesions from different views tends to avoid involving ambiguous or miss-annotated samples in the training set, towards better AL results. In this direction, our strategy selects mammogram pairs with consistent multi-view predictions such that the aforementioned examples are not taken into account in priority by the oracle.
2.3 Active Learning Strategies
The key of AL is to select the most informative samples to optimize a learnable model. However, the definition of informativeness is still an open question. In the common practice of AL, one considers examples with the most uncertainty or examples that are most likely to be wrong as informative examples. However, we need to check if this paradigm remains valid for medical imaging. To this end, we implement three AL strategies: random (rand), best consistency (bestC) and worst consistency (worstC) selections. For each AL cycle, rand strategy randomly selects b mammogram pairs from unlabeled dataset \(D_u\), while bestC (worstC) selects b pairs with the highest (lowest) consistency score S. We visualize Fig. 4 mammogram pairs selected by each AL strategy. Each point represents a CC/MLO pair. Red (green) points are b pairs selected by worstC (bestC). We estimate the linear regression between S and mass segmentation (Fig. 4(a)) or detection (Fig. 4(b)) accuracy. The consistency score appears as a reasonable reference of the prediction quality. Results were obtained during training (i.e. without full convergence) so some points fall in the area of low consistency scores.
3 Experiments
3.1 Implementation Details
We use two publicly-available datasets for our experiments: DDSM-CBIS (Digital Database for Screening Mammography) [25] and INbreast [26], with respectively 1514 and 107 cases containing ground truth mass delineations. For training AL cycles, 586 CC/MLO mammogram pairs are found from DDSM-CBIS and employed to compute the dual-view information consistency. These pairs are divided into a small labeled subset \(D_l\) and a simulated unlabeled pool \(D_u\). For INbreast, all 107 images are employed as the test set since pair-wise data is not mandatory during inference. The original mammogram has a resolution of \(4084\times 3328\) or \(3328\times 2560\), which is computationally expensive. Therefore, we resize images to \(512\times 256\) for all experiments. Mammograms are normalized before feeding into neural networks. Random image rotation, cropping, padding, and flipping operations are applied during the training phase for data augmentation.
The proposed framework was implemented using PyTorch. We use SGD optimizer with a learning rate of 0.1 and a cosine annealing schedule. The proposed MSN (MDN) has 45,705 (80,202) learnable parameters in total and was trained for 2k (6k) iterations with a batch size of 32. Each experiment is repeated 5 times, and we report their average performance and the standard error. Following common practice, we adopt the Dice coefficient and the Average Precision (AP) score to respectively evaluate segmentation and detection performances. Dice coefficient is defined as \(1-L_{dice}\) (Eq. 1) whereas the AP score is calculated by taking the area under the precision-recall curve.
For each AL experiment, we start by training an initial model on a random labeled subset \(D_l\) containing b pairs. During each AL cycle, we adaptively select the next b pairs from DDSM-CBIS using three different AL strategies (rand, bestC or worstC) from unlabeled dataset \(D_u\). These images are assigned with annotations and appended to \(D_l\) for fine-tuning at the next AL cycle. We fix an annotation budget B to end AL cycles. Concretely, we set b to 8 (16 images) for all experiments. Noting that the annotation cost for segmentation is much higher than for detection, we set the annotation budget B to 40 (80 images) for the mass segmentation task and 56 (112 images) for the detection task. In other words, we implement 4 (6) active cycles for segmentation (detection). Each cycle adds 1.37% of labeled data and the whole segmentation (detection) AL process takes 6.83% (9.56%) of labeled data in the training set.
Mass segmentation and detection performance with rand (green), bestC (red) and worstC (blue) AL strategies. Black dashed lines indicate results using the complete training set. We report average Dice score of mass segmentation (a), Dice score standard error (b), average AP score of mass detection (c) and AP score standard error (d). (Color figure online)
3.2 Results
We conducted extensive experiments to evaluate the performance of rand, bestC and worstC AL strategies. Averaged results are shown in Fig. 5. It can be seen that the model performance is improved progressively cycle by cycle, and bestC (\(Dice=37.00\%\), \(AP=52.83\%\)) is consistently better than the other strategies. bestC presents \(1.62\%\) Dice improvement and 4.02% AP gains relative to the rand baseline. Conversely, worstC (\(Dice=34.37\%\), \(AP=43.51\%\)) is not superior to the baseline. From Fig. 5(b) and (d) we observe that both bestC and worstC reduce the performance instability of rand strategy to a certain extent. In particular, with only \(6.83\%\) (\(9.56\%\)) labeling budget for mass segmentation (detection), bestC achieves performance comparable to the fully supervised model (37.00 vs \(37.59\%\) for segmentation, 52.83 vs \(54.33\%\) for detection), showing the great potential of our method in alleviating the annotation burden. Besides, we observe greater performance gaps for detection than segmentation. Since detection annotations only provide sparse box-level supervision, the detection task is more critical in terms of the amount of training images.
In the common practice of traditional AL, examples with high consistency scores provide better prediction quality, and could be seen as well-learned examples which are normally not included in AL cycles. Our results seem to contradict this practice, since pairs with higher consistency seem more useful than those with lower consistency. For these results, we propose some explanations: mammography analysis is actually more difficult than general natural image analysis tasks since it is difficult for humans without clinical knowledge to distinguish masses from surrounding healthy tissues. Medical imaging datasets can also be very biased due to different acquisition conditions. Learning with a small amount of medical images is challenging, especially for the first few AL cycles. For detection, Fig. 5(c) shows an AP drop for the first AL cycle of worstC, indicating that not all labeled data are beneficial when the model does not yet have a full understanding of what masses are. Picking examples with good prediction results helps to consolidate what has been learned while avoiding corner cases.
4 Conclusion
We propose a label-efficient deep learning approach that explores the prediction consistency arising from dual-view mammograms. The main novelty is the combination between multi-view mammogram analysis and active learning, which has not been studied in the field of medical imaging to our knowledge. Our contributions significantly alleviate the burden of manual labeling in breast mass segmentation and detection tasks, which is beneficial to the development of CAD tools. A future possible extension is to integrate existing single-view criteria into our current framework, towards a unified active learning system.
References
Sung, H., et al.: Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. (2021)
Kooi, T., et al.: Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 35, 303–312 (2017)
Singh, V.K., et al.: Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst. Appl. 139, 112855 (2020)
Yan, Y., Conze, P.H., Quellec, G., Lamard, M., Cochener, B., Coatrieux, G.: Two-stage multi-scale mass segmentation from full mammograms. In: IEEE International Symposium on Biomedical Imaging (2021)
Yan, Y., et al.: Cascaded multi-scale convolutional encoder-decoders for breast mass segmentation in high-resolution mammograms. In: IEEE International Engineering in Medicine and Biology (2019)
Dhungel, N., Carneiro, G., Bradley, A.P.: A deep learning approach for the analysis of masses in mammograms with minimal user intervention. Med. Image Anal. 37, 114–128 (2017)
Agarwal, R., Diaz, O., Lladó, X., Yap, M.H., Martí, R.: Automatic mass detection in mammograms using deep convolutional neural networks. J. Med. Imaging 6(3), 1–9 (2019)
Ribli, D., Horváth, A., Unger, Z., Pollner, P., Csabai, I.: Detecting and classifying lesions in mammograms with deep learning. Sci. Rep. 8(1), 1–7 (2018)
Vijayarajan, S.M., Jaganathan, P.: Breast cancer segmentation and detection using multi-view mammogram. Acad. J. Cancer Res. 7(2), 131–140 (2014)
Yan, Y., Conze, P.H., Lamard, M., Quellec, G., Cochener, B., Coatrieux, G.: Multi-tasking siamese networks for breast mass detection using dual-view mammogram matching. In: International Workshop on Machine Learning in Medical Imaging, pp. 312–321 (2020)
Yan, Y., Conze, P.-H., Lamard, M., Quellec, G., Cochener, B., Coatrieux, G.: Towards improved breast mass detection using dual-view mammogram matching. Med. Image Anal. 71, 102083 (2021)
Perek, S., Hazan, A., Barkan, E., Akselrod-Ballin, A.: Mammography dual view mass correspondence. arXiv preprint arXiv:1807.00637 (2018)
Ma, J., et al.: Cross-view relation networks for mammogram mass detection. arXiv preprint arXiv:1907.00528 (2019)
Gu, X., Shi, Z., Ma, J.: Multi-view learning for mammogram analysis: auto-diagnosis models for breast cancer. In: IEEE International Conference on Smart Internet of Things, pp. 149–153 (2018)
Budd, S., Robinson, E.C., Kainz, B.: A survey on active learning and human-in-the-loop deep learning for medical image analysis. arXiv preprint arXiv:1910.02923 (2019)
Shen, H., et al..: Deep active learning for breast cancer segmentation on immunohistochemistry images. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 509–518 (2020)
Li, H., Yin, Z.: Attention, suggestion and annotation: a deep active learning framework for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 3–13 (2020)
Zhao, Yu., Chen, D., Xie, H., Zhang, S., Lixu, G.: Mammographic image classification system via active learning. J. Med. Biol. Eng. 39(4), 569–582 (2019)
Shen, R., Yan, K., Tian, K., Jiang, C., Zhou, K.: Breast mass detection from the digitized X-ray mammograms based on the combination of deep active learning and self-paced learning. Future Gener. Comput. Syst. 101, 668–679 (2019)
He, K., Zhang, X., Ren, S., Sun, J. :Deep residual learning for image recognition. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 234–241 (2015)
Lin, T.Y., Goyal, P., Girshick, R., He, K., Dollár, P.: Focal loss for dense object detection. In IEEE International Conference on Computer Vision, pp. 2980–2988 (2017)
Conze, P.H., et al.: Abdominal multi-organ segmentation with cascaded convolutional and adversarial deep networks. Artif. Intell. Med. (2021)
Pang, J., Chen, K., Shi, J., Feng, H., Ouyang, W., Lin, D.: Libra R-CNN: towards balanced learning for object detection. In: IEEE Conference on Computer Vision and Pattern Recognition, pp. 821–830 (2019)
Lee, R., Gimenez, F., Hoogi, A., Miyake, K.K., Gorovoy, M., Rubin, D.: A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 4, 170177 (2017)
Moreira, I.C., et al.: INbreast: toward a full-field digital mammographic database. Acad. Radiol. (2012)
Acknowledgements
This work was partly funded by France Life Imaging (grant ANR-11-INBS-0006 from the French Investissements d’Avenir program).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Nature Switzerland AG
About this paper
Cite this paper
Yan, Y. et al. (2021). Deep Active Learning for Dual-View Mammogram Analysis. In: Lian, C., Cao, X., Rekik, I., Xu, X., Yan, P. (eds) Machine Learning in Medical Imaging. MLMI 2021. Lecture Notes in Computer Science(), vol 12966. Springer, Cham. https://doi.org/10.1007/978-3-030-87589-3_19
Download citation
DOI: https://doi.org/10.1007/978-3-030-87589-3_19
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-87588-6
Online ISBN: 978-3-030-87589-3
eBook Packages: Computer ScienceComputer Science (R0)